Vol.40,No. 2 JOURNAL OF VIROLOGY, Nov. 1981,p.367-378
0022-538X/81/110367-12$02.00/0
Proteins
Specified
by
Bovine
Herpesvirus
1
(Infectious Bovine
Rhinotracheitis Virus)
VIKIAM MISRA,* ROBERT M. BLUMENTHAL,t AND LORNE A. BABIUK
Departmentof Veterinary Microbiology, WesternCollegeof Veterinary Medicine, University of
Saskatchewan,
Saskatoon,
Saskatchewan,Canada S7N OWOReceived 2February 1981/Accepted 1 July 1981
An electrophoretic analysis of radioactively labeled, purified, "empty" and
DNA-containing infectious bovine rhinotracheitis virions revealed thepresence
of25 to 33 structural (virion) polypeptides. Atotal of11 of these polypeptides
could be labeled with [3H]glucosanine andwere identified as glycoproteins. In
additiontothe25structuralpolypeptides, infectious bovine rhinotracheitis
virus-infected cells also contained atleast 15 nonstructural (nonvirion) polypeptides
thatwerenotpresentinpurified virions. Expression of the viral polypeptides in
infected cellswascontrolled temporally. Thus,mostviralpolypeptidescould be
categorized as "a" (immediate early), ",B" (early), or"y" (late) on the basis of
their order of appearance in infected cells and whether their syntheses were
dependentuponprior viralproteinorDNAsynthesis.Noneof theglycoproteins
belonged tothe a class, although atleast one (GVP11) wassynthesized in the
absence ofviral DNA synthesis. Serum from a cow in which infectiousbovine
rhinotracheitis virus lesionswerereactivated by dexamethasoneprecipitatedboth
structuralandnonstructuralpolypeptides.
Bovineherpesvirus 1,whichis also knownas
infectious bovine rhinotracheitis virus (IBRV),
is similarinstructuretootherherpesvirusesand
possessesalineardouble-strandedDNA genome
which hasamolecularweightofapproximately
10 (J. E.Farley,I. B.Skare,andJ.Skare, Abstr.
Int.Conf.HumanHerpesviruses, 1980).Like the
genomesof otherherpesviruses, thegenome of
IBRV is comprised of L and S components,
althoughonlytwogenomicisomers exist in
prep-aration sinceonlytheScomponentispresentin
both of thetwopossible orientations(Farleyet
al., Abstr. Int. Conf. Human Herpesviruses,
1980). Also like otherherpesviruses, IBRVcan
remain latentinanimals, probablyintrigeminal
ganglions,and can be reactivated with relative
ease(12,21). IBRV isanimportantpathogen of
cattle and can cause severe respiratory
infec-tions, vulvovaginitis, abortions, conjunctivitis, meningoencephalitis, and generalized systemic
infections (9). Thus, IBRV represents a good
modelforstudyingthebiologyandimmunology
of active and latent herpesvirus infections in
natural hosts.
Asmany as 33 virionpolypeptides havebeen
identifiedfor other herpesviruses (4-6, 11,
27-29). In herpesviruses 1 and 2, at least five of these structural polypeptides are glycoproteins.
t Present address: Cold Spring Harbor Laboratory, Cold Spring Harbor,NY11724.
Theseglycoproteinsarealsopresent onthe
sur-faces ofinfectedcells,andatleastsomeof them
areinvolved in immunerecognitionand immune
cytolysis (18).
The herpesvirus proteins can be designated
a, ,B,and-y,dependingonthetemporalorder of synthesis and whether synthesis is dependent
uponthesuccessfulprogressionof certain phys-iological processes in infected cells (24). The transcriptionofa orimmediateearlygenesdoes
notrequire theexpressionof other viral genes,
and in the absence of protein synthesis (e.g.,
after treatment with cycloheximide) mRNA's
from a genes accumulate. If the protein
syn-thetic blockisremoved,aproteinsareproduced
inlargeamounts evenin the absenceof further
mRNA synthesis (24). The synthesis of /3 or
early proteinsisdependentupontheprior
syn-thesis of theaproteinsand inturnleadstothe
cessation ofaproteinsynthesis.
'The
transcrip-tionofy orlategenesrequirestheprior
expres-sionofa and
/B
proteins,aswellastheonsetofviralDNAsynthesis.
There are considerable data available
con-cerning thekineticsof virusreplicationinvitro
andimmunedestructionofIBRV-infected cells
by bovine leukocytes (1, 25). Very little
infor-mationhas beenreported concerningthe
corre-lation of immune destruction to specific
poly-peptide synthesis and expression on host cell
membranes. Inanattempttoidentifythe poly-367
on November 10, 2019 by guest
http://jvi.asm.org/
368 MISRA, BLUMENTHAL, AND BABIUK
peptides involved in immune destruction, the
polypeptides expressed in latently infected cells,
and the temporalpatternof polypeptide
synthe-sis in lytically infected cells, we tried to identify
and characterize allof the polypeptides
synthe-sized in IBRV-infected cells. Here we describe
the identification of more than 25 structural
polypeptides, 11of whichareglycoproteins and
15ofwhicharenonstructural polypeptides. We
also attempted to assign the IBRV-specified
polypeptidestothea,,B,and y temporal classes.
MATERIALS AND METHODS
Virusand cells. Strain P8-2 of IBRVwascultured
inGeorgia bovinekidneyorMadin-Darbybovine
kid-neycells as describedpreviously(1).Briefly,cells were
grown toconfluency inpetridishes (diameter, 60 or
100mm;Corning Glass Works, Corning, N.Y.) in Eagle
minimal essential medium(MEM) supplementedwith
5%fetal bovineserum.
Infection and labeling of cells. Monolayers of
bovine kidney cells were infected with IBRV at a
multiplicity of infection of10PFU per cell.After the
virusabsorbed for1 hat370C,themonolayerswere
washed with MEM andoverlaid with methionine-free
MEM (catalog no. 79-0115; GIBCO Laboratories,
GrandIsland, N.Y.) containing2%fetal bovine serum.
After incubation for6h to permit cessation of host
protein synthesis, 25,LCiof[35S]methionine (catalog
no. SJ204; Amersham Corp., ArlingtonHeights, Ill.)
permlwasaddedtoeach tissue culture dish. The cells
wereharvested20h afterinfection. To label
glycopro-teins, 25,ICiof[3H]glucosamine(catalogno.TRK398;
Amersham)permlwasaddedtothe infected cultures
6hafterinfection.
Toidentify proteins synthesized in the absence of
DNA synthesis, cells were infected and then
main-tained in the presence of50,ugofcytosinearabinoside
(ara-C;Cytosar;TheUpjohn Co., Kalamazoo, Mich.)
perml. At6hafterinfection, the mediumwasreplaced
with5mlof MEMcontaining50,uCiof
[3S]methio-nine per ml and 50,ugof ara-C per ml.
Toidentifyapeptides,cellswereinfected and
main-tained in the presence of50,ugofcycloheximide
(Boeh-ringer MannheimCorp.,NewYork, N.Y.)perml. At
6hafterinfection,thecellswerewashedextensively
with MEM before the addition of MEMcontaining50
gCiof[3S]methionineperml and 2.5,ugof
actinomy-cinD(Sigma Chemical Co., St. Louis, Mo.) per ml.
Purification of virus. [3S]methionine- or
[3H]glucosamine-labeled virus waspurified from the
growth supernatant of infected cells. After the cell
debriswasremovedby centrifugationat 500xgfor
10 min, the virus was pelleted by centrifugation at
100,000xgfor2h. The viruspelletwassuspendedin
1mlofphosphate-buffered saline (PBS) (0.15MNaCl,
2.5mMKCI, 1.5mMKH2PO4,8mMNa2PO4,0.02%
dextrose,pH 7.4) by repeated pipetting and mild
son-ication (1to2 s at alow powersetting). The viruswas
thenlayeredonto an11-ml linear20to50%potassium
tartrategradientin 0.15 MNaCl-0.01MTris-0.001 M
EDTA(pH 7.5) andcentrifugedat80,000xg for1.5
h at 4°C. Fractions (0.2ml) were collected,and the
J. VIROL.
virus-containingfractionswereidentifiedby
measur-ing theradioactivity in each fraction. These fractions
werethenpooledanddiluted withPBS,and the virus
wasrecoveredbypelleting.
Preparation of cell extracts. The cell extracts werepreparedby the method of Purifoy and Powell
(22).Approximately 107 pelleted cellsweresuspended
in1ml ofasolutioncontaining20mMTris(pH 7.5),
2mM,B-mercaptoethanol,and 500 g of bovineserum
albumin per ml. After thispreparationwassonicated
for 2 min at apowersetting of7 in a Sonifier cell
disrupter (Biosonics, Plainview, N.Y.), 1ml of1.7M
NaCl-5 mM EDTAwasadded,andthe mixturekept
on ice for 40 min.The resulting precipitate was
re-movedbycentrifugation at30,000 xgfor20minat
4°C, and the mixturewasdialyzed againstasolution
containing200mMTris(pH7.5),50mMNaCl,1mM
EDTA, 2mM/i-mercaptoethanol, and 10%glycerol.
Thelight precipitatethat formedduring dialysiswas
removedbycentrifugationat100,000xgfor60minat
40C.
PreparationofStaphylococcusaureusstrain A
adsorbant. The Cowan strain of S. aureus was
ob-tained from the American Type Culture Collection
(ATCC12598) andwaspreparedforadsorption by the
technique of Kessler (13). The bacteriaweregrownfor
24 h at 37°C per ml in brain heart infusion broth
(GIBCO)supplementedwith4,ugofniacin per ml and 2Mugofthiamine-hydrochloride perml;then the cells
were harvested by centrifugation and washed twice
with PBS-2 (150 mM NaCl,20mMNa2HPO4, 20 mM
NaH2PO4, pH 7.2) containing 0.05% sodium azide.
After thecellsweresuspendedto aconcentrationof
10% (vol/vol) in PBS-2 containing azide, they were
stirredat roomtemperaturefor1.5h inthe presence
of1.5%Formalin, washed,andsuspendedtothesame concentration in buffer without Formalin. The cells
were then stirred for 5 min at 80°C, and this was
followed by rapid cooling on ice. After two washes
with PBS-2containing azide, the bacteria were
sus-pended to afinal concentration of 6% (vol/vol) and
storedat4°C.Before adsorption,the cellswere
pel-leted, suspended to a concentration of 10% in 0.5%
Nonidet P-40(Shell)in NETbuffer(150mMNaCl,5
mM EDTA, 50mM Tris, pH 7.4, 0.02% azide),and
incubatedat roomtemperaturefor15min.They were
then washed once and resuspended in fresh buffer
containing0.5% Nonidet P-40tothesamevolume. Forimmunoprecipitation,the bovine anti-IBRV
im-munoglobulin G fraction and the cell extract were
incubated inoptimumproportions for1h at37°C; this
wasfollowed by the addition of200
pl
ofanS. aureuscellsuspension.After thecells stood for90minat4°C
and were washed once with NETbuffer containing
Nonidet P-40, they were suspended in 50
pl
of 2%sodiumdodecyl sulfate (SDS)-6 M urea, incubated for
15 min at 45°C, heated at 100°C for 3 min, and
analyzed bySDS-polyacrylamide gel electrophoresis
(PAGE).
Analysis ofproteins by PAGE. Samples were
electrophoresed in the presence of SDSthrough 7.5
and 10%polyacrylamide gels (15) by using a 15-cm
verticalelectrophoresis apparatus (Richter Scientific,
Vancouver, Canada). Gels containing'S-labeled
sam-pleswere dried and autoradiographed on Kodax
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 40, 1981
OMAT-R film. 'H- and 14C-containing gels were
soaked in En3Hance (New England Nuclear Corp.,
Lachine, Canada) and fluorographed on preflashed
film.
The molecular weights of the radioactive bands
weredetermined by comparing theRfvalues of these
bands with theRfvalues of markers of known
molec-ularweights(high-andlow-molecular-weight
calibra-tion kits; Pharmacia, Uppsala, Sweden)
electropho-resed on the samegel. Molecular weight markerswere
visualized by staining the gels with Coomassie blue
beforedrying.
Two-dimensional gel electrophoresis. The
technique of O'Farrell (19) was used, with the
modifi-cations described by Garrels (8). Virus pellets were
suspended in
100-pd
volumes of a solutioncontaining0.24% SDS and 1.4M /3-mercaptoethanol; 2 ul of0.5
M Tris(pH8.8)buffercontaining1mgof DNaseI,0.5
mg of RNase A per ml, and 0.1 M MgCl2 was then
added. After thepreparationstoodonice for5 min,
100 mgof solidurea wasadded, followedby15ml of
lysis buffer (10 M urea, 4% Nonidet P-40, 0.5 mM
lysine-hydrochloride,0.1Mdithiothreitol, 0.25% SDS,
4.5%glycerol, 0.05 ml of ampholytes per ml).
Polypep-tideswereseparated along the first dimensioningels
PROTEINS SPECIFIED BY IBRV 369
containing9partsofBiolyte (pH5to7)and1partof
Biolyte (pH3to10) andalong the second dimension
bySDS-PAGEon10%gels. The gelswere thensoaked
inEn3Hance (NewEnglandNuclear), dried,and
fluo-rographed.
RESULTS
Purificationof IBRV. Theaccurateanalysis
of virionproteins requires thatthevirus
prepa-ration be essentially free of cellular and
non-structuralviral proteins. Potassium tartrate
gra-dients wereusedtopurify IBRV since they have been used to purify other herpesviruses (5, 14) and give good yields ofpure virus. To reduce
contamination with host cell proteins further,
the virus was purified from the extracellular
medium rather than fromacelllysate.
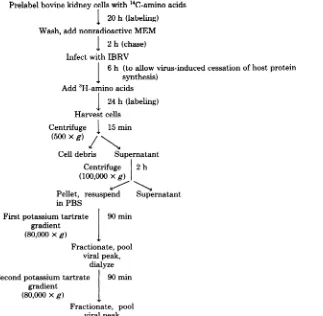
The efficacy of potassium tartrate gradients
in the purification of IBRV was evaluated by
labeling the purifying virus (Fig. 1). At every
stage of the purification process, samples were
treated with trichloroacetic acid, and the
amounts of 3H-labeled (viral) protein and
14C-Prelabel bovine kidneycells with 14C-amino acids
I 20h(labeling)
Wash, add nonradioactive MEM
l 2h (chase)
Infect withIBRV
J 6 h (toallow virus-induced cessation of hostprotein
synthesis)
Add3H-amino acids
I 24 h(labeling)
Harvestcells
Centrifuge 15min
(500xg) /
Celldebris Supernatant
Centrifuge 2h
(100,000xg)
Pellet, resuspend Supernatant
inPBS
Firstpotassiumtartrate 90min
gradient
(80,000xg)
Fractionate, pool viral peak,
dialyze Secondpotassiumtartrate 90min
gradient
(80,000xg)
Fractionate, pool
viralpeak
FIG. 1. Evaluationofpotassiumtartrategradientsas ameansofpurifyingIBRV.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.497.99.422.330.646.2]370 MISRA, BLUMENTHAL, AND BABIUK
labeled (cellular) proteinweredetermined.
Ta-ble1showsthatsimplecentrifugation at 100,000
x g resulted in significant purification since
morethan90% of the'4Cremained inthe
super-natant, whereas almost all of the 3H (added to
the cell culturesafter cessation of host protein
synthesis) wasremoved inthe pellet. Through
thenext twostepsofpurification, the ratio of 3H
to 14C remained relatively constant, although
therewasaconsiderableloss oftotal
radioactiv-ity. In subsequent experiments the virus was
centrifugedthrough onlyonetartrategradient.
To determine whether the small amount of
14C that remained associated with the virus in
the experiments described above was due to a
breakdown of cellular proteinsfollowed by
rein-corporation of the released label into viral
pro-teins or due tocontamination by cellproteins,
we performed the following experiment. Cells
were grown for 20 h in the presence of
[tS]-methionine, washed,grownforanadditional2h
in isotope-free MEM, and then infected with
IBRV. No radioactiveprecursor wasadded after
infection. Virus was purified from the cell-free
growth supernatantasdescribed above. The
vis-ible virus band was recovered from the first
tartrate gradient, pelleted, and analyzed by
SDS-PAGE along with labeled IBRV marker
proteins. The smallamountofradioactivity
as-sociated with the viruswasdistributedevenlyin
all viral polypeptides, suggesting that amino
acids fromprelabeled cellproteinswere
proba-bly released andreincorporated. In any case,it
seemed clear that thepurification protocol
de-scribed above yielded IBRV virions that were
essentiallyfree of host cellproteins.Virus
puri-fied in thismanner was also free of
contamina-tion withnonstructuralviralproteins since
non-TABLE 1. Purification ofIBRV6
Trichloroacetic acid-precipitable Sample radioactivity (cpm)
3H 14C
Cell-freegrowthsuper- 4.7 x106(100)b 9.2 x104(100) natant
100,000-x-gsuperna- 1.7x105(36) 8.6 x104(93) tant
100,000-x-gpellet 4.9 x106(100) 3.0 x104(32)
Virus bandafterfirst 1.42 x106(30) 1 xl04(10)
tartrategradient
Virus band aftersecond 2.5 x105(4.3) 8.6 x102(0.9)
tartrategradient
'Georgiabovinekidney cellswerelabeledwith"4C-amino
acids before infectionandwith3H-amino acidsafter infection
with IBRV.Virus was purifiedfromgrowthsupernatantfrom
which the cell debris hadbeenremovedbycentrifugationat 500xgfor15min.
bNumbersinparentheses arepercentages ofthetrichlr roaceticacid-precipitable radioactivityin thecell-freegrowth
supernatant.
J. VIROL.
structural viral peptidespresentin theinfected
cells inrelativelyhigh concentrations werenot
apparent in the virus preparation even after
prolonged autoradiography ofthe gel (data not
shown).
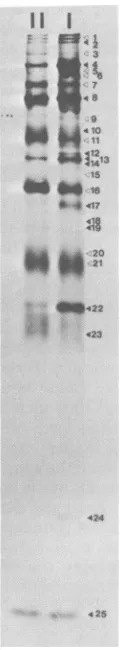
IBRV structural proteins. In potassium
tartrate gradients IBRVwasresolved intotwo
distinctpeaks (Fig. 2A). [3S]methionine-labeled
IBRVwasrecovered from the radioactivepeaks
andanalyzed bySDS-PAGE,followedby
auto-radiography andfluorography.Thegelsrevealed
25 bands, which were designated GVP1, VP2,
GVP3, VP4, GVP5, GVP6, GVP7, VP8, GVP9,
VP10, GVP11, VP12, VP13, VP14, GVP15, GVP16, VP17, VP18, VP19, GVP20, GVP21, VP22, VP23, VP24, and VP25. These bands
*0
CE)
x
U)
0
±
(l.)
FRACTION#
FIG. 2. Analysis ofIBRV onpotassium tartrate
gradients.(A)[3Slmethionine-labeledIBRVwas
re-coveredfrom thegrowth supernatant by
centrifuga-tion. Thepelletwassuspended in1 mlofPBS and
layeredonto a 20 to50%potassiumtartrategradient.
Aftercentrifugation for90minat80,000xg, 0.2-mi
fractionswerecollected, and theradioactivity
asso-ciated with eachfractionwasdetermined.Fractions
14 to 20 were combined intopool I (peak I), and
fractions22 to 26 were combined intopoolII(peak
II). Viruses were recovered from thesepools and
analyzedby PAGE (see Fig. 3). (B)Infectedcellswere
labeled with either [3H]thymidine or "C-amino
acids.At 24 hafter infection.th-c,, uwthsupernatants
were combined, o"I ahe virus was recovered and
analY7pAJ . a20to50%potassiumtartrategradient.
2,actions12to40 wereprecipitated with
trichloro-acetic acid, and the amountsof"C and 3H in the
macromoleculesofeachfractionweredetermined.
I
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.497.264.461.245.493.2] [image:4.497.62.256.478.621.2]PROTEINS SPECIFIED BY IBRV 371
ranged in molecular weight from more than
330,000 toless than 14,000(Fig.3and Table2).
A total of 11 of these bands (GVP1, GVP3,
GVP5, GVP6, GVP7, GVP9, GVP11, GVP15,
GVP16,GVP20,andGVP21)wereglycoproteins
since they were labeled with
[3H]glucosamine
(Fig.4). GVP9 and GVPllwere presentin
rel-atively large amounts. Some of the remaining
bands were lighter and were visible only after
prolonged exposure. GVP9, GVP5, GVP6, and
GVP15 either hadverylow methioninecontents
or wererelatively highlyglycosylatedsincethey
were not labeledextensively with
[3S]methio-nine.
Peak II, the more slowly sedimenting peak,
differed from peak I in that it lacked VP17,
VP22, and VP24, but the glycoproteins were
present atroughlythesamelevelsin bothpeaks.
3 - 1413 15 4417 418 419 2 0 422 M423 424 .425
FIG. 3. Analysis of
r35SJmethionine-labeled
virusby electrophoresison a10%polyacrylamidegel.
Vi-ruses were recovered from peaks I and II of the
potassium tartrategradient shown in Fig. 2A. The
open arrowheads indicate viralglycopeptides, and
the solid arrowheads indicate
[image:5.497.256.440.75.497.2]non-glycosylatedpoly-peptides. Viralpolyl GVP VP2 NSa GVP NSb NSc VP4 GVP GVP NSd NSe GVP' NSf VP8 NSg NSh NSi GVP vPl1 GVP VP12 VP13 VP14 NSJ NSk GVP GVP VP17 VP1E VP19 GVP NS1 GVP NSm NSn VP22 NSo VP23 VP24 VP25
TABLE 2. Viralpolpeptidesa
Mol wt cal- Mol wt
cal-culated from cal-culated firom
peptide 10% gel 7.5% gel
(X103) (X103) 1 >330 263 185 -3 182 182 166 141 5 138 6 130 126 120
7 104.7 105
93
89.1 91
89.1 91
81.2 87
83
9 77.6 82
74.1 79
'11 67.6 74
63.9 69
ib 63.4 65
1 62.9 62
58.2 60
56.1 57
'15 54.9 55
16 53.9 54
3b 20 21 52.8 50.9 50.0 49.1 48.2 42.2 38.4 36.3 34.8 29.4 26.2 16.2 13.9 48 41 39 35 32 Cate-gory Y 9 9 Y a a ?
11
? Y ? a Y a p ? /1 /1 p Y ? 'Y p 9 9 9 ? ? /1? ? 11 /3aPolypeptideswerenumberedinorder of
decreas-ing molecularweight. Structural polypeptides were
designated VP (virionpolypeptide)orGVP
(glycosyl-ated virionpolypeptide),and nonstructural
polypep-tides were designated NS andwere assigned
lower-caseletters.
bThesespeciesmay becomposedofmorethanone
polypeptide since they produced multiple spots on
two-dimensionalelectrophoresisgels.
Therefore,it isunlikelythatpeakIIrepresented
nucleocapsids and peak I contained complete
particles.To determine the nature of these two
IBRV peaks, infected cells were labeled with
either [3H]thymidine or
"4C-amino
acids([3H]-thymidine and
"4C-amino
acids labeled DNAand proteins, respectively). When most of the
cells had detached from thegrowthsurface,the
VOL. 40,1981
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.497.109.169.272.597.2]372 MISRA, BLUMENTHAL, AND BABIUK
FIG. 4. Analysis ofpurified[aH]glucosamine-and
[35S]methionine-labeled IBRV. 3H- and 35S-labeled
viruseswerepurifiedonpotassiumtartrategradients andanalyzed by electrophoresison10%(lanes 1 and
2) and 7.5%(lanes3through 5)polyacrylamidegels. Lanes 1, 3, and5, [3H]glucosamine-labeled IBRV (theselanescontaineddifferentamountsof radioac-tivity andwereexposed for varying lengths oftime to
illustrate all radioactive bandsmoreclearly); Lanes
2and4,[35S]methionine-labeledIBRV.
culture supernatantsweremixed, and the virus
was pelleted and then centrifuged through a
potassium tartrategradientasdescribed above.
Macromoleculesinfractionsof thegradientwere
precipitated with trichloroacetic acid, the
pre-cipitatesweretrappedonglassfiberfilters, and
the amounts of 3H and 14Cineachfractionwere
determined. AsFig.2Bshows,peakIcontained
both 3H and '4Candprobably represented
com-pleteparticles. Ontheotherhand, peakII
con-tainedonly '4C andwasprobably comprised of
particlesthat lackedDNA.
Analysisof IBRV structural proteins by two-dimensionalgelelectrophoresis.To
re-solve IBRV structuralproteinsfurther, weused
two-dimensionalelectrophoresistoseparate
pro-teinsalongonedimensiononthebasisofcharge
differences by isoelectric focusing and along the second dimension on thebasis of size on
SDS-polyacrylamide gels (19). This technique has
been used recently to resolve vaccinia virus
structuralpolypeptidesintomorethan100spots
(7)and herpes simplex virus1-infectedcell
poly-peptides into200spots (L. Haarr and H.
Mars-den, Abstr. 4thCold SpringHarborSymp.
Her-pesviruses, p. 109,1979).
Figure 5 shows theresolutionof IBRV
struc-turalpolypeptidesontwo-dimensionalgels.The
approximatepositionsof the IBRVpolypeptides
separated by SDS-PAGE alone are indicated.
Thisfigure alsoshowsthepositions of the spots
that consistently appeared when the analysis
wasrepeated. Electrophoretic streaking was
al-waysobserved for thepolypeptides which were
probably VP4 and VP8. Similar heterogeneity
has also been observed with the 60,000- and
62,000-dalton major virion peptides of vaccinia
virus (7) and the phosphorylated major coat
protein (VP1) of simian virus40(20).Spotsthat
possessed electrophoretic mobilities through
10% SDS-acrylamide gels which were
compa-rable to the mobilities ofglycoproteins GVP1,
GVP5, GVP6, GVP7, and GVP16 appeared as
multiplespots.
Infected cell proteins. Bovine kidney cells
were infected with IBRV at a multiplicity of infection of10PFU/cell.At 6hafterinfection,
[35S]methionine
wasaddedtothecells,and20h after infectionthecellswere harvested andan-alyzed bySDS-PAGEon10and 7.5%
polyacryl-amidegels(Fig. 6A and B, lanes 2). The analysis of the viral proteinsin infected cellswas made easier because withamultiplicityofinfection of
5 to 10 PFU/cell, by6h afterinfection all host
proteinsynthesiswasgreatlyreduced and there-fore theradioactive amino acid precursors that
were added at that time were incorporated
al-most exclusively into viral proteins. As Fig. 6
shows, the bands in the infected cell sample
(lanes2) didnot have thesameelectrophoretic
mobilitiesasthe bands in the mock-infected cell
sample (lanes 1). Even the dark 46,000-dalton
host bandwas almost completely missing from lanes 2. In additionto containing the 25 IBRV structural polypeptides, the infected cells also contained12nonstructuralproteins,whichwere
designated NSc, NSd, NSe, NSf, NSg, NSh,
NSj, NSk, NSl, NSm, NSn, and NSo. These
polypeptides ranged in molecular weight from
182,000to 32,000(Table 2).
Todetermine thetemporalorder ofsynthesis
of thesestructuralandnonstructuralproteinsin
infectedcells, bovinekidneycellswere infected with IBRV andlabeled with[35S]methioninefor
2-h periods at 0, 2, 4, 6, 10, and 12 h after
infection. Mock-infected cellswerelabeled for2 J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.497.87.233.78.397.2]PROTEINS SPECIFIED BY IBRV
A
+ .w
,I< <7
48
:* 4,13
41.
<116
422
[image:7.497.122.373.78.323.2]425
FIG. 5.Analysis of[3Slmethionine-labeledIBRVby two-dimensionalelectrophoresis.Thepolypeptides of
[3S]methionine-labeledIBRVwereseparated alongthehorizontal axisbyisoelectricfocusingandalongthe
vertical axisonthe basisofsizebyPAGE. A and B indicate the acidic and basic endsofthefirstdimension
(approximately pH3.5 and8.5,respectively).Thearrowsindicate spots and streaks thatconsistentlyappeared
whendifferent preparationswereanalyzed. Thearrowheadsontherightindicate theapproximate positions
oftheIBRVpolypeptidesseparated byone-dimensionalelectrophoresis; glycopeptidesareindicatedbyopen
arrowheads.
hatthe timeof mock infection. The cells were
harvestedatthe endofthelabelingperiodand wereanalyzed by SDS-PAGE(Fig. 7). No
radio-active viral polypeptides were observed inthe
first 2 h afterinfection, probably because viral proteins at this early stage were produced in
insufficient quantitiestobeobservedagainst the heavy background ofhost polypeptides. How-ever,between2 and 4 h afterinfection, polypep-tides with electrophoretic mobilities similar to
those of VP4, NSb, NSc, NSd, NSi, and NSj
were observed against a slightly less intense
background of hostproteins. Between4and 6 h afterinfectionGVP7, VP8, NSf, NSl, NSm, and
NSnwerealso synthesized.Inaddition, the
in-tensity ofpeptide NSj beganto decrease. The
remaining viral polypeptides were labeled
be-tween6 and 8 h afterinfection.Almostallofthe
viral polypeptides that were labeled in the
2-and 4-hlabelingperiodswerealsolabeledat6h, althoughthe intensities of bands NSdand NSi decreasedandNSjdisappeared.Inaddition,
ces-sation of host polypeptide synthesis, as
deter-minedbythealmostcomplete absence of
radio-activity in the46,000-dalton host band, occurred
at6 hpostinfection.
Thepattemoflabeling remained unchanged
in the 10- and 12-hlabelingperiods.
Herpesviruses are generally believedto
pos-sess acomplexsystem ofgeneregulation,where
expression ofyorlategenesrequires prior syn-thesisofaand,B proteins,aswellastheonsetof
viralDNAreplication (16, 29, 30). Therefore,we
examined the pattern of protein synthesis in
IBRV-infected cells in which DNA synthesis had been inhibited bytreatment with 50,ugof
ara-Cperml. This concentration was 10 times
thatrequiredtoshut down viral DNAsynthesis in infected cells almost completely (data not
shown).
Immediate early (a), early (8), and late (y)polypeptides. Toidentifyimmediateearly, early, and lateviralproteins,bovinekidneycells
weremock-infectedorinfectedwithIBRV; then
these cells were incubated in the presence of
either50,ugofcycloheximide permlor50,ug of ara-Cperml, ortheywereleftuntreated. At 6
h afterinfection, the cycloheximide-treated
cul-tureswere washed extensively with MEM, and
the growthmediumwasreplacedwith medium
lackingcycloheximide.FurthermRNAsynthesis inonebatch ofcultureswasprevented by adding
VOL. 40, 1981
B
373
'*
on November 10, 2019 by guest
http://jvi.asm.org/
374 MISRA, BLUMENTHAL, AND BABIUK
9 8 7 6 5
t 2
a.
4,
.;
_ ...._
1
i:>
F5F: 1
2 a 3.b
-4
-d -e -7 _f
-8.9
-h
-9.1
-10
-11
-12 13 -14
-i
-k
-16
Bmw9 8 7 6 5 4 3
7
I4
aa|~
-'
60* ei
480
236* 4 4
30* _ 0
-I2 1-3,b
de
-7
-8 :~1o
-11
.-1213
-i
-k -16
1--m
.n
* 22
0
.. 0
I
- -m
f -n
-2
'l-
-020*
4*.
[image:8.497.69.450.81.411.2]14.4*
FIG. 6. Analysis ofIrSlmethionine-labeledmock-infectedandIBRV-infected ceU proteinson7.5 and10%
polyacrylamide gels.Lanes1,Mock-infectedbovinekidney cells; lanes2,IBRV-infectedcellslabeled 6to20
h after infection;lane3,purified IBRV;lanes4,mock-infectedcellstreated with50pgofara-Cpermland
labeled 6to20 hpostinfection;lanes5,IBR V-infectedcellstreated with 50pgof ara-Cperml;lanes 6,
mock-infectedcells labeledafterreleasefrom50pgof cycloheximideperml;lanes 7, IBRV-infectedcellslabeled
after releasefrom cycloheximide; lanes8, mock-infected cells labeled in a medium containing 25 pg of
actinomycin Dpermlafterreleasefromcycloheximide; lanes9, IBRV-infectedcells labeledinamedium
containing actinomycinDafterreleasefromcycloheximide.Inlane 5 theopencirclesindicatepolypeptides
thateitherwerenotsynthesizedor weresynthesizedinreducedamountsin thepresenceofara-C. Thearrows
inleft marginindicatethepositions ofmolecularweight (MW)markers.Viralpeptidesandglycosylatedviral
polypeptidesarenumbered,andnonstructuralpolypeptidesaredesignatedwith lower-caseletters.
2.5 ,ug of actinomycin D per ml of medium.
[3S]methioninewasaddedtoallcultures. At 20
h afterinfection, the cells were harvested, and
theproteinswereanalyzedbySDS-PAGE.
yPolypeptides. Althoughmost of theviral
structuraland nonstructuralpolypeptideswere
synthesizedininfectedcells treated withara-C,
atleast five polypeptides (GVP1, GVP3, NSe, VP8, and VP13) were missing or were
synthe-sizedingreatlyreduced amounts (Fig. 6B,lane
5). Thus, thesewerethetrue 'yorlate
polypep-tides since they were not synthesized in the
absence of viral DNA synthesis. ara-C-treated cells contained an additional band, designated
NSi, which was not observed inuntreated
in-fected cells.NSimayhaverepresenteda precur-sor that was not processed in the absence of
DNAsynthesis, oritmayhave been a,
poly-peptide which was not observed in untreated
cellslabeled 6 h after infection because its syn-thesiswas inhibitedstrongly by they
polypep-tides.
aPolypeptides. Threeproteins appearedto
besynthesizedinlargeamountsafter the cyclo-heximide was removed if actinomycin D was
addedtothewashed cultures. In the absence of
actinomycin D,all of the viralpolypeptides
ob-served in the ara-C-treated cellswere
synthe-A
MW 330 *220 *
-24
::tg
-25
.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
PROTEINS SPECIFIED BY IBRV 375
[3H]glucosamine and analyzed by SDS-PAGE
(Fig. 8).No radioactive bandswereobserved in
the cells released from thecycloheximide block,
andonlyoneglycopeptide (GVP11)was
synthe-sized inara-C-treated cells.
/3 Polypeptides. All viral polypeptides
syn-thesized in the absence ofviral DNA synthesis
(50
jig
of ara-Cperml)weredesignated,Bpoly-peptides (Fig.6,lane5,and Table 2).
FIG. 7. Time courseofIBRVpolypeptide
synthe-sis. IBRV-infectedceUs werelabeled with
[9S]me-thionineat0 to2(lane 2),2 to 4(lane3),4 to6(lane
4),6to 8(lane 5),10 to 12(lane6), and12to 14(lane
7) hafter infection.Mock-infectedcellswerelabeled 0 to2hafter mock-infection (lane1).Atthe endof
thelabeling period,thecellswereharvested,andthe
proteins were analyzed by electrophoresis on 7.5%
polyacrylamidegels.
sized(Fig.6A andB, lanes 7). Thiswasprobably
because in the absence of actinomycin D, /3
transcriptsweresynthesizedand translated after
initialsynthesis ofa peptides; /8geneproducts
theninhibitedfurther translation ofamessages.
Threeapolypeptideswereidentifiedininfected
cellsafter thecycloheximide wasremoved, and
these weredesignatedNSb, NSc,andNSg (Fig.
6B, lane 9).Noneoftheviralglycoproteins could
beidentified asa proteins,and theonly
glyco-protein made in appreciable amounts in the
absence ofviralDNAsynthesisappearedtobe
GVP11. These observations were confirmed
when infectedcellsthat were treated withara-C
or released from the cycloheximide block and treated with actinomycin D were labeled with
FIG. 8. a, ,B, and y viral glycopeptides.
Mock-in-fected or IBRV-infected cells were treated as
de-scribed in the testand in thelegendtoFig. 6. The
cellswerethenlabeled with[3H]glucosamine. At20
h postinfection the cells were harvested, and the
proteins were analyzed by electrophoresis on 7.5%
acrylamidegels.Radioactivebandswerevisualized
by fluorography.LaneA,Mock-infectedcells; lane B, IBRV-infected cells; lane C, mock-infected cells
treated with ara-C; lane D, IBRV-infected cells
treated withara-C; laneE, mock-infected cells
la-beled in a medium containing actinomycin D after
releasefrom cycloheximide; lane F, IBRV-infected
cells labeledinamediumcontaining actinomycinD
after releasefrom cycloheximide. Thenumbered
ar-rowheads indicate viralglycopeptides.
VOL. 40,1981
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.497.76.214.81.413.2] [image:9.497.277.418.187.520.2]376 MISRA, BLUMENTHAL, AND BABIUK
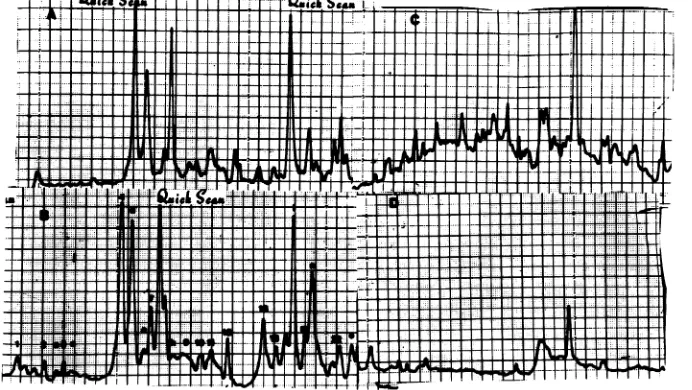
Immunoprecipitation of infectedcell
pro-teins. To determine whether animals with
re-currentIBRV infections had antibodies against
structural and nonstructural IBRV
polypep-tides, immunoglobulinwaspurified fromacow
thathad been infected intranasallywith IBRV.
Recurrent IBRV lesions were induced in this
animal by administering dexamethasone (21).
The immunoglobulin preparation had a serum
neutralization titerof 1:1,000. Cell-free extracts
from infected or uninfected cells were mixed
with theimmunoglobulin preparation in optimal
proportions, and the antigen-antibody
com-plexeswerethenprecipitated with S.aureus and
analyzed bySDS-PAGE. With the exception of
a small amount of actin (molecular weight,
46,000), the antibodies did not precipitate
sig-nificant amountsofanymock-infectedcell
pro-teins.On the other hand,all
'S-labeled
polypep-tides observed in infected cells appeared tobe
precipitated (Fig. 9).
DISCUSSION
Electrophoresis ofIBRV virions on 7.5 and
10% SDS-polyacrylamide gels which resolved
polypeptides with molecular weights ranging
from 12,000 to 330,000 revealed 25 distinct
J. VIROL.
bands.Fiveof these bandsmayhave contained
two orthreeproteins,asrevealedby
two-dimen-sional electrophoresis (isoelectric focusing and
SDS-PAGE), although some ofthese multiple
spots wereprobablyartifactualinorigin(Fig. 5).
Thus,IBRVvirions containatleast25
polypep-tides, andmorelikely28to33polypeptides.This
is in goodagreementwithstudies of other
her-pesviruses; 33 polypeptideshave beenfoundin
herpes simplexvirus (28),20polypeptideshave
been foundin pseudorabies virus (29),26
poly-peptides have been found in murine
cytomega-lovirus (5, 13), and 33 polypeptides have been
found inEpstein-Barr virus (6). Of the33herpes
simplex virion proteins, 5 are glycosylated (2,
18), whereas 11 of the 28 to 33 IBRV virion
proteinscould be labeled with[3H]glucosamine
andare thought to beglycoproteins. Although
11 of28 to 33 seemshigh compared with5 of
33,theputative glycoproteins arewell resolved
inmolecularweight, andmostgive characteristic
patterns on two-dimensional gels. However, it
remainstoberigorously demonstrated thatall
11 are actually distinct glycoproteins. On
two-dimensional gels, spots that possessed
electro-phoretic mobilitiesthrough 10%
SDS-acrylam-ide gels comparable to those of glycoproteins
GVP1, GVP5, GVP6, GVP7, and GVP16
ap-__'''''
.
1
.. ....11
4iTX
&1.. ... ~~~~~~~~~...
-...i.... ,XF .
FIG. 9. Immunoprecipitation of mock-infectedorIBRV-infectedcellextractswithimmunoglobulin from
cowswithreactivated IBRV lesions. Cellextractsfrom mock-infectedandIBRV-infectedbovinekidneycells
wereimmunoprecipitatedwiththeimmunoglobulin fraction fromserumobtainedfromacowwith reactivated
IBRV lesions(19). S.aureusstrain Aadsorbantwasusedtofacilitate immunoprecipitation.
Immunoprecip-itateswerewashed andelectrophoresedon7.5%polyacrylamide gels,and theradioactive bandswerelocated
byautoradiography.AutoradiographswerescannedwithaHelenaQuickScan Jr.densitometer. (A)
IBRV-infectedcells,whole-cellextract.(B)IBRV-infected cells, immunoprecipitate. (C) Mock-infected Madin-Darby
bovinekidneycells, whole-cellextract.(D) Mock-infected Madin-Darbybovinekidneycells,
immunoprecipi-tate.
F F
on November 10, 2019 by guest
http://jvi.asm.org/
[image:10.497.89.435.378.573.2]VOL. 40, 1981
peared as multiple spots. Each set ofmultiple
spots may representmolecules of thesame
gly-coprotein that differ in the extent of
glycosyla-tion. Similar multiple glycoprotein spots were
observed inherpessimplexvirus1-infectedcells
by Haarr and Marsden (Abstr.4th ColdSpring
Harbor Meet.Herpesviruses, p. 109, 1979) who
showed bypulse-chase experiments that these
spotsrepresentedstepsin thepost-translational
modification process. The presence ofmultiple
glycoprotein spots in virus preparations may
indicate either that post-translational modifica-tionoccursinthe virusorthat the virus contains
forms of thesameglycoproteinsthatare
glyco-sylated to different extents. Our analysis also
indicated that certain bandsonone-dimensional
SDS-PAGE gelsmay represent more than one
polypeptide.Twoor morespots were present at
the positions occupied by bands VP12, VP13,
VP18, VP19 and VP22, and the results of a
simple SDS-PAGE analysis of these proteins
mustbeinterpretedwith caution.
We found that IBRV virions were resolved
into two distinct bands on potassium tartrate
gradients (Fig. 2A). An SDS-PAGE analysis of
the proteins indicated that the lower,
DNA-bearing virions containedproteinsVP17, VP22,
and VP24 which were absent from the empty
virions. Bothcomponents were probably
enve-loped since the viral glycoproteinswere
associ-ated with both of them. Asimilarphenomenon
has been observed with herpes simplex virus. Thus, nucleocapsids of herpes simplex virus band as two separate components on sucrose
gradients. The faster-sedimenting component
comprisesparticlesthat containDNA, whereas the more slowly sedimenting peak consists of
emptycapsids. In additiontocontainingDNA,
the fullparticlesalso containafew
DNA-asso-ciatedpolypeptidesthatare notpresentin the
emptyparticles(10).
Ouranalysisofviral proteinsininfected cells
wasmadesimplerby the drastic decreaseinthe
rate of host cell protein synthesis caused by
IBRV infection, such that radioactive precursors
added to the infected cells 6 h after infection
wereincorporated almost exclusivelyinto
virus-induced polypeptides. In addition to the 25
structuralpolypeptides, infectedcellscontained
atleast15nonstructuralpolypeptides.Although
atleastsomeoftheseareprobably
virus-speci-fied enzymes, others may represent precursors
of other structural and nonstructural
polypep-tides.
Herpesviruses, whichpossess the capacity to
code for more than 100 genes, have aparticularly
complex temporal pattern of gene expression.
Thus, herpesvirus-specified genes, transcripts,
PROTEINS SPECIFIED BY IBRV 377
and polypeptides can be classified as a or
im-mediateearly,,B orearly, and-y orlate,
depend-ing upon the order of expression during the
infectious cycle.InthecaseofmostDNAviruses
andatleastsomeherpesviruses, suchashuman
cytomegalovirus (30), murine cytomegalovirus
(16; J. K. Chantler, personal communication),
and pseudorabies virus (3), the synthesis ofy
polypeptides also requires the prioronsetof viral
DNAsynthesisintheinfectedcells.In contrast
tohumancytomegalovirus, murine
cytomegalo-virus, andpseudorabies virus, inhibitors of DNA
synthesis donotpreventthesynthesisofy
poly-peptides and theassemblyofempty
nucleocap-sids in herpes simplex virus-infected cells (23).
In this respect IBRV more closely resembles
humancytomegalovirus, murine
cytomegalovi-rus,and pseudorabies virus. Atleast five
poly-peptides (GVP1, GVP3, NSe, VP8, and VP13)
eitherwere missingfromara-C-treatedcellsor
weremade indrasticallyreducedamounts.
Sim-ilar resultswereobtained when viral DNA
syn-thesis was inhibited in infected cells by
phos-phonoformic acid or bromovinyldeoxyuridine
(datanotshown).
Wealso identified threeapolypeptides (NSb,
NSc, and NSg), whichweresynthesized inlarge
amountsaftercycloheximidewasremoved from
cultures of infected cells.Although NSg has the
sameelectrophoretic mobilityonSDS-PAGEas
VP8,webelieve that it isadifferentpolypeptide
sinceVP8wasidentifiedasaypolypeptideand
was probably not synthesized until after the
onsetof viralDNAsynthesis.
The glycoproteins specified byherpesviruses
are ofparticular interest since they are social
proteins that are involved in interactions
be-tween virions and host cells, between infected
cells and the immunesystem, and between
in-fectedcells. Theherpessimplexvirus
glycopro-teins gC, gD,gA, andgB (allyclass) have been
shown to be involved in immune recognition,
andantibodies directedagainst these
glycopro-teinscanmediate immunocytolysisin
conjunc-tion withcomplementonmononucleareffector
cells (16). None of theIBRV glycoproteins
be-longed to the a class, but surprisingly at least
oneIBRVglycoprotein (GVP11)behaved likea
,Bpolypeptide since itwassynthesized (labeled
with [3H]glucosamine) in the absence of viral
DNAsynthesis. We are now studyingthe roles
played bythe various IBRVglycoproteins, and
in particular GVP11, in immune recognition.
TheabilityofGVP11 toparticipatein immune
cytolyticprocesses would havespecial
implica-tions in interacimplica-tions with non-productivity or
latently infectedcellsin which viral expression
ispresumablyrestricted to the a and,Bclasses.
on November 10, 2019 by guest
http://jvi.asm.org/
378 MISRA, BLUMENTHAL, AND BABIUK
ACKNOWLEDGMENTS
We thank Kris Komendant for her excellent technical assistance.
Funds from the Medical Research Council of Canada made thisproject possible.
LUMRATURE CITED
1. Babiuk, L. A.,R.C. Wardley,and B.T. Rouse. 1975. Defensemechanisms against bovine herpesvirus: rela-tionships of virus-hostcell events tosusceptibility to antibody-complementcelllysis. Infect. Immun. 12:958-963.
2. Baucke,R.B., and P. G.Spear.1979.Membrane protein specified byherpessimplexvirus.V.Identification of anFc-binding glycoprotein. J. Virol. 32:779-789.
3. Ben-Porat, T., and A. S. Kaplan. 1973. Replication biochemicalaspects, p. 164-216. In A.S.Kaplan (ed.), Theherpeaviruses.AcademicPress, Inc.,New York.
4. Cassai, E.N.,M.Sarmiento,and P.Spear.1975. Com-parison of the virion proteinsspecified by herpes sim-plexvirus types 1 and2.J.Virol. 16:1327-1331.
5. Chantler,J.K., andJ. B.Hudson. 1978. Proteins of murine cytomegalovirus: identification of structural and nonstructural antigens in infected cells. Virology 86:22-36.
6. Dolyniuk,M., R.Pritchett,and E.Kieff.1976.Proteins ofEpstein-Barrvirus. I. Analysis of thepolypeptidesof purifiedenveloped Epstein-Barrvirus. J. Virol. 17:935-949.
7. Essani, K.,and S. Dales.1979.Biogenesisofvaccinia: evidence for more than 100polypeptidesin the virion. Virology95:385-394.
8. Garrels, J. I. 1979. Two dimensional gel electrophoresis and computer analysis of proteins synthesized by clonal cell lines. J. Biol. Chem. 254:7961-7977.
9. Gibbs,E. P.J.,and M. M.Rweyemamu.1977.Bovine herpesviruses.I.Bovineherpesvirus-1. Vet. Bull. (Lon-don)47:317-343.
10.Gibson, W.,and B.Roizman.1972.Proteinsspecified byherpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2.J. Virol. 10:1044-1052.
11.Heine, J. W., R. W. Honess, E. Cassai, and B. Roiz-man. 1974.Proteinsspecifiedby herpessimplexvirus. XII.The virionpolypeptidesof type 1 strains. J. Virol. 14:640-651.
12.Homan,E.J.,and B.C.Easterday. 1980.Isolation of bovineherpesvirus-1fromtrigeminal gangliaof clini-cally normalcattle. Am. J. Vet. Res.41:1212-1213. 13.Kessler, S. W. 1975. Rapid isolation of antigens from cells
with astaphylococcal protein A-antibody adsorbant: parametersof the interaction ofantibody-antigen com-plexeswithprotein A. J. Immunol. 115:1617-1642. 14.Kim, K.S.,V.L.Supienza,R. I.Carp,and H. M. Moon.
1976.Analysisof thestructural proteins of murine cy-tomegalovirus. J. Virol. 17:906-915.
15.Laemmli, U. K. 1970. Cleavage of structural proteins
J. VIROL.
during the assembly of the head of bacteriophage T4. Nature(London) 227:680-685.
16. Misra, V., M. T.Muller, J. K. Chantler, and J. B. Hudson.1978.Regulationof murinecytomegalovirus geneexpression. I. Transcriptionduring productive in-fection. J.Virol. 27:263-268.
17. Misra,V., M. T.Muller,and J. B. Hudson.1977.The enumeration of viral genomes in murine cytomegalovi-rusinfected cells.Virology 83:458-461.
18. Norrild, B., S. L Shore, and A. J. Nahmias. 1979. Herpes simplex glycoproteins: participation of individ-ual herpes simplex virus 1 glycoprotein antigens in immunecytolysis and their correlation with previously identified glycopeptides. J. Virol. 32:741-748. 19. O'Farrell,P. H.1975.Highresolution two-dimensional
electrophoresis of protein. J. Biol. Chem. 250:4007-4021.
20.O'Farrell,P.H.,and H. M. Goodman.1976.Resolution ofSV40 proteins in wholecellextracts by two dimen-sional electrophoresis: heterogeneity of the major cap-sid protein.Cell9:289-298.
21. Pastoret, P.P.,L. A.Babiuk,V.Misra,andP. Grie-bel. 1980. Reactivation oftemperature-sensitive and non-temperature-sensitive infectiousbovine rhinotra-cheitis vaccine virus with dexamethasone. Infect. Im-mun.29:483-488.
22. Purifoy,D. J.M.,and K.L.Powell.1976. DNAbinding proteins induced by herpes simplex type 2 in HEp 2 cells.J. Virol. 19:717-731.
23. Roizman,B. 1978. Theherpesviruses, p.769-48.In D. P.Nayak (ed.), The molecular biology ofanimalviruses, vol. 2. Marcel Dekker, New York.
24. Roizman,B., M.Kozak,R. W.Honess,and G. Hay-ward. 1974.Regulation of herpesvirus macromolecular synthesis:evidence for multi-levelregulation of HSV-1 RNA andprotein synthesis. ColdSpringHarborSymp. Quant.Biol. 39:687-701.
25. Rouse,B.T.,and L. A.Babiuk. 1978. Mechanisms of recovery from herpesvirus infections. Can. J. Comp. Med.42:414-427.
26. Spear,P.G. 1976. Membraneproteinsspecifiedby herpes simplex viruses. I. Identification offour glycoprotein precursorsand theirproductsin type1-infected cells.J. Virol. 17:991-1008.
27. Spear, P. G., J. M. Keller, and B. Roizman. 1970.
Proteins specified byherpes simplex virus. II. Viral glycoproteins associated with cellular membranes. J. Virol.5:123-131.
28. Spear,P.G.,and B.Roizman.1972.Proteinsspecified by herpessimplexvirus. V.Purificationand structural proteinsof theherpes virion. J. Virol. 9:143-159.
29. Stevely,W. S.1975.Virus-inducedproteinsin pseudo-rabies-infectedcelLs.II. Proteins of the virion and nu-cleocapsid.J.Virol. 16:944-950.
30. Stinsky,M. F. 1978. Sequence ofprotein synthesis in cellsinfectedby humancytomegalovirus:early and late virus-inducedpolypeptides. J. Virol.26:686-701.
on November 10, 2019 by guest
http://jvi.asm.org/

![FIG. 4.2)[35S]methionine-labeledLanes2virusesandtivity(theseillustrate and and Analysis ofpurified [aH]glucosamine- and IBRV](https://thumb-us.123doks.com/thumbv2/123dok_us/1471244.99738/6.497.87.233.78.397/methionine-labeledlanes-virusesandtivity-theseillustrate-analysis-ofpurified-glucosamine-ibrv.webp)
![FIG. 5.[3S]methionine-labeledofwhenvertical(approximately the Analysis of[3Slmethionine-labeled IBRV by two-dimensional electrophoresis](https://thumb-us.123doks.com/thumbv2/123dok_us/1471244.99738/7.497.122.373.78.323/methionine-labeledofwhenvertical-approximately-analysis-slmethionine-labeled-dimensional-electrophoresis.webp)

![FIG. 7.polyacrylamideproteins4),thionine0thesis.7) to h 6 Time course of IBRVpolypeptide synthe- IBRV-infected ceUs were labeled with [9S]me- at 0 to 2 (lane 2), 2 to 4 (lane 3), 4 to 6 (lane to 8 (lane 5), 10 to 12 (lane 6), and 12 to 14 (lane after infec](https://thumb-us.123doks.com/thumbv2/123dok_us/1471244.99738/9.497.76.214.81.413/polyacrylamideproteins-thionine-thesis-course-ibrvpolypeptide-synthe-infected-labeled.webp)