Baculovirus FP25K Localization: Role of the Coiled-Coil Domain
Tyler A. Garretson, Jason C. McCoy,*Xiao-Wen Cheng Department of Microbiology, Miami University, Oxford, Ohio, USA
ABSTRACT
Two types of viruses are produced during the baculovirus life cycle: budded virus (BV) and occlusion-derived virus (ODV). A particular baculovirus protein, FP25K, is involved in the switch from BV to ODV production. Previously, FP25K from the model alphabaculovirusAutographa californicamultiple nucleopolyhedrovirus (AcMNPV) was shown to traffic ODV envelope pro-teins. However, FP25K localization and the domains involved are inconclusive. Here we used a quantitative approach to study FP25K subcellular localization during infection using an AcMNPV bacmid virus that produces a functional AcMNPV FP25K-green fluorescent protein (GFP) fusion protein. During cell infection, FP25K-GFP localized primarily to the cytoplasm, particu-larly amorphous structures, with a small fraction being localized in the nucleus. To investigate the sequences involved in FP25K localization, an alignment of baculovirus FP25K sequences revealed that the N-terminal putative coiled-coil domain is present in all alphabaculoviruses but absent in betabaculoviruses. Structural prediction indicated a strong relatedness of AcMNPV FP25K to long interspersed element 1 (LINE-1) open reading frame 1 protein (ORF1p), which contains an N-terminal coiled-coil do-main responsible for cytoplasmic retention. Point mutations and deletions of this dodo-main lead to a change in AcMNPV FP25K localization from cytoplasmic to nuclear. The coiled-coil and C-terminal deletion viruses increased BV production. Further-more, a betabaculovirus FP25K protein lacking this N-terminal coiled-coil domain localized predominantly to the nucleus and exhibited increased BV production. These data suggest that the acquisition of this N-terminal coiled-coil domain in FP25K is important for the evolution of alphabaculoviruses. Moreover, with the divergence of preocclusion nuclear membrane break-down in betabaculoviruses and membrane integrity in alphabaculoviruses, this domain represents an alphabaculovirus adapta-tion for nuclear trafficking of occlusion-associated proteins.
IMPORTANCE
Baculovirus infection produces two forms of viruses: BV and ODV. Manufacturing of ODV involves trafficking of envelope pro-teins to the inner nuclear membrane, mediated partly through the FP25K protein. Since FP25K is present in alpha-, beta-, and gammabaculoviruses, it is uncertain if this trafficking function is conserved. In this study, we looked at alpha- and betabaculovi-rus FP25K trafficking by its localization. Alphabaculovibetabaculovi-rus FP25K localized primarily to the cytoplasm, whereas betabaculovibetabaculovi-rus FP25K localized to the nucleus. We found that an N-terminal coiled-coil domain present in all alphabaculovirus FP25K proteins, but absent in betabaculovirus FP25K, was critical for alphabaculovirus FP25K cytoplasmic localization. We believe that this rep-resents an evolutionary process that partly led to the gain of function of this N-terminal coiled-coil domain in alphabaculovirus FP25K to aid in nuclear trafficking of occlusion-associated proteins. Due to betabaculovirus breakdown of the nuclear mem-brane before occlusion, this function is not needed, and the domain was lost or never acquired.
I
nsect-specific baculoviruses in the familyBaculoviridaehave a circular, double-stranded DNA genome of 88 to 180 kb with the capacity to code for 90 to 180 putative proteins (1). Based on the insect hosts from which these baculoviruses were isolated and their biological characteristics, they are divided into four genera: Alphabaculovirus,Betabaculovirus,Gammabaculovirus, and Delt-abaculovirus(2). The alpha- and betabaculoviruses are composed of lepidopteran-specific nucleopolyhedroviruses (NPVs) and granuloviruses (GVs), whereas the gamma- and deltabaculovi-ruses are composed of hymenopteran- and dipteran-specific baculoviruses, respectively (2).Baculoviruses, with the exception of gammabaculoviruses, un-dergo a biphasic virus life cycle during insect cell infection, ini-tially involving the production of extracellular viruses or budded viruses (BVs) and later involving the production of intracellular viruses or occlusion-derived viruses (ODVs) (3, 4). After entry into the cell, early gene transcription by the host RNA polymerase II starts in the nucleus (3). Following viral DNA replication, late gene transcription by the viral RNA polymerase begins to produce viral structural proteins (5). Viral structural proteins and genomic DNA assemble to produce nucleocapsids in the
viro-genic stroma in the nucleus (6). Initially, nucleocapsids egress primarily through the nuclear pore complexes to the cytoplasm and acquire an envelope from the plasma membrane to bud out to produce BV (7,8). In contrast, nucleocapsids destined to remain in the nucleus acquire an envelope from blebbing of the inner nuclear membrane (INM) to form ODVs (8,9). This membrane-blebbing process is partially facilitated by a particular 25-kDa
nu-Received25 June 2016 Accepted3 August 2016
Accepted manuscript posted online10 August 2016
CitationGarretson TA, McCoy JC, Cheng X-W. 2016. Baculovirus FP25K localization: role of the coiled-coil domain. J Virol 90:9582–9597.
doi:10.1128/JVI.01241-16.
Editor:G. McFadden, University of Florida
Address correspondence to Xiao-Wen Cheng, Chengx@miamioh.edu.
*Present address: Jason C. McCoy, Department of Molecular Genetics, Biochemistry and Microbiology, College of Medicine, University of Cincinnati, Cincinnati, Ohio, USA.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
crossmark
on November 7, 2019 by guest
http://jvi.asm.org/
cleocapsid protein, FP25K (8,10,11). During this late phase of infection, occlusion bodies (OBs) are formed around ODVs with either the matrix protein polyhedrin or granulin, in the case of alpha- and gammabaculoviruses or betabaculoviruses, respec-tively (3,4,8,9). No polyhedrin/granulin orthologues are present in the deltabaculoviruses (4). Unlike other baculoviruses, the oc-clusion process of betabaculoviruses occurs after the breakdown of the nuclear envelope in a cytoplasmic-nuclear mix (3,12,13). The process of INM blebbing, facilitated by FP25K and other ODV envelope proteins, is thought to be a general switch from BV to ODV production (14–16). This multifunctional FP25K protein is not required for infectionin vitro, but it is necessaryin vivofor proper ODV envelopment, virion occlusion, and insectper os in-fectivity (10,16–19).
Since it is not essentialin vitro, and inactivation provides a selective advantage, thefp25kgene of baculoviruses readily under-goes mutation by transposition or replication slippage errors re-sulting in a base insertion or deletion (indel) during infection in cell culture (14,20–22). These mutations lead to an evident few-polyhedra (FP) phenotype, characterized by decreased produc-tion of OBs, fewer ODV envelope proteins localized to the nu-cleus, and increased BV titers (18,22–25). Therefore, it has been proposed that FP25K may be involved in associating with, sorting, and trafficking ODV envelope proteins to the INM and viral en-velope, including ODV-E66 and ODV-E25 (17, 26, 27). Even though FP25K is not absolutely necessary for OB formation and ODV envelopment, its function yields more efficient localization of ODV envelope proteins to the nucleus, and it has been pro-posed that this nuclear trafficking is performed by other proteins, including E26 and importin-␣-16 (26). In addition, there is a sig-nificant reduction in the major alphabaculovirus structural pro-tein polyhedrin at both the transcriptional and propro-tein levels in fp25kmutant infections (28). This suggests that FP25K may also play a role in the regulation of viral gene expression at the tran-scriptional level. Recent evidence also supports FP25K as a nega-tive factor of BV infectivity due to increased GP64 abundance in fp25kmutant BV in addition to its ability to prohibit viral genome incorporation (29).
Most studies to date on FP25K have focused on its function in the context of alphabaculovirus infection rather than orthologues from other baculovirus genera. FP25K orthologues are present in alpha-, beta-, and gammabaculoviruses and absent in deltabacu-loviruses (30). To gain insight into functional differences in alpha-and betabaculovirus FP25K proteins, Nakanishi et al. examined phenotypic changes between recombinant Bombyx mori NPV (BmNPV) expressing FP25K fromAutographa californicamultiple NPV (AcMNPV),Spodoptera lituramultiple NPV (SpltMNPV), or Xestia c-nigrum GV (XecnGV) (19). They found that XecnGV FP25K was unable to rescue the defects in oral infectivity and postmortem host degradation observed in the BmNPV FP25K deletion mutant. It is possible that XecnGV FP25K is incompatible in this BmNPV; however, data from that study may also suggest that thefp25kfunctionalities of alpha- and betabaculoviruses are different. Even thoughfp25korthologues were found in gamma-baculoviruses, thesefp25ksequences have low homology to alpha-and betabaculovirusfp25ksequences, with 8% identity and 14% similarity, and were therefore not considered for our localization studies (23,31). The relatedness of this gammabaculovirusfp25k orthologue is still uncertain.
Previous studies on AcMNPV FP25K localization suggested
that it is present near the endoplasmic reticulum (ER); however, to date, baculovirus FP25K localization and its amounts in the cytoplasm and nucleus have been inconclusive (26). Using immu-nogold transmission electron microscopy (TEM) analysis, Harri-son and Summers revealed that AcMNPV FP25K localized to the cytoplasm but were unable to ascertain its presence in the nucleus due to the presence of electron-dense structures (32). Also, FP25K was confined to large “amorphous structures,” making its local-ization difficult to determine by traditional biochemical methods (32). In immunofluorescence studies, Braunagel et al. also found that FP25K was abundant in the cytoplasm, but its presence in the nucleus was not evident (17).
In order to study FP25K localization, we cloned anfp25k-gfp fu-sion gene into anfp25kinsertion mutant bacmid virus
[AcFP-GFP(ProPH)]. During AcFP-GFP(ProPH) infection, AcMNPV
FP25K localized primarily to the cytoplasm, with a small percent-age being localized in the nucleus. FP25K is predicted to contain a highly conserved coiled-coil domain (17). During infection with an N-terminal coiled-coil domain deletion virus (⌬L13:L44), FP25K localization significantly changed to be primarily nuclear. WhenPlutella xylostellagranulovirus (PxGV) FP25K, which lacks the coiled-coil domain, was fused to green fluorescent protein (GFP) and cloned into thefp25kmutant bacmid, its infection also resulted in FP25K being localized predominantly to the nucleus. This suggests a critical role for this coiled-coil domain in main-taining alphabaculovirus FP25K cytoplasmic localization.
MATERIALS AND METHODS
Insect cell lines, viruses, and primers.Trichoplusia niBTI-TN-5B1-4 (Tn5), Spodoptera frugiperda IPLB-SF21AE (Sf21), and Sf21-specific clone (Sf9) cells were cultured in TNM-FH medium (Invitrogen, Carls-bad, CA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA) at 27°C. The AcMNPV bacmid (AcBacmid) viruses were produced by transfection of Tn5 cells using polyethylenimine (PEI) (Polysciences Inc., Warrington, PA) with bacmid DNA extracted from
Escherichia colistrain DH10Bac (Invitrogen) (33). BV was harvested by collection of culture medium, followed by centrifugation at 500⫻gfor 5 min to remove cell debris. To generate a virus that produces nonfused GFP, thegfpcoding sequence (CDS) was cloned from pBlueGFP into the pFastBac1 vector, and the resulting construct, pFastBac1-GFP, was trans-formed into competent DH10Bac bacteria to produce AcBacGFP in insect cells (10). Titers of all recombinant viruses, including viruses used for the setup of the time course study described below, were determined by end-point dilution assays (50% tissue culture infective dose [TCID50]) as de-scribed previously (34). PxGV granules were provided by Q. Qin of the Chinese Academy of Science. Genomic DNA of PxGV was purified ac-cording to previously reported procedures (34). All primers referred to in this study are shown inTable 1.
Generation of AcBacmid with inactivated endogenousfp25k. Wild-type AcBacmid (bMON14272) DNA was isolated from the respective bac-terial stocks and transfected into Sf21 cells. AcBacmid BV from transfec-tion was harvested and subsequently passaged 10 times into fresh Sf21 cells by using the previously passaged BV as the inoculum. Viral DNA was extracted from passaged AcBacmid BV. PCR was performed on passaged AcBacmid BV DNA by usingfp25kgene-specific primers fp25k-F and fp25k-R (Table 1) with expected 1.2-kb (wild-type) and 1.5-kb (mutant)
fp25kproduct sizes (10). The BV of the passage that showed a prominent 1.5-kb PCR product (passage 10) was used for viral DNA extraction and eventual transformation ofE. coliDH10B cells. Screening and selection of AcBacmid clones with an inactivated endogenousfp25k(AcBac-fp::287) were performed by colony PCR with the above-describedfp25kPCR primers and conditions. Thefp25klocus of AcBac-fp::287 was mutated through the insertion of a 287-bp piece of Sf21 host DNA into thefp25k
on November 7, 2019 by guest
http://jvi.asm.org/
coding sequence at the TTAA site at nucleotide (nt)⫹425, as previously reported (10). With a premature stop codon, this FP25K mutant sequence contains a 73-amino-acid (aa) deletion at the C terminus. fp25k::287 was sequenced, and the sequence is available under GenBank accession num-berKX284746. The resulting AcBac-fp::287 bacterial clone was trans-formed with helper plasmid pMON7124 to produce DH10Bac-fp::287 cells for ease of generation of recombinant viruses in our AcBac-fp::287 shuttle vector.
Generation of AcFP-GFP(ProPH), PxFP-GFP(ProPH), AcFP-GFP(ProFP), and GFP-AcFP recombinant viruses.To generate AcFP-GFP(ProPH)and PxFP-GFP(ProPH), expressing GFP-tagged FP25K from AcMNPV and PxGV, respectively, thefp25kgene CDSs from AcMNPV and PxGV were PCR amplified by using the high-fidelityPfuenzyme with genomic DNAs of the two viruses as the templates and cloned separately into pGEM-T Easy (Promega, Madison, WI) by using primers fuse-F and AcFP-fuse-R for AcMNPV and primers PxFP-fuse-F and Px-AcFP-fuse-R for PxGV (Table 1). To generate AcFP-GFP(ProFP), expressing GFP-tagged AcMNPV FP25K from its nativefp25kpromoter, thefp25kgene CDS and itsfp25kpromoter from AcMNPV were PCR amplified and cloned into pGEM-T Easy by using primers AcFPpromoter-F and AcFPpromoter-R. For AcFP-GFP(ProPH)and PxFP-GFP(ProPH), the C-terminal ends of each of thefp25kCDSs were translationally fused togfpin the pFastBac1 vector by BamHI/XbaI digestion of thefp25kcassettes and XbaI/XhoI digestion of thegfpcassette from pUC19-GFP (a clone collection in our laboratory), followed by subcloning into the BamHI/XhoI sites of pFastBac1, resulting in pFB1-AcFP-GFP(ProPH)and pFB1-PxFP-GFP(ProPH).
The fp25k promoter-driven bacmid [AcFP-GFP(ProFP)] was con-structed differently. Thefp25kCDS with the nativefp25kpromoter was translationally fused togfpas detailed above; however, the BamHI/XhoI site of pFastBacHTa was used for subcloning, resulting in pFBHTa-AcFP-GFP(ProFP). In order to drive expression from only the nativefp25k pro-moter, thepolyhedrin(polh) promoter from this construct was deleted by SfoI/SnaBI digestion and self-ligation with T4 DNA ligase (New England BioLabs Inc., Ipswich, MA).
For the GFP-AcFP virus,gfpwas translationally fused to the N termi-nus of AcMNPVfp25k. To do this, thegfpgene was PCR amplified from the pUC19-GFP template and cloned into pGEM-T Easy by using primers GFP-F-EcoRI and GFP-R-BamHI. The pFastBac1-GFP-AcFP vector was
produced by EcoRI/BamHI digestion of thegfpcassette from pGEMT-GFP and BamHI/XbaI digestion of thefp25k cassette from pGEMT-Acfp25k (from a clone collection in our laboratory, which contains the AcMNPVfp25kopen reading frame), followed by subcloning into the EcoRI/XbaI sites of pFastBac1.
All of these constructs were confirmed by sequencing and transformed into DH10Bac-fp::287 cells. The recombinant bacmid DNA produced was used to transfect Tn5 cells, and the resulting BV was harvested. Each of the resulting recombinant BVs was amplified only once in Tn5 cells to reduce any unwanted recombination of thefp25k-gfpfusion constructs.
Generation of the pIE1-AcFPGFP construct for expression of AcMNPV FP25K-GFP from a plasmid.AcMNPVfp25kwas transla-tionally fused togfpas described above but subcloned into the pIE1 vector (an expression vector containing theie-1promoter). The AcMNPVfp25k
fragment was recovered from pGEMT-Acfp25k⌬TAA (produced as de-scribed above) by BamHI/XbaI digestion. Thegfpfragment was cut out of pUC19-GFP by XhoI/Klenow digestion and then XbaI digestion. These fragments were subcloned into the XbaI/Klenow and BamHI sites of pIE1. The resulting construct, pIE1-AcFPGFP (at 1g), was transfected into Tn5 cells by using PEI and analyzed by confocal microscopy at 48 h post-transfection.
[image:3.585.43.546.78.314.2]Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting of FP25K-GFP and GFP proteins.Tn5 cells (5⫻105) and Sf9 cells (1⫻106) in 35-mm tissue culture dishes were infected with AcBacGFP and AcFP-GFP(ProPH)at a multiplicity of infection (MOI) of 1 and harvested at 48 h postinfection (hpi). Cells were rinsed in phosphate-buffered saline (PBS), harvested in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]), and soni-cated. The amount of protein in each sample was quantified by a Bradford assay (35). Each sample (30g) was heated at 100°C for 10 min in loading buffer (50 mM Tris-Cl [pH 6.8], 2% SDS, 0.1% bromophenol blue, 10% glycerol, and 100 mM dithiothreitol) and then subjected to SDS-poly-acrylamide gel electrophoresis (PAGE). SDS-PAGE gels were either stained with Coomassie brilliant blue or transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH) for Western blotting. The membrane was blocked with 5% nonfat dried milk (NFDM) in Tween plus Tris-buffered saline (TTBS) (150 mM NaCl, 50 mM Tris-HCl [pH TABLE 1List of primers
Primer Sequence (5=–3=) (restriction site)a
Fp25k-F TTTACGCACCATATACGC
Fp25k-R ATGCATAGCAATGTCTTC
AcFP-fuse-F GGATCCATGGATCAATTTGAACAGTTG (BamHI)
AcFP-fuse-R TCTAGATAAATTAAATTTTGAAGCATTTTTTCGC (XbaI)
PxFP-fuse-F GGATCCATGGACGACTGTGTGG (BamHI)
PxFP-fuse-R TCTAGATAATAAAACGATAAATATTTGATATCC (BamHI)
AcFPpromoter-F GGATCCAGTGTCTGTAAACTTGTTGGTCT (BamHI)
AcFPpromoter-R TCTAGATAAATTAAATTTTGAAGCATTTTTTCGC (XbaI)
pp34-F GTTGGAAACCGCTAAAGATGT
pp34-R AAGTCGGGTAACGAGTCTGTAA
AcFP-L13:L44-F CAGAGACACGTTAATCAACT
AcFP-L13:L44-R ACAGACAGCGTTGAGATATA
AcFP-G52:L139-F GTATATCTCAACGCTGTCTGTG
AcFP-G52:L139-R CAGCGGAGCAAAAGCTG
AcFP-K142:S206-F TTTAAGTAACAGCTTTTGCTC
AcFP-K142:S206-R AGCGAAAAAATGCTTCAAAATTTAA
AcFP-L36P-F CAATATCAAGTCGATGAGCGAAAAACCAAAAAGGC
AcFP-L36P-R ATTGTCGTATTCTAGCCTTTTTGGTTTTTCGCTC
AcFP-D22P-F AGTCGTTGATCAAAACGCAAATCCCCGAAAATGTG
AcFP-D22P-R CTTGATATTGTCCGACACATTTTCGGGGATTTGCG
GFP-F-EcoRI GAATTCATGGTGAGCAAGGGCGAG
GFP-R-BamHI GGATCCGGAACCACCACCACCCTTGTACAGCTCGTCCATG
aUnderlining indicates the location of the restriction site.
Garretson et al.
on November 7, 2019 by guest
http://jvi.asm.org/
7.6], and 0.05% Tween 20). The membrane was treated with an anti-GFP antibody at a 1:2,000 dilution (Affinity Bioreagents, Golden, CO) over-night (16 h) at 4°C, washed three times with TTBS (without NFDM), and incubated with horseradish peroxidase (HRP)-linked anti-rabbit IgG at a 1:1,000 dilution (Cell Signaling, Danvers, MA) at room temperature (23°C) for 1 h. The membrane was washed three times with TTBS (with-out NFDM) and developed with HRP color development reagent (Bio-Rad, Hercules, CA). The same procedure was used for PxGV FP25K-GFP, although only Tn5 cells were infected with PxFP-GFP(ProPH) and AcBacGFP.
BV assays.A time course was set up as follows: Tn5 cells (1⫻105) were infected in triplicate at an MOI of 1 with appropriate viruses, and BV was harvested at 12, 24, 36, 48, 60, and 72 hpi. The titers of all of the harvested BV samples were determined simultaneously by a modified quantitative real-time PCR (qPCR) method, as described previously (36). To convert the resulting viral copy number to PFU per milliliter, a stan-dard virus, AcBacGFP, was used for qPCR. The titer of this stanstan-dard virus was determined by a TCID50assay. Instead ofie-1-specific primers for qPCR, we used primers pp34-F and pp34-R, which were resistant to prim-er-dimer formation (Table 1).
Confocal microscopy and localization of GFP.For all confocal anal-yses, either Tn5 cells (5⫻105) or Sf9 cells (1⫻106) were adhered to specialized 35-mm glass-bottom dishes (MatTek Co., Ashland, MA), and these cells were either left uninfected or infected with AcBacGFP or AcFP-GFP(ProPH)viruses at an MOI of 1. At 12, 24, 36, and 48 hpi, the cells were stained with a live, permeable nuclear stain (Hoechst 33342; Thermo Fisher, Waltham, MA) at a 1:1,000 dilution for 1 h at room temperature (23°C) and then washed twice and incubated with TNM-FH medium. Samples were imaged by using a confocal laser scanning microscope (FV500; Olympus) with a 40⫻or 100⫻oil immersion objective lens and FluoView software. To prevent bleed-through, sequential scans of the GFP and 4=,6-diamidino-2-phenylindole (DAPI) channels were used. Composite images were assembled by using Adobe Photoshop CS5 soft-ware. For z-stack imaging, 40 scans of both GFP and DAPI channels were taken at increments of a 500-nm depth.
To determine the percentages of GFP localized to the nucleus and cytoplasm, individual cells in the confocal images were manually analyzed by using ImagePro software. A total of 30 GFP-positive cells from each infection were chosen for quantitative measurements. Cells were selected based on their middle position within the focal plane. The middle position can be approximated by the near-round shape of the nuclei and conden-sation of DNA as seen in Hoechst 33342 dye-only images. By using Im-agePro software, the total GFP sum intensity or the sum of all the pixels in a given region of each individual cell was determined in the GFP-only image. In the Hoechst 33342 dye-only image, a nuclear boundary was made for each cell based on the blue stain. This boundary was used in the GFP-only image to quantify the GFP sum intensity in the nucleus of that cell. The total GFP sum intensity of the cell was subtracted by the quantity in the nucleus to determine the amount of GFP in the cytoplasm. There-fore, the amount of GFP in either the nucleus or cytoplasm was divided by the total amount of GFP in the cell to determine the percentage in each cell compartment.
Determination of AcMNPV FP25K-GFP expression levels by GFP measurements.Tn5 cells (1⫻105) and Sf9 cells (5⫻105) were seeded into 24-well plates and infected with AcFP-GFP(ProPH) and AcFP-GFP(ProFP)at an MOI of 1 in triplicate. Infected cells were harvested at 48 hpi. Cells were rinsed once with PBS and lysed with 0.1% SDS at 27°C for 15 min. Lysis was confirmed by microscopy, and supernatants were transferred to 96-well black polystyrene plates (Greiner Bio-One, Mon-roe, NC). GFP measurements were taken at an excitation wavelength of 485 nm and an emission wavelength of 535 nm on a Filtermax F5 micro-plate reader (Molecular Devices, Sunnyvale, CA). The amount of GFP for each sample is represented by the change in fluorescence units (⌬FU), or the mean fluorescence units (FU) of the infected sample over the mean FU of the uninfected sample.
Alignment of baculovirus FP25K sequences.All available baculovi-rus genomes from GenBank (as of September 2014) were uploaded to the CLC Bio Main Workbench. These genomes were manually searched for FP25K/open reading frame 61 (ORF61) homologues. The amino acid sequences of these FP25K/ORF61 homologues from 61 baculovirus ge-nomes were extracted, compiled, and aligned by using CLC Bio Main Workbench.
Coiled-coil prediction. The amino acid sequences of wild-type FP25K and both the D22P and L36P mutants were entered into the COILS server (http://www.ch.embnet.org/software/COILS_form.html), and the coiled-coil structure of these proteins was predicted (37). The output for this prediction is represented by graphical plots, with all resi-due-scanning windows (14,21,28) containing the probability of a coiled-coil structure based on similarity to structures in a database of known coiled-coil proteins. High-probability peaks in these plots indicate the likelihood of a coiled-coil structure in that region.
Generation of recombinant FP25K deletion and point mutation vi-ruses.Inverse PCR and site-directed mutagenesis, as described above, were used to producefp25kdeletions andfp25kpoint mutations, respec-tively, in the template pFBHTa-AcFP-GFP(ProFP).
Inverse PCR was performed on pFBHTa-AcFP-GFP(ProFP)to produce
⌬L13:L44,⌬G52:L139, and⌬K142:S206 deletions within FP25K. The fol-lowing primers were used: AcFP-L13:L44-F and AcFP-L13:L44-R for ⌬L13:L44, AcFP-G52:L139-F and AcFP-G52:L139-R for⌬G52:L139, and AcFP-K142:S206-F and AcFP-K142:S206-R for⌬K142:S206 (Table 1).
Site-directed mutagenesis was performed on pFBHTa-AcFP-GFP(ProFP) to produce D22P and L36P mutations within FP25K by using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The following primers were used: AcFP-L36P-F and L36P-R for the L36P mutation and D22P-F and AcFP-D22P-R for the D22P mutation (Table 1).
Both the deletions and point mutations produced were confirmed by sequencing. The resulting deletion and point mutation constructs were transformed into DH10Bac-fp::287 cells. The recombinant bacmid DNAs produced were used to transfect Tn5 cells, and the resulting BV was har-vested.
The GenBank nucleotide sequence accession number for the AcM-NPVfp25kand 287-bp Sf21 host DNA fusion sequences isJX569226.
RESULTS
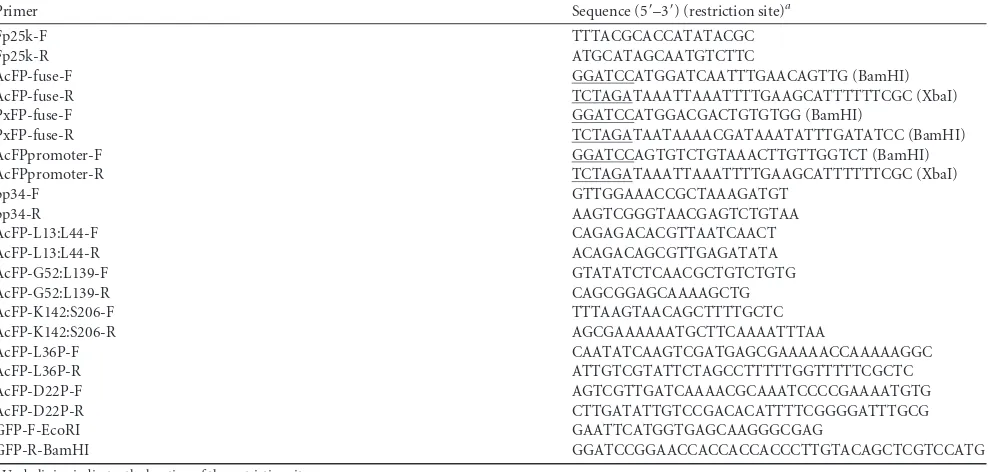
Construction, expression, and functionality of the AcMNPV FP25K-GFP fusion during infection.Previous work on FP25K localization was largely inconclusive due to inconsistent results of fractionation and immunogold TEM studies (32). Therefore, we determined FP25K localization using a more quantitative strategy by the production of a virus with an AcMNPV FP25K-GFP fusion for quantitative measurement of GFP levels during infection of cells (Fig. 1B). In order to accomplish this, we first inactivated the endogenousfp25kcopy of AcBacmid by passaging AcBacmid BV 10 times in Sf21 cells, followed by PCR screening (Fig. 1A, left). fp25kmutants with an inserted 287-bp piece of host cellular DNA (GenBank accession numberKX284746) were prominently de-tected at passage 10 (Fig. 1A, left). As determined by Southern blot analysis of the viral genome using this 287-bp host DNA as a probe, this host DNA was inserted only once in the viral genome at thefp25klocus (data not shown) and resulted in the introduction of a premature stop codon in the 287-bp host DNA sequence. The AcBacmid DNA from passage 10 was then introduced intoE. coli cells by transformation. A clone (C9) carrying the mutantfp25k gene and termed AcBac-fp25k::287 was isolated and confirmed by PCR (Fig. 1A, right). Next, the AcMNPVfp25k-gfpcassette was introduced into the polh locus of AcBac-fp25k::287 to create AcFP-GFP(ProPH). Fusion protein production was confirmed by
on November 7, 2019 by guest
http://jvi.asm.org/
immunoblotting of AcFP-GFP(ProPH)-infected Tn5 and Sf9 cells (Fig. 1C). Relative to the 27-kDa molecular mass of GFP from infection with the nonfused AcBacGFP control, it is clear that a full-length AcMNPV FP25K-GFP fusion protein was produced, which ran at the predicted fusion molecular mass of 51 kDa in both cell lines (AcMNPV FP25K, 24 kDa; GFP, 27 kDa). There was a small percentage of the AcMNPV FP25K-GFP fusion protein that was cleaved during infection of both cell lines. Previous stud-ies also observed full-length 25-kDa and cleaved 23-kDa FP25K products from cell lysates of AcMNPV E2 infections, in addition to cleaved FP25K from Helicoverpa armigera nucleopolyhedrovi-rus (HearNPV) infections (16, 17). It was noted previously by Braunagel et al. (17) that the cleavage was likely caused by degra-dation during harvesting; however, we found that harvesting at 48 hpi yielded considerably fewer cleavage products than those at 72 hpi, suggesting that this process occurred during infection (Fig. 1Cand data not shown).
Finally, in order to confirm the loss of function of the endog-enousfp25kcopy of AcBac-fp25k::287 and rescue by AcMNPV FP25K-GFP at the polhlocus, we performed a BV production assay (Fig. 1D). Since AcBac-fp25k::287 is a mutant clone result-ing from virus passage in cells and served as the viral background for the production of the fusion proteins in this study, it was important to test the loss of functionality by the mutant and com-plementation by the addition of the fusion protein. As infection progressed, particularly after late times (⬎24 hpi), the AcBac-fp25k::287 mutant produced more BV than did the AcBacGFP wild-type control, which has a functionalfp25kgene, as previously reported (16,18). At the same time, the AcMNPV FP25K-GFP fusion protein from AcFP-GFP(ProFP)complemented the mutant fp25kcopy of AcBac-fp25k::287 since the level of BV production of AcFP-GFP(ProFP)was similar to that of the AcBacGFP control during cell infection. Since AcFP-GFP(ProFP)demonstrated nor-mal levels of BV production, it was used for quantitative FP25K FIG 1Generation of an AcMNPV FP25K-GFP fusion protein for FP25K localization studies. (A) AcMNPV-based bacmid (AcBacmid) transfection and subsequent continuous BV passaging in Sf21 cells lead to insertional mutagenesis of thefp25klocus. (Left) PCR of thefp25klocus from AcBacmid passages. Purified AcBacmid DNA from bacteria was transfected, and BVs from prior infections were used as the inoculum for sequential passaging in Sf21 cells 10 times. BV DNA was extracted, and thefp25klocus was amplified by PCR usingfp25k-specific primers (Table 1). (Right) PCR confirmation of anfp25kmutant AcBacmid (AcBac-fp25k::287) clone. BV DNA from AcBacmid passage 10 was transformed intoE. colicells. A particularfp25kmutant clone (C9) was isolated, and thefp25k
locus was amplified by PCR using the samefp25k-specific primers. NTC, nontemplate control. (B) Diagram of AcFP-GFP(ProPH)virus construction. All subsequent figures depict a similar construction utilizingE. coliDH10Bac-fp25k::287 cells. AcMNPVfp25kandgfpcoding sequences were fused and introduced into the donor vector pFastBac1. The resulting pFB1-AcFP-GFP(ProPH)plasmid was transformed intoE. coliDH10Bac-fp25k::287 cells. This cell line also contains the pMON7124 helper plasmid that encodes the Tn7transposase allowing recombination between the mini-Tn7attachment sites on bacmid bMON14272 at the
polyhedrin(polh) locus and pFastBac1 (boxed). Bacmid bMON14272 in this cell-specific line contains thefp25kinsertional mutant generated as described above for panel A. ProPH,polhpromoter; L, linker sequence betweenfp25kandgfpCDSs. (C) Detection of the full-length AcMNPV FP25K-GFP fusion protein from AcFP-GFP(ProPH)-infected Tn5 and Sf9 cells by Western blotting. Tn5 and Sf9 cells were infected with the unfused AcBacGFP control virus and AcFP-GFP(ProPH). At 48 hpi, cell lysates were subjected to Western blot analysis using anti-GFP polyclonal antibody. (D) AcMNPV FP25K-GFP shows functionality in terms of reduced BV production during AcFP-GFP(ProPH)infection in the budding assay. Tn5 cells were infected with AcBacGFP (wild-type control), AcBac-fp25k::287-GFP (mutant), and AcFP-AcBac-fp25k::287-GFP(ProFP)at an MOI of 0.1. At different time points postinfection, BV was harvested, and titers were determined by qPCR. Statistical significances were determined by using the Studentttest and are indicated by asterisks (*,P⬍0.05; **,P⬍0.005).
Garretson et al.
on November 7, 2019 by guest
http://jvi.asm.org/
[image:5.585.42.537.66.372.2]localization studies. Overall, these BV production assay results confirm the inactivation of endogenous fp25k in the AcBac-fp25k::287 mutant and the rescue of FP25K functionality in the virus producing the AcMNPV FP25K-GFP fusion [AcFP-GFP(ProFP)]. It is possible that the insertional mutant AcBac-fp25k::287 may encode a partial FP25K protein; nonetheless, this virus does not have full functionality in the context of BV production (Fig. 1D).
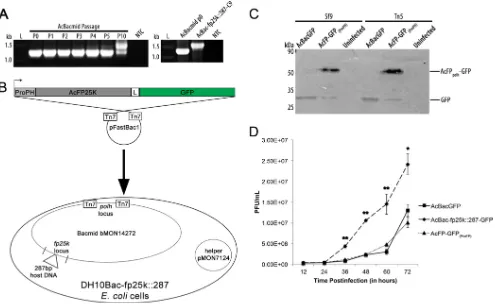
Quantitative analysis of AcMNPV FP25K localization to the nucleus and cytoplasm.In order to investigate where AcMNPV FP25K was localized during infection, Tn5 and Sf9 cells were in-fected with AcFP-GFP(ProPH)and monitored by confocal micros-copy at 12, 24, 36, and 48 hpi (Fig. 2A). A small quantity of AcMNPV FP25K-GFP was produced starting at 24 hpi and was found entirely in the cytoplasm. By 36 and 48 hpi, about 4% of the total FP25K was present in the nucleus, but it still accumulated predominantly in the cytoplasm, at 96% (Fig. 2BandC). In addi-tion, at these times, a large proportion of FP25K localized to the cytoplasm, and several GFP-labeled amorphous structures were observed in many cells (Fig. 2A, white arrowheads). Previously, these large FP25K amorphous structures were observed during AcMNPV infection (18). This localization appears to be FP25K specific, because the GFP localization was uniform across both cell lines (as seen for AcBacGFP control infection) (Fig. 2A). Also, the percentages of FP25K found in the nucleus and cytoplasm are almost identical between infections of Sf9 and Tn5 cells (Fig. 2B andC). The same localization was also observed with an N-termi-nal GFP-AcFP25K fusion (data not shown) and an FP25K anti-body (38).
In immunogold TEM studies, Harrison and Summers ob-served FP25K in electron-dense structures in the nuclear periph-ery, thought to be chromatin (32). We also found a small quantity of FP25K present in the nucleus (Fig. 2A). It is important to verify this finding because the role of and/or interactions with FP25K in the nucleus could be different from those in the cytoplasm. To confirm that AcMNPV FP25K-GFP was indeed found in the nu-cleus and was not from signal bleed-through, z-stack analysis was performed during AcFP-GFP(ProPH)infection of Tn5 and Sf9 cells (Fig. 2D). FP25K was found across multiple depths of the nucleus for both Tn5 and Sf9 cell infections. FP25K was found both in the center of the nucleus and near the nuclear envelope. Previously reported evidence supports the localization of membrane-associ-ated FP25K to the INM surface because of its association with trafficking ODV envelope proteins from the outer nuclear mem-brane to the INM (26).
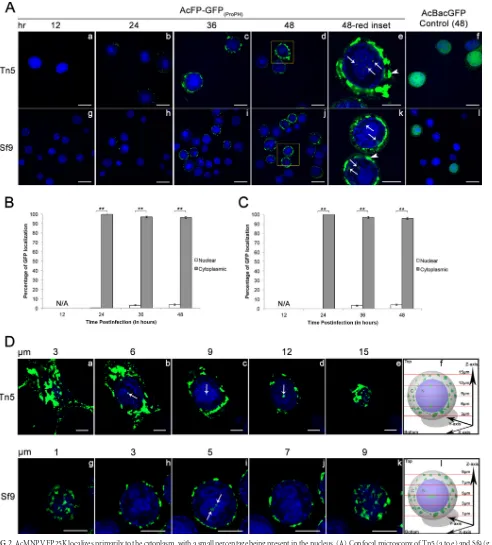
Localization of AcMNPV FP25K is independent of expres-sion levels.Overexpression of proteins can alter subcellular local-ization (39–42). In AcFP-GFP(ProPH) infection, the AcMNPV FP25K-GFP fusion protein is under the control of the highly active polhpromoter. Therefore, it was necessary to verify that overex-pression of AcMNPV FP25K-GFP did not alter its localization within the cell. To achieve this, we produced the AcFP-GFP(ProFP) virus that expresses AcMNPVfp25k-gfpunder the control of the nativefp25kpromoter (Fig. 3A). First, the promoter activities of AcFP-GFP(ProPH)and AcFP-GFP(ProFP)were compared by mea-suring the amount of GFP being produced during infection of Tn5 and Sf9 cells (Fig. 3B). During infection of both Tn5 and Sf9 cells, AcFP-GFP(ProPH)resulted in a ⬎4-fold increase in GFP levels compared to those with AcFP-GFP(ProFP)infection. The subcellu-lar localization of AcMNPV FP25K-GFP from infection of Tn5
cells with AcFP-GFP(ProPH)and AcFP-GFP(ProFP)was analyzed by confocal microscopy at 48 hpi (Fig. 3C). Considerably less overall FP25K-GFP and fewer and smaller amorphous structures were observed in AcFP-GFP(ProFP)-infected cells. However, as seen with AcFP-GFP(ProPH), some AcMNPV FP25K-GFP was also found to localize to the nucleus of AcFP-GFP(ProFP)-infected cells. Overall, the percentages of detectable FP25K in the nucleus (4% versus 5%) and cytoplasm (96% versus 95%) were very similar between the two viruses (Fig. 3D). These data support the notion that over-expression has no impact on the nuclear and cytoplasmic localiza-tion of FP25K in infected insect cells.
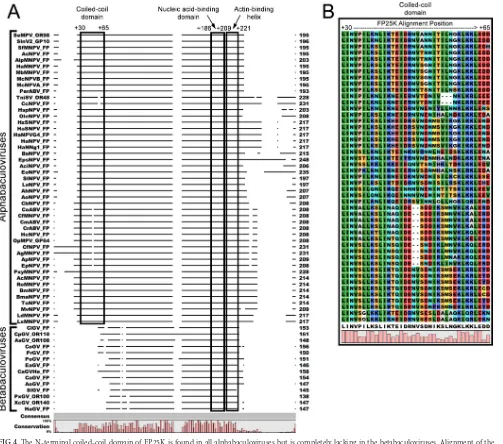
An N-terminal coiled-coil domain is present in alphabaculo-virus FP25Ks but not betabaculoalphabaculo-virus FP25Ks.We observed lo-calization of FP25K primarily to the cytoplasm. However, to date, the genetic determinant(s) of FP25K localization is uncertain. Braunagel et al. reported the association of FP25K with several ODV proteins and, using microsomal membranes, presented ev-idence suggesting that this may be occurring at the ER membrane (26). Protein domain searches indicated that AcMNPV FP25K is composed of a conserved N-terminal coiled coil, a putative actin binding helix, and a nucleic acid binding motif (43,44). We hy-pothesized that because of its role in protein-protein interactions, the N-terminal coiled-coil domain of FP25K plays a role in its predominant cytoplasmic localization by associating with ODV proteins. To understand its conservation among baculoviruses, the FP25K or ORF61 homologue amino sequences were extracted from all available (as of 2014) baculovirus genomic sequences and aligned (Fig. 4A). By comparing the entire aligned sequences, it is evident that all alphabaculovirus FP25K sequences contain an N-terminal coiled-coil domain and a small fragment (5 to 52 aa in length) at the C-terminal end, both of which are lacking in the betabaculovirus FP25K sequences (Fig. 4A). This N-terminal coiled-coil domain sequence spanned from around alphabaculo-virus amino acid position⫹5 to position⫹40 (or alignment po-sitions⫹20 to⫹55). Among the alphabaculovirus FP25Ks, the N-terminal coiled-coil domain sequence shows a high level of conservation, with a score of 98 out of 100 from a T-Coffee align-ment (Fig. 4B) (45).
To investigate whether this coiled-coil domain is necessary for protein localization in other related proteins, the AcMNPV FP25K amino acid sequence was run through a protein struc-ture prediction server. Sequence-based similarity searches (e.g., BLAST) for FP25K have been largely unsuccessful in identifying proteins with similar sequences in order to predict the function(s) of FP25K and its corresponding domains. We employed a protein structure prediction search using the more sensitive HHpred pro-gram to identify more remotely related proteins (46). Almost the entire AcMNPV FP25K protein (aa 10 to 189 out of 214) displayed 99.9% structural similarity and a strong E value of 3.0E⫺23 with long interspersed element 1 (LINE-1) open reading frame 1 pro-tein (ORF1p). FP25K from betabaculovirus PxGV also had 99.5% structural relatedness and an E value of 7.4e⫺14 with LINE-1 ORF1p. However, because PxGV FP25K does not contain the pu-tative N-terminal coiled-coil domain, it aligned to only part of ORF1p. LINE-1 is an autonomous retrotransposon found in mammalian genomes, and its ORF1 protein functions as a nucleic acid chaperone (47). Like the alphabaculovirus FP25Ks, this ORF1p has a conserved N-terminal coiled-coil domain. ORF1p also contains a highly conserved noncanonical RNA recognition motif (RRM) and a basic carboxy-terminal domain (CTD).
on November 7, 2019 by guest
http://jvi.asm.org/
FIG 2AcMNPV FP25K localizes primarily to the cytoplasm, with a small percentage being present in the nucleus. (A) Confocal microscopy of Tn5 (a to e) and Sf9 (g to k) cells infected with AcFP-GFP(ProPH)and AcBacGFP viruses, as indicated, at an MOI of 1. At 12 hpi (a and g), 24 hpi (b and h), 36 hpi (c and i), and 48 hpi (d to f and j to l), AcFP-GFP(ProPH)- and AcBacGFP-infected Tn5 and Sf9 cells were treated with the permeable nuclear stain Hoechst 33342 (blue, nucleus), and confocal images were taken. The yellow insets from samples from AcFP-GFP(ProPH)infection at 48 hpi (d and j) are shown in panels e and k. White arrows indicate locations where GFP is present in the nucleus. White arrowheads designate the described amorphous structures. Bars, 25m (a to d, f, g to j, and l) and 10m (e and k). (B and C) Quantitative analysis of AcMNPV FP25K-GFP localization in the cytoplasm and nucleus by confocal microscopy of AcFP-GFP(ProPH)-infected Tn5 (B) and Sf9 (C) cells. The amounts of GFP are based on 30 GFP-positive cells from each infection. Values are reported as a percentage of the total cellular AcMNPV FP25K-GFP in either the cytoplasm or the nucleus. N/A signifies that no AcMNPV FP25K-GFP was found at 12 hpi. Error bars represent the standard deviations of data from the 30 cells analyzed. Statistical significance is indicated by asterisks (**,P⬍0.005). (D) z-stack analysis of Tn5 (a to e) and Sf9 (g to k) cells infected with AcFP-GFP(ProPH)virus at an MOI of 1. In the
zaxis, images were taken every 3m for Tn5-infected cells and every 2m for Sf9-infected cells. Cells were treated with the nuclear stain Hoechst 33342 (blue, nucleus). White arrows indicate GFP in the nucleus. The relative depth of the image in micrometers is indicated on top of each image. Bars, 10m. Panels f and l show schematics summarizing data from all the images taken in thezplane. The red lines indicate where the images were taken on thezaxis.
Garretson et al.
on November 7, 2019 by guest
http://jvi.asm.org/
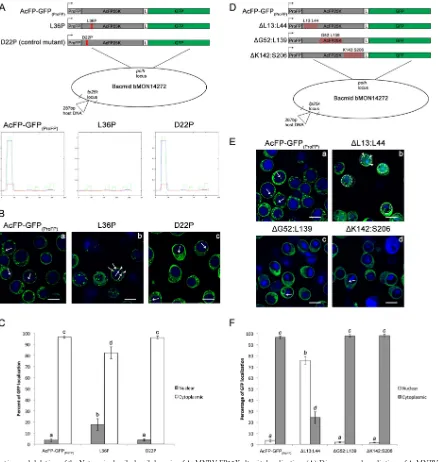
[image:7.585.47.539.66.611.2]Goodier et al. found that the N-terminal one-third of ORF1p is necessary for cytoplasmic retention and protein localization (48). Interestingly, they also discovered that ORF1p coimmu-noprecipitated and colocalized with several RNA binding pro-teins known to be part of cytoplasmic stress granules. This could offer some insight into the amorphous FP25K structures that we, and others, have observed in the cytoplasm of AcM-NPV-infected cells (Fig. 2and3) (32). This demonstrates that this putative N-terminal coiled-coil domain present in alpha-baculovirus FP25Ks could be involved in protein localization. FP25K coiled-coil domain mutations change localization.In order to address the role of the N-terminal coiled-coil domain in FP25K localization, a set of proline point mutations were made in the coiled-coil domain of the AcMNPV FP25K-GFP fusion
pro-tein at amino acid positions 22 and 36 (Fig. 5A, top). As predicted by the COILS program, a leucine or isoleucine at position 36 is highly conserved and critical for the formation of the coiled-coil structure, and the introduction of a proline here disrupted its predicted structure (Fig. 5A, bottom) (37). On the other hand, the aspartic acid at position 22 is variable and not required for the coiled-coiled structure, so a proline substitution here should not and did not impact its structure (Fig. 5A, bottom). Localization of AcFP-GFP(ProFP) (control), the L36P mutant (coiled-coil mu-tant), and the D22P mutant (control mutant) during infection of Tn5 cells was monitored by confocal microscopy at 48 hpi (Fig. 5B). More amorphous structures and overall GFP were observed in the nuclei of L36P mutant-infected cells than in the nuclei of AcFP-GFP(ProFP)- and D22P mutant-infected cells (Fig. 5B, white FIG 3polhandfp25kpromoter-driven expression does not alter AcMNPV FP25K-GFP localization. (A) Diagrams indicating the differences between the AcFP-GFP(ProPH)and AcFP-GFP(ProFP)viruses. Both viruses were constructed as described in the legend ofFig. 1. However, for AcFP-GFP(ProFP), the AcMNPV
fp25kcoding sequence (in gray) containing its respective promoter sequence upstream (in light gray) was fused togfp. ProPH,polhpromoter; ProFP,fp25k
promoter; L, linker sequence betweenfp25kandgfpCDSs. (B) Confirmation of lower AcMNPV FP25K-GFP expression levels infp25kthan inpolh promoter-driven virus. Tn5 and Sf9 cells were infected with AcFP-GFP(ProFP)(gray bars) and AcFP-GFP(ProPH)(black bars) viruses at an MOI of 1. The level of GFP expression was determined at 48 hpi and is represented by the change in fluorescence units (⌬FU) in the infected sample relative to the uninfected sample. (C) Confocal microscopy at 48 hpi of Tn5 cells infected with AcFP-GFP(ProPH)and AcFP-GFP(ProFP)viruses at an MOI of 1. Cells were treated with the nuclear stain Hoechst 33342 (blue, nucleus). The yellow insets from the 48-hpi samples (a and c) (bar⫽25m) are shown in panels b and d (bar⫽10m). White arrows indicate locations where GFP is present in the nucleus. White arrowheads designate the described amorphous structures. (D) Quantitative analysis of AcMNPV FP25K-GFP localization in the cytoplasm and nucleus by confocal microscopy of AcFP-GFP(ProPH)- and AcFP-GFP(ProFP)-infected cells. The amounts of GFP were based on 30 GFP-positive cells from each infection. Values are reported as a percentage of the total cellular AcMNPV FP25K-GFP level in either the cytoplasm or nucleus. Error bars represent the standard deviations of data from the 30 cells analyzed. Statistical significance is indicated by the asterisks (**,P⬍
0.005).
on November 7, 2019 by guest
http://jvi.asm.org/
[image:8.585.41.541.63.420.2]arrowheads). This was confirmed by quantitative analysis, where
⬃18% of GFP from L36P mutant infection localized to the nu-cleus, resulting in a⬎5-fold increase compared to levels from AcFP-GFP(ProFP)and D22P mutant infections (Fig. 5C).
Since these point mutations had only a moderate effect on phenotype, we decided to confirm the role of the coiled-coil do-main in localization by deleting portions of the FP25K protein (Fig. 5D). A series of three deletions in FP25K were made:⌬L13: L44 (coiled-coil domain deletion),⌬G52:L139 (middle deletion), and⌬K142:S206 (C-terminal deletion) (Fig. 5D). Next, confocal microscopy was performed on Tn5 cells infected with these three deletion viruses, in addition to the full-length AcFP-GFP(ProFP) FP25K control (Fig. 5E). In contrast to the localization observed
for the other deletions (⌬G52:L139 and⌬K142:S206) and the full-length control, it is clear that deletion of the N-terminal coiled-coil domain (⌬L13:L44) dramatically changed FP25K localiza-tion to a largely nuclear localizalocaliza-tion (Fig. 5E). Approximately 76% of GFP from⌬L13:L44 infection localized to the nucleus, whereas⬍4% of GFP was nuclear in AcFP-GFP(ProFP),⌬G52: L139, and⌬K142:S206 infections (Fig. 5F).
The difference in phenotype between⌬L13:L44 and the other viruses was more robust and apparent than that of the point mu-tations. Even though the L36P point mutation altered the pre-dicted coiled-coil structure, it is possible that this FP25K point mutant could still maintain some weak association with ODV proteins or other partners in the cytoplasm. Also, it appears that FIG 4The N-terminal coiled-coil domain of FP25K is found in all alphabaculoviruses but is completely lacking in the betabaculoviruses. Alignment of the baculovirus FP25K amino acid sequences was performed by using CLC Main Workbench. The FP25K or ORF61 homologues were extracted from currently availableBaculoviridaegenomes. Shown is a representative wide view of the whole FP25K protein (A), and the coiled-coil domain is boxed (B). The alpha- and betabaculovirus genera are indicated, along with the predicted coiled-coil domain, nucleic acid binding domain, and actin-binding helix at the top. The amino acid positions according to the alignment are also designated.
Garretson et al.
on November 7, 2019 by guest
http://jvi.asm.org/
[image:9.585.42.545.65.509.2]FIG 5Mutation and deletion of the N-terminal coiled-coil domain of AcMNPV FP25K alter its localization. (A) Diagrams and prediction of AcMNPV FP25K coiled-coil formation in each point mutation construct. (Top) Location of the point mutations within the N-terminal coiled-coil domain of the AcMNPV FP25K-GFP fusion protein. The red “X” indicates the position of the point mutation in each of the constructs. These constructs were introduced into the mutant bacmid, as described in the legend ofFig. 1. (Bottom) Graphical output for prediction of AcMNPV FP25K coiled-coil formation from the primary sequence of wild-type FP25K and the L36P and D22P mutants by using the COILS program (http://www.ch.embnet.org/software/COILS_form.html). Theyaxis represents the probability of coiled-coil formation obtained by the program, and thexaxis shows the position of the residue. Strong, high peaks by window 14 (green), window 21 (blue), and window 28 (red) represent a high probability of coiled-coil formation. (B) Confocal microscopy at 48 hpi of Tn5 cells infected with AcFP-GFP(ProFP), L36P, or D22P virus at an MOI of 1. Cells were treated with the nuclear stain Hoechst 33342 (blue, nucleus). Bars, 25m. White arrows indicate locations where GFP is present in the nucleus. White arrowheads designate the described amorphous structures. Dotted white circles indicate the position of the nucleus in cells with a diffuse GFP localization. (C) Quantitative analysis of AcMNPV FP25K-GFP localization in the cytoplasm and nucleus by confocal microscopy of AcFP-GFP(ProFP), L36P, or D22P samples. Samples were based on 30 GFP-positive cells from each infection. Values are reported as a percentage of the total cellular AcMNPV FP25K-GFP level in either the cytoplasm or the nucleus. Error bars represent the standard deviations of data from the 30 cells analyzed. Different letters indicate statistical significance between the localizations of the constructs (P⬍0.05). (D) Diagrams showing the location of the deletions within the AcMNPV FP25K-GFP fusion protein. The red squares indicate the position of the deletion in each of the constructs. (E) Confocal microscopy at 48 hpi of Tn5 cells infected with AcFP-GFP(ProFP),⌬L13:L44,⌬G52:L139, and⌬K142:S206 viruses at an MOI of 1, as described above for panel B. (F) Quantitative analysis of AcMNPV FP25K-GFP localization in the cytoplasm and nucleus by confocal microscopy of AcFP-GFP(ProFP),⌬L13:L44,⌬G52:L139, and⌬K142:S206 samples, as described above for panel C.
on November 7, 2019 by guest
http://jvi.asm.org/
[image:10.585.75.515.65.527.2]even without the coiled-coil domain, some FP25K remained in the cytoplasm. However, we believe that this is attributed mainly to the time of infection at which microscopy was performed, because at later time points (⬎72 hpi), almost all of the FP25K-GFP made it to the nucleus during⌬L13:L44 infection (data not shown).
It appears that in addition to localization changes seen with the
⌬L13:L44 virus, infections with the⌬G52:L139 and⌬K142:S206 mutants resulted in less FP25K amorphous structure formation (Fig. 5E). The strong punctate FP25K-GFP structures seen with wild-type AcFP-GFP(ProFP) infection appeared to be reduced considerably, leading to more homogeneous localization across the cytoplasm with the⌬G52:L139 and⌬K142:S206 mu-tants (Fig. 5E).
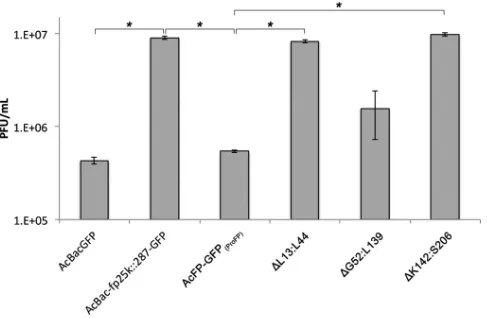
The coiled-coil domain and C-terminal region of AcMNPV FP25K regulate BV production.Previous studies showed that FP25K may play a key role in regulating the switch between and the ratio of the two virion phenotypes BV and ODV (18,38). In fp25kmutants, fewer occlusion bodies and fewer ODVs per occlu-sion body are formed, but more BV is produced (16,18,23,49). Therefore, to functionally investigate the role of the FP25K N-ter-minal coiled-coil domain in BV production, we performed a bud-ding assay on the AcFP-GFP(ProFP), ⌬L13:L44,⌬G52:L139, and
⌬K142:S206 viruses (Fig. 6). Since the largest difference in BV production was observed at 72 hpi, as shown inFig. 1D, this time point was represented inFig. 6and8. As expected, and as shown in Fig. 1D, AcBac-fp25k::287-GFP (bacmid mutant) had 21- and 17-fold-higher levels of BV production than did AcBacGFP (wild-type bacmid) and AcFP-GFP(ProFP)(FP repair control), respec-tively. The coiled-coil ⌬L13:L44 and C-terminal ⌬K142:S206 viruses had 15- and 18-fold-higher BV titers than did AcFP-GFP(ProFP), respectively. However, the⌬G52:L139 virus did not show an increase in BV production. This finding suggests that the N-terminal coiled-coil domain and C-terminal region of FP25K are critical for the budding process of the virus.
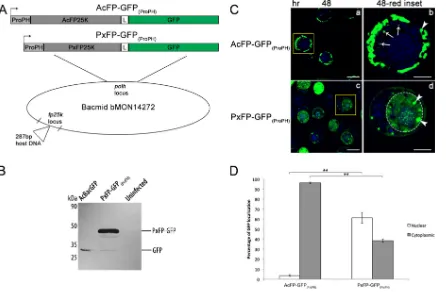
PxGV FP25K localizes to the nucleus.The alignment of bac-ulovirus FP25K sequences yielded an intriguing observation: none of the betabaculovirus FP25Ks contained an N-terminal
coiled-coil domain (Fig. 4). It was reported previously that XecnGV FP25K can rescue some of the phenotypic defects of the BmNPV fp25kmutant, but it is uncertain how betabaculovirus FP25K lo-calizes during infection (19). Based on our findings that the N-ter-minal coiled-coil domain determines the fate of FP25K in the cytoplasm, we hypothesized that since betabaculovirus FP25Ks do not contain this coiled-coil domain, PxGV FP25K would localize to the nucleus. To test this hypothesis, a PxGV FP25K-GFP fusion protein was produced and tracked during infection (Fig. 7A). First, the PxGVfp25k-gfpfusion cassette was introduced into the polhlocus of AcBac-fp25k::287 C9, and fusion protein production was confirmed by immunoblotting of PxFP-GFP(ProPH)-infected Tn5 cells (Fig. 7B). Relative to the 27-kDa nonfused GFP product from AcBacGFP control infection, there was a full-length PxGV FP25K-GFP fusion protein being produced at the predicted mo-lecular mass of 42 kDa (PxGV FP25K, 15 kDa; GFP, 27 kDa). As shown in Fig. 1C, there are some cleavage products detected, which were reported previously (17). This cleavage between FP25K and GFP may represent a conserved process across bacu-lovirus FP25Ks, but more studies need to be done in order to confirm this result. The subcellular localization of PxGV FP25K was resolved by performing confocal microscopy on Tn5 cells infected with AcFP-GFP(ProPH)and PxFP-GFP(ProPH)viruses at 48 hpi (Fig. 7C). There was a stark contrast between the primarily cytoplasmic localization of AcMNPV FP25K-GFP and the more prominent nuclear localization of PxGV FP25K-GFP in AcFP-GFP(ProPH)- and PxFP-GFP(ProPH)-infected cells, respectively. In PxFP-GFP(ProPH)infection, 61% of PxGV FP25K-GFP localized to the nucleus, which was 17-fold higher than the amount of nuclear AcMNPV FP25K-GFP (Fig. 7D). This fold difference in nuclear localization was similar to that detected between⌬L13: L44 virus (coiled-coil deletion)- and AcFP-GFP(ProFP) control-in-fected cells (Fig. 5F). PxGV FP25K-GFP also localized primarily to the nucleus at 24 hpi (data not shown), which indicates that be-tabaculovirus FP25K localizes to the nucleus soon after expression and likely before nuclear membrane breakdown (3, 12, 13). In addition, it appears that FP25K amorphous structure formation was sustained during PxFP-GFP(ProPH)infection, as shown by the punctate FP25K structures observed in both the cytoplasm and nucleus (Fig. 7C, white arrowheads).
To verify that sequence and, more specifically, nuclear local-ization signal (NLS) differences were not the primary drivers of localization changes between AcMNPV FP25K and PxGV FP25K and that the presence or absence of the coiled-coil domain was the main contributor, the PxGV and AcMNPV FP25K sequences were analyzed for a putative NLS. According to NLS Mapper (a pro-gram that predicts nuclear localization signals), both PxGV FP25K and AcMNPV FP25K sequences have similar scores, indi-cating the presence of NLSs in both proteins (50). Therefore, it can be concluded that betabaculovirus FP25Ks localize to the nucleus because of their lack of coiled-coil domain structure and the pres-ence of NLSs. The NLS present in alphabaculovirus FP25Ks does not drive its cytoplasmic localization, because it is likely hidden or masked by the structure or protein-protein interactions guided by the coiled-coil domain.
PxGV FP25K is unable to restore BV production.When BV production of the deletion viruses was assessed (Fig. 6), the result-ing levels of BV from infections with the⌬L13:L44 (coiled-coil deletion) and ⌬K142:S206 (C-terminal deletion) viruses were similar to those from infections with thefp25kmutant. In order to FIG 6BV production is altered by deletion of the N-terminal coiled-coil
domain and C-terminal region of AcMNPV FP25K. Tn5 cells were infected with AcBacGFP (wild-type control), AcBac-fp25k::287-GFP (mutant), AcFP-GFP(ProFP)(deletion background),⌬L13:L44,⌬G52:L139, and⌬K142:S206 viruses at an MOI of 0.1. At 72 hpi, BV was harvested, and titers were then determined by qPCR. Statistical significance is indicated by asterisks (*,P⬍
0.05). Garretson et al.
on November 7, 2019 by guest
http://jvi.asm.org/
[image:11.585.41.285.66.225.2]confirm that the N-terminal coiled-coil domain of alphabaculo-virus FP25K plays a role in BV production, we determined the level of BV production of PxFP-GFP(ProFP)in Tn5 cells (Fig. 8). As with AcFP-GFP(ProFP), the PxFP-GFP(ProFP)virus expresses PxGV fp25k-gfpunder the control of its nativefp25kpromoter. PxFP-GFP(ProFP)infection yielded 7-fold-higher BV titers than those of AcFP-GFP(ProFP)infection. The amount of PxFP-GFP(ProFP)BV (which contains PxGVfp25k-gfpin thepolhlocus of the fp25k::287 bacmid) produced approached and was not significantly different from that of the AcBac-fp25k::287 mutant. Therefore, PxGV FP25K is unable to rescue BV production to the wild-type level. This provides further evidence that the N-terminal coiled-coil do-main of alphabaculovirus FP25Ks plays an important role in BV production and that betabaculovirus FP25Ks may not require this domain for the process of budding in betabaculoviruses.
DISCUSSION
The FP (few-polyhedra) phenotype was first observed over 40 years ago and is characterized by fewer OBs, a decrease in virion occlusion, and an increase in BV production, which provides a
FIG 7PxGV FP25K localizes primarily to the nucleus. (A) Diagrams indicating the differences between AcFP-GFP(ProPH)and PxFP-GFP(ProPH)viruses. The fusion constructs were introduced into the mutant bacmid, as described in the legend ofFig. 1. In PxFP-GFP(ProPH), the PxGVfp25kcoding sequence (which does not contain an N-terminal coiled-coil domain) was fused togfp. L, linker sequence betweenfp25kandgfpCDSs. (B) Detection of the full-length PxGV FP25K-GFP fusion protein from PxFP-GFP(ProPH)-infected Tn5 cells by Western blotting. Tn5 cells were infected with the unfused AcBacGFP control virus and PxFP-GFP(ProPH). At 48 hpi, cell lysates were subjected to Western blot analysis using an anti-GFP polyclonal antibody. The GFP protein is 27 kDa. The predicted molecular mass of the PxGV FP25K-GFP fusion protein is 42 kDa. (C) Confocal microscopy at 48 hpi of Tn5 cells infected with AcFP-GFP(ProPH)and PxFP-GFP(ProPH)viruses at an MOI of 1. Cells were treated with the nuclear stain Hoechst 33342 (blue, nucleus). The yellow insets from the 48-hpi samples (a and c) (bar⫽25m) are shown in panels b and d (bar⫽10m). White arrows indicate locations where GFP is present in the nucleus. White arrowheads designate the described amorphous structures. Dotted white circles indicate the position of the nucleus in cells with a diffuse GFP localization. (D) Quantitative analysis of AcMNPV FP25K-GFP localization in the cytoplasm and nucleus by confocal microscopy of AcFP-GFP(ProPH)- and PxFP-GFP(ProPH)-infected samples. Samples were based on 30 GFP-positive cells from each infection. Values are reported as a percentage of the total cellular AcMNPV FP25K-GFP level in either the cytoplasm or the nucleus. Error bars represent the standard deviations of data from the 30 cells analyzed. Statistical significance is indicated by asterisks (**,P⬍0.005).
FIG 8PxGV FP25K does not restrict virus budding to wild-type levels. Tn5 cells were infected with AcBacGFP (wild-type control), AcBac-fp25k::287-GFP (mutant), AcFP-AcBac-fp25k::287-GFP(ProFP), and PxFP-GFP(ProFP)viruses at an MOI of 0.1. At 72 hpi, BV was harvested, and titers were then determined by qPCR. Statistical significance is indicated by asterisks (*,P⬍0.05).
on November 7, 2019 by guest
http://jvi.asm.org/
[image:12.585.71.517.67.359.2] [image:12.585.298.544.516.674.2]selective advantage for this phenotype in cell culture (18,23,25, 51,52). This FP phenotype has been observed in multiple group I and II alphabaculoviruses and is often associated with mutations in thefp25kgene (16, 18,23,49,51, 53, 54). Two localization studies on AcMNPV FP25K yielded disparate results (17,32). By way of immunogold electron microscopy of AcMNPV-infected cells, Harrison and Summers found that FP25K localized to large cytoplasmic amorphous structures and electron-dense structures in the nucleus, thought to be chromatin (32). Contrary to these results, Braunagel et al. observed the presence of FP25K only in the cytoplasm (17). Using a virus that produced a functional AcM-NPV FP25K-GFP fusion protein during infection (Fig. 1D), we detected 96% of AcMNPV FP25K in the cytoplasm, present mostly in large amorphous structures; however, 4% localized to the nucleus (Fig. 2AtoC). Nuclear AcMNPV FP25K was con-firmed by z-stack analysis and was localized to both the center and the periphery of the nucleus (Fig. 2AandD). Despite being under the control of the strongpolhpromoter, the expression levels of the FP25K-GFP fusion protein did not change its lo-calization (Fig. 3).
Our localization data led us to ask, what is the genetic determi-nant(s) of FP25K localization? Braunagel et al. reported that membrane-associated FP25K, along with BV/ODV-E26 and im-portin-␣-16, traffic and sort viral proteins across the INM (26). Several ODV envelope proteins associate with FP25K and/or BV/ ODV-E26, such as ODV-E66 and ODV-E25 (26). From domain prediction, it is apparent that AcMNPV FP25K has conserved N-terminal coiled-coil, putative actin binding helix, and nucleic acid binding domains. Additionally, a protein structure prediction search using the AcMNPV FP25K amino acid sequence in HH-pred resulted in an extremely high level of structural relatedness (99.9%) to LINE-1 ORF1p, which also contains an N-terminal coiled-coil domain previously shown to be necessary for cytoplas-mic retention (46,48). Based on its established function in pro-tein-protein interactions and structural similarity to the domain in LINE-1 ORF1p, we hypothesized that the N-terminal coiled-coil domain of AcMNPV FP25K is responsible for its prominent cytoplasmic localization. When baculovirus FP25K amino acid sequences are aligned, the N-terminal coiled-coil domain is found in all alphabaculoviruses but none of the betabaculoviruses (Fig. 4). Both the coiled-coil domain point mutation to disrupt struc-ture (L36P) and a complete deletion (⌬L13:L44) resulted in a shift of AcMNPV FP25K localization from the cytoplasm to the nu-cleus (Fig. 5). In addition, when PxGV FP25K localization was tested, it was found primarily in the nucleus (Fig. 7). This suggests that the N-terminal coiled-coil domain present in all alphabacu-lovirus FP25K sequences plays a key role in their localization to the cytoplasm, possibly by way of protein-protein interactions with ODV envelope proteins, as previously reported for AcMNPV FP25K (17). Additionally, many of these ODV envelope proteins contain N-terminal domains abundant in hydrophobic residues that are critical for FP25K or ODV-E26 interactions, and hydro-phobic domains often associate with coiled-coil domains (55,56). Based on its lack of an N-terminal coiled-coil domain and nuclear barrier during betabaculovirus occlusion, we propose that the roles of interaction and trafficking of ODV envelope proteins within the betabaculovirus life cycle may not be performed by their FP25K homologues (3,12,13).
Coiled-coil regions within cellular and viral proteins are criti-cal for protein locriti-calization, such as the Golgi protein giantin (57),
the T-cell activation scaffold protein CARMA1 (58), the transcrip-tion factor promyelocytic leukemia retinoic acid receptor alpha (PML-RAR␣) (59), foamy virus Gag protein (60), and the integral membrane glycoprotein tetherin (61). At times, the coiled-coil domain function in subcellular localization is associated with ho-mo- or hetero-oligomerization of these proteins. Another prom-inent example of these properties is LINE-1 ORF1p, which has strong structural relatedness to the FP25K orthologues in alpha-baculoviruses and betaalpha-baculoviruses. LINE-1 ORF1p contains an N-terminal coiled-coil domain, a noncanonical RRM, and a basic CTD, all of which are highly conserved (47). These predicted fea-tures are found in alphabaculovirus FP25Ks and, with the excep-tion of the N-terminal coiled-coil domain, betabaculovirus FP25Ks. Like the alphabaculovirus FP25Ks, the N-terminal coiled-coil domain of ORF1p was also found to be a critical deter-minant of its cytoplasmic retention and protein localization (48). Additionally, the N-terminal coiled-coil domain of ORF1p is also important for its oligomerization, which is necessary for ORF1p RNA binding activity (47). Not only did ORF1p localize to the cytoplasm, it also was present near large cytoplasmic foci identi-fied as stress granules. Intriguingly, we, and others, observed that the alphabaculovirus FP25K protein studied also localized to large cytoplasmic amorphous structures (32). Stress granules are cyto-plasmic aggregates assembled in response to stress conditions like heat shock, oxidative stress, and viral infection and are necessary for cellular maintenance of gene expression, homeostasis, and cy-topathology (62). As with other cellular processes, these stress granules can be controlled and manipulated by viruses to promote viral replication, viral translation, and antiviral evasion. Recently, Kotani et al. found that the BmNPV nucleic acid binding protein baculovirus repeated open reading frames B/E (BRO-B/E) local-ized to amorphous cytoplasmic foci and associated with a cellular translation regulator, T-cell intracellular antigen 1 homologue (BmTRN-1), a known component of stress granules (63). They also determined that BRO-B/E expression led to decreased protein synthesis and proposed that this may be an additional mechanism for controlling protein synthesis in baculoviruses. Since alpha-baculovirus FP25Ks appear to localize to similar amorphous cy-toplasmic structures with or without infection (data not shown) (structures are smaller in uninfected cells), and regardless of the expression level, we believe that these structures are part of the virus life cycle and are controlled by baculoviruses for their benefit through posttranscriptional regulation, perhaps in part through FP25K. Further studies are needed to investigate these FP25K cy-toplasmic foci and their potential identity as stress granules along with the predicted activity of nucleic acid binding in FP25K and possibly FP25K oligomerization involved in this process.
The phenomenon of higher-level BV production, less virus occlusion, and OB production in alphabaculovirusfp25kmutants is well established in the literature (8,16,18,23,24,49,54). We looked at BV production by the AcMNPV FP25K deletion viruses (⌬L13:L44, ⌬G52:L139, and ⌬K142:S206) and found that the coiled-coil domain (⌬L13:L44) and C-terminal domain (⌬K142: S206) deletions resulted in higher levels of BV production, reach-ing levels similar to those of thefp25kmutant (AcBac-fp25k::287) (Fig. 6). Since the FP25K C-terminal deletion (⌬K142:S206) un-couples localization changes and increased BV production, it would be intriguing to investigate its role in FP25K function. The PxFP-GFP(ProFP)virus also increased levels of BV production to nearlyfp25kmutant levels (Fig. 8). This is slightly dissimilar from Garretson et al.