0022-538X/78/0028-0314$02.00/0
Copyright © 1978 AmericanSocietyforMicrobiology Printed inU.S.A.
Evidence for
an
Adenovirus
Type
2-Coded
Early
Glycoprotein
YUN-HUAJENG,WILLIAMS.M.WOLD,* AND MAURICEGREEN
Institutefor Molecular Virology, St.Louis University Schoolof Medicine, St. Louis, Missouri63110
Receivedfor publication28February 1978
Wehaveidentifiedanadenovirus type2(Ad2)-induced early glycopolypeptide
with an apparent molecularweightof 20,000 to 21,000 (20/21K), as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The 20/21K poly-peptide could belabeledin vivowith[3H]glucosamine.[35S]methionine- and
[3H]-glucosamine-labeled 20/21K polypeptides bound to concanavalin A-Sepharose
columns and were eluted with 0.2 M methyl-a-D-mannoside. The pulse-labeled
polypeptide appearedasasharp band withanapparentmolecular weight of 21K,
butafterachase it convertedtomultiplebands with an average molecular weight of 20K. Thisvariabilityinelectrophoretic mobility is consistent with glycosylation
or deglycosylation of the 20/21Kpolypeptide. Analysis of the pulse and
pulse-chase-labeled formsby using partial proteolysis indicated that the polypeptides
werehighlyrelatedchemically, butnotidentical. Most of the 20/21Kpolypeptide
islocalized in thecytoplasm fraction of infected cells lysed by Nonidet P-40. The 20/21K polypeptide and a 44K polypeptide, labeled with [35S]methionine or
[3H]glucosamineinAd2-infected humancells, were precipitated by a rat
antise-rumagainstanAd2-transformedratcell line (T2C4),butnotby antisera against
three other Ad2-transformed rat cell lines, or by serum from nonimmune rats. The partial proteolysis patterns of the 20/21K and the 44K polypeptides were
indistinguishable, indicating that the two polypeptides are highly related, and
suggestingthat the 44Kpolypeptide might beadimerof the 20/21K polypeptide.
The 20/21K polypeptide was also induced in Ad2-early infected monkey and hamstercells. These results imply that the 20/21K polypeptide is synthesized in infected human, monkey, and hamster cells, and in one but not all Ad2-transformedratcells.Thus, the 20/21K polypeptide is probably viral coded rather thancell coded and viral induced.
Polypeptides induced in cells during early
stages (i.e., before viral DNA replication) of infectionby human adenovirus type2(Ad2)are
of great interest becausetheymayregulateviral DNA replication, transcription, and cell
trans-formation (34). We have attempted to identify Ad2-induced early polypeptides by labeling in-fected and mock-inin-fected cells with [35S]methi-onine,subjectingtheproteinextracts to
electro-phoresis through sodium dodecyl
sulfate-poly-acrylamideslabgels(SDS-PAGE),and compar-ing the infected and mock-infectedpolypeptide bands seen in autoradiograms. We have also identified Ad2 early polypeptides by immuno-precipitation, using antisera against
Ad2-trans-formed rodent cells. Prominent early poly-peptide bands with the following approximate apparent molecularweights have been detected: 73,000 (73K), 53K, 21K, 19K, 15K, 11.5K, and 1lK (8, 10, 14). Additional minor polypeptide
bands of about 17.5K, 15.5K, 13.5K, 13K, 12K, 8.8K, and 8.3K can be observedin some gels, or
canbe immunoprecipitated by antisera against
certain lines ofAd2-transformed ratcells (Wold
and Green, unpublished data). Similar in
vivo-labeled polypeptides have been identified by
otherlaboratories (12,23). Similar
polypeptides
havealso been observedbycell-free translationofpolyribosomal RNA fromAd2-earlyinfected
cells andbyimmunoprecipitation, usingarabbit antiserumagainstearlyinfected HeLa cells(23).
Six polypeptides (72K, 44 to 50K, 19K, 15.5K,
15K, and
11K)
have been mapped by cell-free translation ofearlymRNA purified by hybridi-zation to restriction endonuclease DNA frag-ments(18).Relatively little isknownabout thechemistry
of these polypeptides or their biological func-tions. The73Kpolypeptideisaphosphoprotein
(14, 17, 19,22) that is viral coded (9, 18, 30), that binds to single-stranded DNA (24, 27, 29), and
thatapparentlyfunctionsinviralDNA
replica-tion (28,31). This protein hasbeen purified to
homogeneity and partially characterized (19,
27). The53K (8, 16) and the 15K polypeptides
(8) are candidate transformation proteins be-causetheyare immunoprecipitated byantisera
against Ad2- or Ad5-transformed rodent cells.
314
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 28,1978
Genescoding polypeptides of 44 to 50K and 15K polypeptides have been mapped in the trans-forming region of the Ad2 genome (18). The 11K polypeptide is localized mainly in the nuclear fraction (3, 23; our unpublished data). The 73K, 21K, 15K, 11K, and possibly 8.3Kpolypeptides arepresent inasolublecomplex thatsynthesizes Ad2 DNA, suggesting a possible role for these in viral DNA replication (21).
Ishibashi and Maizel (13) reported that an Ad2-induced early 19K polypeptide could be labeled in vivo with [3H]glucosamine. In this report, we present evidence that our 21K
poly-peptide is an Ad2-coded earlyglycopolypeptide, in that it can be labeled with[3H]glucosamine,
it binds to concanavalin A (ConA)-Sepharose,
itsmobilitychangesinpulse-chase experiments,
it is immunoprecipitated by antisera to
Ad2-transformed cells (T2C4), and it issynthesized
in Ad2-infectedmonkey and hamster cells.
MATERIALS AND METHODS
Chemicals.
L-['S]methionine
(Met) (400 Ci/mmol) and [3H]glucosamine (20 Ci/mmol) were purchased from New England NuclearCorp.;
1-/i-D-arabinofuranosylcytosine (ara-C), ConA-Sepharose, and methyl-a-D-mannoside were purchased from Sigma ChemicalCo.; acrylamidewaspurchased from Eastman Kodak Co.; Nonidet P-40 was purchased from Shell ChemicalCorp.; N,N-methlenebisacrylam-ide,N,N,N',N'-tetramethyladenediamine, and ammo-niumpersulfatewerepurchased from Bio-Rad Labo-ratories; andStaphylococcusaureusV8 proteasewas purchased from MilesLaboratories, Inc.
Cell culture and virus infection.Suspension
cul-turesof human KB cellsweregrown inEagle minimal
essential medium(MEM) containing5%horseserum. Cells at aconcentration of6x 106/mlwereinfected with100PFUof Ad2(strain 38-2) per cell in medium without horse serum. After 1 h ofadsorption, cells weresuspendedat aconcentrationof3.5x 105/mlin medium with 5% horseserum.Cyclohexamide (CH;25
,Lg/ml)wasaddedto someculturesat 1hpostinfection (p.i.). Ara-C (20,ug/ml) wasaddedat4hp.i. At 9 h p.i., cellswerewashed andsuspendedeither inwarm Met-free medium or medium with 10% the normal glucoseconcentration; both media contained5%horse
serumand20,ugof ara-C per ml.Cells,at a
concentra-tion of3.5 x 105/ml, werelabeled with[3S]Met (10
t,Ci/ml) or [3H]glucosamine (10
liCi/ml)
forvarioustimes. At the end of the labeling period, cells were centrifuged and washed twice with cold
phosphate-bufferedsalinelackingCa2'and
Mg2e.
Mock-infectedcellsweresimilarly labeled,except that viruswas not added.
CV-1(monkey) cell monolayersweremaintained in 75-cm2 plastic flasks containing MEM with 5% calf serum,and in anatmosphere of 5%CO2.Nearly con-fluent monolayers were infected (or mock infected)
with Ad2 in MEM without serum. After 2 hof
adsorb-tion,MEMwith5% calf serum(withorwithout 25yg
of CH perml) wasadded to 25 ml, and incubation continued.Ara-C (20,ug/ml)wasaddedat 4hp.i. Cells
315
werelabeled from 9 to 20 hp.i. with [3S]Met in Met-free MEM containing 20
jig
of ara-C per ml.Cell fractionation. Infectedormock-infected cells labeled with [3S]Met were washed and then sus-pended in isotonic high pH buffer (33). All steps were at 0 to 4°C, and all buffers contained 1 to 2 mM phenylmethylsulfonyl fluoride. Cells were lysed by using 0.5% Nonidet P-40 for 5 min. Lysis was moni-tored by phase-contrast microscopy until 99% com-plete. Nuclei were pelleted by centrifugation at 200 x gfor3min.The cytoplasmic supernatant was obtained by centrifugation at 12,000xg for 20min to remove mitochondria. Isolated nuclei were resuspended in phosphate-buffered saline, and treated with 0.86% Tween 40 and 0.43% sodium deoxycholate to remove
outerand innernuclear membranes (11). The
nuclear-detergent suspension wasvigorously mixed in a Vortex mixer for 30 s, and the nuclei werecollected by cen-trifugation at 500 x g for 3 min. The pellet was resuspended in cold phosphate-buffered saline with the addition of 1% Triton X-100 and disrupted by sonic treatmentfor 10 min in a Raytheon sonic oscil-lator. The nucleoplasmsupernatant and nuclear pellet wereseparated bycentrifugation at 12,000xg for 20 min.Portions of subfractionated samples were precip-itated with 10% coldtrichloroacetic acid for analysis oflabeledpolypeptides.
ConA-Sepharosecolumnchromatography. Af-finity chromatography of glycoproteins on ConA-Sepharose columnswas carried out asdescribed by Stohlmanetal.(26),withthefollowing modifications. All stepswere at4°C. The column (1.5x 25cm) was firstequilibrated with10bed volumes of50 mM Tris-hydrochloride (pH 7.5), 50mM NaCl, 1 mM MnCl2,
and 2 mM phenylmethylsulfonyl fluoride (bufferA)
containing 0.2Mmethyl a-D-mannoside, followedby
buffer A containing 0.2% Nonidet P-40 and 0.2% so-dium deoxycholate (buffer A-detergent). The
cyto-plasmic protein fraction (no CH pretreatment) was diluted 25-fold with bufferA,andsodiumdeoxycholate
wasaddedto afinal concentration of 0.2%. Thesample wasdisrupted by sonic treatment(see above), centri-fuged at 12,000 x gfor20min, dialyzed extensively
against bufferA-detergent,and loadedontothe ConA-Sepharose column. The column was washed with bufferA-detergent untilnosignificantradioactive ma-terialwaseluted. The adsorbedproteinswereeluted withbufferAcontaining0.2Mmethyl-a-D-mannoside.
Thecolumnflow-through,wash,and
methyl-a-D-man-noside-elutedfractionswereconcentratedby
precipi-tation with cold 10%trichloroacetic acid andwashed
with acetone to removetrichloroacetic acid and
deter-gent.
Radioimmunoprecipitation ofproteins. T2C4
cells, fromanAd2-transformedratcell line(6),were
obtained from P. H. Gallimore. Antiserum was
pre-paredin ratsagainstextracts ofT2C4cells, and the
immunoglobulin G (IgG) fraction was purified (8).
Infected andmock-infected labeled cellextracts were
assayedforT2C4-specificpolypeptides bythe double-antibodyimmunoprecipitationprocedure,asdescribed earlier(8).Portionscontainingequalcountsper min-ute from infected and mock-infected samples were
incubated with T2C4ornonimmune rat IgG at4°C
for 18 h. Thengoatserumanti-ratIgGwasadded,and
thesamplewasincubated for2hat37°C.Precipitates
on November 10, 2019 by guest
http://jvi.asm.org/
werecollectedbycentrifugationfor90 sat23,000xg
in a Beckman 152microfuge,washed three times with
phosphate-bufferedsalinecontaining0.5M urea, 0.5%
Nonidet P-40, and 1% sodiumdeoxycholate,suspended
in anequalvolume ofgelelectrophoresis samplebuffer
without 2-mercapthoethanol, and analyzed by
SDS-PAGE.
SDS-PAGE and autoradiography. SDS-PAGE
was carried out as described elsewhere (14). Unless
statedotherwise,8to21%gradient acrylamide 10-cm
slab gels were used. Radiofluorography as described
by Bonner and Laskey (1) was used to detect
[1H]-labeledpolypeptides. The absorbance of bands in au-toradiogramswasmonitored byusingaJoyce-Loebel densitometer. The area under the absorbance tracing was calculated as the quantitative estimation of the intensityof bands.
Partial proteolysis. The relationship between
[;35S]Met-labeled 44K and 20/21K polypeptides
im-munopreciptatedby T2C4antiserumwasinvestigated, usingthe partial proteolysis procedure essentiallyas
describedby Cleveland andcolleagues (4).The
poly-peptidesimmunoprecipitated byT2C4 antiserum were
resolvedbySDS-PAGE, thegelsweredried, and the
fluorographs were developed. The 44K and 20/21K
bands were cut from thegel, andthe gel slices were
insertedinto wells (4 mm wide, 0.75 mm thick,
con-taining soaking buffer which consisted of: 0.125 M
Tris-hydrochloride [pH 6.8], 0.1% SDS, and 1 mm
EDTA) of a second slabgel,andhydratedbysoaking
for 30 min. The proteolysis gel consisted of a 4-cm
stacking gel of9% acrylamide (pH 6.8) and a 5-cm
runninggelof17% acrylamide (pH8.8). S. aureus V8
protease was diluted to 2 mg/ml in soaking buffer
containing10%glyceroland0.025%bromophenol blue,
and 10
Al
wasaddedtowellscontaining the44Kand20/21Kgel slices. Electrophoresiswascarried out with
the gel apparatus connected to a circulating water
bath at 20°C. The sample was subjected to electro-phoresis (25 mA pergel) until the bromophenolblue
band had almost reached the bottom of thestacking
gel. The powerwas shut off for30 minto allow the
protease to digest the
[K5S]Met-labeled
polypeptidesthat hadmigratedfrom thegel slices. Electrophoresis
wasthen continued until thedyereached the bottom
ofthe runninggel,and thegels were dried and fluo-rographed.
RESULTS
Identification of Ad2-induced early
poly-peptidesanddemonstration of
electropho-retic mobility changes in a 20/21K
poly-peptide in pulse-chase experiments. The
electropherographinFig. 1illustrates
[35S]Met-labeled Ad2-infected and mock-infected cell
polypeptides. Inthis and allotherexperiments,
ara-C (20
,ig/ml)
was added at 4 h p.i. to inhibit viral DNA replication. Therefore, cells were in early stagesofinfection. Lanes A to H in Fig. 1 showpolypeptides from cells incubated with CH before labeling, a procedure that enhances the synthesisofAd2-specific early polypeptidesrel-ative to hostpolypeptides (10). Six Ad2-induced
polypeptides areclearly visible: DBP (the single-stranded DNA binding protein of about 73K daltons), 20/21K, 19K, 15K, 11.5K, and 11K. Lanes A and B and lanes C and D show infected and mock-infected polypeptides labeled 9 to 10 and9 to 12 hp.i., respectively. In these lanes(A and C), the 20/21K polypeptide appeared as a fairly sharp band of about 21K daltons.
How-ever, when cells were labeled for long periods,
e.g., 9to 24 h p.i. (lanes E and F), or9to 12 h p.i. followed bya 12-h chase in complete MEM (lanes G and H), the intensity of the 21Kband decreased, and new bands ofroughly 20K
ap-peared. Lanes ItoL illustratepolypeptides from cells labeled without preincubation with CH (only DBP and the 20/21K bands are visible). Again, a distinct 21K band was apparent in cell
extractslabeled 9to 12hp.i. (lanesI andJ),but this band became reduced in intensity and seemed to increase in mobility in cells labeled for 9 to 24 hp.i. (lanes K and L). These results provide initial evidence that the 20/21K poly-peptide was modified after translation. Since glycosylation ofpolypeptides affects their SDS-PAGEmobility,weconcentratedonthe20/21K
polypeptide as acandidateglycopolypeptide.
Figure 2 illustrates the analysis ofthe pulse (30 min) 21K polypeptide and the pulse-chase (30 min followed by a 15-h chase) 19K to 20K forms of this polypeptide by the partial prote-olysisprocedure. Lane A shows the pulse-labeled 21K polypeptide before chromatography on
ConA-Sepharose, and lanes B toJ show
pulse-and pulse-chase-labeled forms (purified on
ConA-Sepharose). In this experiment, the
chasedpolypeptideformedabroad bandranging from apparentmolecularweightof 19Kto20K. LanesC, F,and I represent the 20Kregion,and lanesD,G, and J represent the 19Kregion.
Mostof thepartialproteolysis polypeptidesof
thepulseandpulse-chaseformsclearly coincide,
confirming that the 21K and 20K are highly
related.However,thepolypeptidesare not
iden-tical,because the chased forms, especially 19K,
contained at least one band (e.g., second from
bottom inlanes D and G) notfoundamongthe
proteolysis products of thepulse form. Note that in lanes A to G some of the polypeptide has
spontaneously"polymerized" to 44K. Thiswas
areproducible phenomenon.
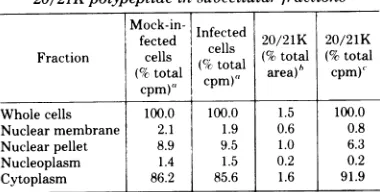
Cellularlocalization of the 20/21K
poly-peptide. To further analyze the 20/21K
poly-peptide, we established which cellular fraction contained themajority of thepolypeptide. Ad2-infected and mock-infected cells were labeled
with[35S]Metfrom9 to 12hp.i.,lysedwith0.5%
NonidetP-40,and thenfractionatedintonuclear
pellet, nuclearmembrane, nucleoplasm,and
cy-toplasm. The vast majority of 20/21K
on November 10, 2019 by guest
http://jvi.asm.org/
Ad2 EARLY GLYCOPROTEIN
317
A
B
C
D
E
F
G
H
II
Ias
I hi
t .*_
6.r 14
I
A_
'._
.d.o
_.,.I
"
I
In
&I
UW
I
J
K
L
4-l :.
on-__n:~~~~~~~
-_
w a"
,sn
*
* f
flC_l so nM AV.b.
40 -o"P
20/21
K-
4
19K
15K-
-11.5K
IIK
w
_n
[image:4.500.54.451.66.573.2]- _.
FIG. 1. Identification ofAd2-inducedearlypolypeptides with andwithout CHpretreatment, and
pulse-chaseexperiments demonstrating mobility changesina20/21Kpolypeptide.Ad2-infectedandmock-infected
cellswereprepared.CH(25pg/ml)wasaddedtosomeculturesat1hp.i. (lanesAtoH).Ara-C(20pg/ml)was
addedto allcultures at4 hp.i.. At 9hp.i., allcultureswere washed andresuspendedin warmMet-free
medium containing ara-C and then labeled with[3S]Met forvarious timeperiods. Proteinextracts were
preparedandwereanalyzedby SDS-PAGE. Anautoradiograph ofadried 17-cmgelis shown. (A) Infected,
labeled9 to 10 hp.i.; (B)mockinfected,labeled9 to 10 hp.i.; (C, I)infected,labeled9to12 hp.i.; (D, J)mock
infected,labeled9 to12 hp.i.; (E, K) infected,labeled 9to24hp.i.; (F, L)mockinfected,labeled9 to24hp.i.; (G)infected,labeled9 to12hp.i.,andchasedfor12h;(H)mockinfected,labeled 9to12 hp.i.,and chasedfor 12 h.
DBP
VOL. 28,1978
on November 10, 2019 by guest
http://jvi.asm.org/
- 4
FIG. 2. Partialproteolysis (S. aureus V8protease)ofpulsed21K andpulse-chased 35S-labeled19to 20K
forms of the 20/21Kpolypeptide. Infectedcells werepulse-labeled with[35S]Met for30minorfor30min followed bya15-hchase.Cytoplasmextractswerepreparedandwerepurified by chromatographyon ConA-Sepharosefollowed bySDS-PAGE. The 21Kpulseand20Kor19K chasepolypeptidebandswerecutfromthe gelandanalyzed by partial proteolysisasdescribed in thetext,using 0.5, 5,or50yg ofproteaseperlane. (A)
Pulse-labeled 21Kpolypeptide before chromatography on ConA-Sepharose; (B, E, H) pulse-labeled 21K
polypeptide purifiedonConA-Sepharose; (C, F, I)pulse-chase-labeled20Kafter ConA-Sepharose; (D, G, J) pulse-chaselabeled19Kpolypeptide after ConA-Sepharose.
peptide was present in the cytoplasm fraction
(Table 1).
Labeling of the 20/21K polypeptide with
[3H]glucosamine, and immunoprecipitation
with antiserum directed against an
Ad2-transformed rat cell line (T2C4). Infected
and mock-infected cellswerelabeled with [3H]-glucosamineto testwhether the labelwas incor-porated intothe20/21K polypeptide.Asshown
in Fig. 3, the only Ad2-specific polypeptide la-beledwith[3H]glucosamine had about thesame electrophoretic mobility as the 20/21K poly-peptide (lane B). Antiserum against T2C4cells
precipitated the
'H-labeled
20/21Kpolypep-tides, aswell asa second polypeptide ofabout
44K(lane D).The 20/21Kand44Kpolypeptides
werenotprecipitatedfrom infected cellextracts
bynonimmuneratsera (laneE),orfrom mock-infected cellextractsby either T2C4 or
nonim-mune ratsera (lanesFandG). The T2C4 anti-serum also immunoprecipitated viral specific early[35S]Met-labeled polypeptidesof 53K, 44K,
20/21K, 19K, 18K, 15K, 14.5K, 13.5K, 12K, and 11.5K (Wold and Green, unpublished data).
The 20/21K and 44K polypeptides precipi-tated by the T2C4 antiserum were assayed by
the partial proteolysis procedure (4) to test whethertheywerechemicallyrelated.
[35S]Met-labeled polypeptides were assayed instead of
TABLE 1. Distributionofradioactivityand the
20/21Kpolypeptideinsubcellularfractions
Mock-in-fected Infectedcells 202120/21Ktotl 20/21K
/21tta
Fraction cells (%total cptoa (%total (%ctotal area)h CPM),
cpm), cpm),
Whole cells 100.0 100.0 1.5 100.0 Nuclear membrane 2.1 1.9 0.6 0.8 Nuclearpellet 8.9 9.5 1.0 6.3
Nucleoplasm 1.4 1.5 0.2 0.2
Cytoplasm 86.2 85.6 1.6 91.9
'Total ¢'S countsper minute in the infected whole-cell preparationwas278x10',and in themock-infected prepara-tionwas280x10.
Infected and mock-infected proteins were resolved by SDS-PAGE, andautoradiogramsweredeveloped.The auto-radiogramswerescannedbyusingaJoyce-Loebel densitom-eterto obtain the totalarea represented byeach fraction. These values indicate the percentage of the totalscan area
represented bythe 20/21K polypeptide,minuscontribution frommock-infectedpolypeptides.
'Calculationsarebasedon avalue of 100% for the esti-mated20/21Kcountsper minute in thewhole-cellfraction.
["H]glucosamine-labeled
polypeptides, because larger amounts of radioactivity could be ob-tained.[35S]Met-labeled
polypeptidesimmuno-precipitated by the T2C4 antiserum were
re-solved by SDS-PAGE, and fluorograms were
developed. The 20/21K and 44K bandswerecut
eV -w so
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.500.260.450.388.484.2]Ad2 EARLY GLYCOPROTEIN 319
A
B
C
D
E
F
G
DBP
I
20/
21
K
15K
11.5K
I
IK
FIG. 3. Identification of polypeptides labeled in vivo with [3H]glucosamine andimmunoprecipitated by antiserumagainst 72C4 cells. Ad2-infected and mock-infected cells were labeled with[3H]glucosamine.Total
cellextractswereprepared and immunoprecipitated by using T2C4 IgG and nonimmune ratIgG. Equal
volumesof each precipitate were analyzed by SDS-PAGE. (A) Marker polypeptides:[3S]Met-labeled,early infectedpolypeptidesprepared by the CH enhancement procedure. (B-G),[3H]glucosamine-labeledextracts. (B)Infected, before immunoprecipitation; (C) mock infected, before immunoprecipitation; (D) infected extract
versusT2C4IgG; (E) infected extract versus nonimmune rat IgG; (F) mock-infected extract versus72C4IgG;
(G)mock-infectedextract versusnonimmune rat IgG.
from the driedgelandanalyzed bypartial
pro-teolysis as described above. The partial
prote-olysis slab gel is shown in Fig. 4. The same
polypeptide bands were generated by protease
digestion of the 20/21K and44K polypeptides,
indicating that they are highly related
chemi-cally. Proteolysis ofthe 20/21K and 44K
poly-peptidesby usingotherproteaseconcentrations
also indicated thattheyarehighly related.
Binding of the 20/21K polypeptide to
ConA-Sepharose and its elution by
methyl-a-D-mannoside. To obtain further evidence
that the 20/21Kpolypeptide isa
glycopolypep-tide, 35S-labeled proteinextracts weresubjected
toaffinity chromatographyoncolumns of
ConA-Sepharose. Onlycertain carbohydrate moieties
(a-D-glucopyranosides, and
a-N-acetyl-D-glucos-aminides) bindstronglytoConA(20), so that if
the20/21Kpolypeptide contains thesesugars it
should bind to the ConA-Sepharose column.
Infected and mock-infected cells were labeled
with
[35S]Met
for15h, andcell
cytoplasmswerepreparedand thenloadedonto
ConA-Sepharose
columns. The columns were washed and then
eluted with 0.2 M
methyl-a-D-mannoside.
Thecolumnflow-through, wash,andeluted fractions
were analyzed by SDS-PAGE. Only the
man-noside-elutedinfected cell fraction
(lane
F)con-tained detectable 20/21K polypeptide
(Fig.
5).These results suggest that the 20/21K
poly-peptide contains sugar residues
specific
forConA.
The20/21K
polypeptide
issynthesized
inAd2-infected
monkey
cells.Theimmunopre-cipitation of both
[3H]glucosamine
and[35S]-Met-labeled 20/21K
polypeptide by
the T2C4antiserum suggests that the 21K
polypeptide
is viral coded rather than cell coded and viral induced. The20/21Kpolypeptide(and
theDBP,11K, 8.8K, 8.3K polypeptides) was
synthesized
inAd2-early infected
monkey
(CV-1)
cells(Fig.
6). The 21K, DBP, and 11K were also
synthe-sized in Ad2-early infected hamster cells (notshown). Theseresults
provide
further evidencethat the 21K, DBP, and 11K
polypeptides
areviral coded.
VOL. 1978
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.500.99.422.57.331.2]320 JENG, WOLD, AND GREEN
.- nP
'U
44
I,*
iK I~~~~~~~~~~~~~~~~~~~~7
4KM
IFIG. 4. Partialproteolysis (S.aureus V8protease) of the 44K and20/21K[35S]Met-labeledpolypeptides immunoprecipitatedbyT2C4 antiserum(A) 44K, (B) 21K. Bothpolypeptides were treated with 5 pLg of protease.
DISCUSSION
Our results
provide
strong evidence that theAd2-induced 20/21K
polypeptide
is aglycopo-lypeptide. The
polypeptide
canbelabeled with[3H]glucosamine,
and it binds toConA-Sepha-rose columns and is eluted with
methyl-a-D-mannoside. When labeled with
[35S]Met
forshort periods (30 min to 3 h), it appears as a
single, relatively sharp band with an apparent
molecular weight of about 21K. However, if
la-beled for long periods, or if pulse-labeled and
thenchased, the polypeptideappears as multiple
bands with apparent molecularweights of 20K
to21K. Partial proteolysisdataindicatethat the
pulse-labeled 21K and the pulse-chase-labeled
20K polypeptides are highly related, although
notidentical. Both pulse (30min)and pulse (30
min)-chase (12 h in complete MEM) forms of
[35S]-labeled
20/21K polypeptidebind toConA-Sepharose, and therefore apparently are
glyco-sylated. The multiple bands of the chased
20/21K polypeptide may indicate heterogeneity in sugar content, because glycosylation affects
(usually decreases) the SDS-PAGE mobility of
polypeptides.We cannotexclude the possibility
that there is another distinct polypeptide of
A
B
C
C
'.
-1
F
[image:7.500.90.192.77.295.2]-~~~~~~~~.
FIG. 5. ConA-Sepharose column chromatography: identification of[3S]Met-labeled Ad2-infected and
mock-infected polypeptides in the flow-through, wash, and methyl-a-D-mannoside eluted fractions.
Ad2-infectedandmock-infectedcellswerelabeled with[3S]Met from9to24 hp.i. Thecytoplasm fractionwas
prepared andchromatographedonConA-Sepharose.Appropriate column fractionswerepooled and analyzed
bySDS-PAGE. (A)Markerpolypeptides: Ad2-infectedKBcells extracts labeled with [3SJMet by the CH
enhancementprocedure; (B) infected, flow-through fraction; (C)mock infected, flow-through fraction; (D)
infected,washfraction; (E)mockinfected, wash fraction; (F) infected, methyla-D-mannosideeluate; (G) mock
infected,methyl-a-D-mannosideeluate.
..)
R
... s_1;lcx
.2
)-t
.5K-~
5K
I; ....on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.500.116.396.347.591.2]Ad2 EARLY GLYCOPROTEIN 321
Ad2
Infecti
CV-1
Cell
A
B
C
ft It _ ir
t
I
;b
DBP(79K)-21K
--IlK
._Auh
tF~~~~~~~
i. (.- If.
4.0* k 1
a
_i
8.8K-8.3K
about 20K to 21K that cannot be detected in thesesingle-dimensiongels.
Our results also strongly suggest that the
20/21K
polypeptide
is codedby
anearly
Ad2gene, and is not a cellcodedpolypeptideinduced
by
Ad2. Both[3H]glucosamine
and[35S]Met-labeled 20/21K polypeptide were immunopre-cipitated by rat antiserum against T2C4 cells. T2C4 is a line ofAd2-transformed rat cells (6) that contains all four of the Ad2 early gene
blocks(7) andsynthesizes RNA derived fromall
four blocks (5). Thepolypeptidewas not precip-itatedby nonimmune rat serum, orbyrat
anti-sera against three other Ad2-transformed rat
cells (notshown). Therefore, itisprobable that
the Ad2 sequences present in T2C4 cells (but not in the other 3 Ad2-transformed rat lines) synthesize the 20/21K polypeptide. A 20/21K
polypeptide was also induced in Ad2-early
in-fectedmonkey and hamster cells. These results suggest that the 20/21K polypeptide is viral coded, because it isimprobable (but not
impos-sible) that a cell-coded 20/21K polypeptide
would be synthesized by Ad2-infected human,
monkey, andhamstercells,andbyonebut not
allAd2-transformed rat cells.
We are currently attempting to purify the
20/21K polypeptideforfurther chemical,
phys-ical,
andbiological
characterization.Theprotein
seemstoexist in
multiple
charge
forms,
becauseit is present in fractions eluted from
ion-ex-change
resinsby
salt concentrations of from 10mM to 0.4 M.Also,it seems to exist in
multiple-size forms, as reflected by its behavior on gel
filtration columns. An exampleof this is shown in Fig. 3, where the 20/21K polypeptide
spon-taneously polymerizedto44K,the 44K
sponta-neouslyconverted to21K,andboth the 44K and
the20/21K polypeptide polymerizedtospecies
ofhighmolecularweight. Thistype of apparent size andchargeheterogeneityistypicalofmany
glycoproteins.We have notattemptedtoreduce
7* and alkylate the 20/21K
polypeptide
todeter-minewhethersulfhydralgroups areinvolvedin
the polymerization of thepolypeptide.
Chin and Maizel (2) reported that a
35S-la-beled
polypeptide
(E2)was acomponent of theplasma membrane. E2, apparently, is the same
polypeptide that Ishibashi and Maizel (13)
re-portedtobelabeledwith[3H]glucosamine. The
20/21Kpolypeptide probably correspondstoE2, because we have seen no indication that any otherearly proteinisglycosylated. Ourfinding
I
I
M MCH
CH CH
CH+
FIG. 6. [35S]Met-labeledpolypeptides induced in Ad2-early infected monkey (CV-1) cells, with and without CH(25pg/ml)pretreatment. (A)Infected, no
CH;(B)infected,withCH; (C) mock infected, no CH; (D) mockinfected, with CH.
VOL. 28,1978
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.500.38.221.78.661.2]JENG, AND GREEN
that the 20/21K polypeptide is located mainly in the cytoplasm of Nonidet-P40-lysed cells would be consistent with thepossibility that this polypeptide is a membrane component.
How-ever,wehavealso found the20/21Kpolypeptide in a complex purified from lysed nuclei that synthesizes Ad2 DNA (21). The 20/21K poly-peptide can also be immunoprecipitated by
T2C4 antiserum from the nucleoplasm of cells. Therefore, anuclear role forthispolypeptide is notexcluded.
Glycoproteinsarewidelydistributed innature
and serve a variety of structural, lubricating, enzymatic, hormonal, and plasma membrane-as-sociatedfunctions (15, 25,32). The finding that
anAd2-earlygeneproduct isaglycoprotein is of
interest,especially if it is foundtoserve a regu-latory function. Studies on the 20/21K poly-peptide should be ofinterestnotonly regarding Ad2 replication, but also the general role of glycosylation in protein function.
ACKNOWLEDGMENTS
We thank H. Thorntonforassistance in cell culture andin
preparation ofthe T2C4antiserum,and C.Devine for
tech-nical assistance.WearegratefultoP.H.Gallimoreforagift ofthe T2C4cells.
Thisworkwassupported byPublic Health Servicegrants Al01725-19from the National InstituteofAllergyand
Infec-tious Diseases and CA 21824-01 from the National Cancer
Institute, and by contract NOI CP 43359 from the Virus
Cancer Program within the National Cancer Institute.
W.S.M.W. waspartially supported byafellowshipfrom the
Medical ResearchCouncil of Canada.M.G. is therecipientof
aResearchCareer Award(5K06AI 04739)fromthe National
InstituteofAllergyand InfectiousDiseases.
LITERATURE CITED
1. Bonner,W.M.,and R. A.Laskey.1974.Afilmdetection method fortritium-labeledproteinsandnucleicacidsin polyacrylamide gels.Eur.J. Biochem. 46:83-88. 2. Chin,W.W.,and J. V.Maizel,Jr. 1976.Polypeptides
ofadenovirus. VII. Furtherstudies ofearlypolypeptides invivo and localizationofE,andE2Atothecellplasma membrane.Virology 71:518-530.
3. Chin,W.W.,andJ. V.Maizel,Jr. 1976. The polypep-tidesofadenovirus. VIII. The enrichment ofE3(11,000) inthe nuclear matrix fraction.Virology 76:79-89. 4. Cleveland, D. W., S. G. Fischer, M.W. Kirschner,
andU. K. Laemmli. 1977.Peptide mapping bylimited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis.J.Biol. Chem. 252:1102-1106. 5. Flint, S. J., P. H. Gallimore, andP. A.Sharp. 1975.
Comparison ofviral RNAsequencesinadenovirus 2-transformed and lytically infected cells. J. Mol. Biol. 96:47-68.
6. Gallimore, P. H.1974.Interactions of adenovirustype2 withratembryo cells. Permissiveness, transformation andinvitrocharacteristicsofadenovirustransformed
ratembryo cells. J. Gen. Virol. 25:263-273.
7. Gallimore,P.H.,P.A.Sharp, andJ.Sambrook. 1974. Viral DNA in transformed cells. II. A study of the
sequencesofadenovirus 2 DNAin ninelinesof
trans-formed rat cellsusing specific fragments of the viral
genome.J. Mol. Biol.89:49-72.
8. Gilead, Z., Y.-H. Jeng,W.S. M.Wold, K. Sugawara, H.M.Rho,M.L.Harter, and M. Green. 1976.
Im-munological identification oftwoadenovirus 2-induced earlyproteinspossibly involvedincelltransformation. Nature(London) 264:263-266.
9. Grodzicker, T., C. Anderson, J. Sambrook, and M. B. Mathews. 1977. The phvsical locationsofstructural
genesinadenovirus DNA. Virology 80:111-126. 10. Harter, M. L.,G. Shanmugam,W. S. M. Wold, and
M. Green. 1976. Detection of adenovirus type 2-in-duced earlypolypeptides using cycloheximide
pretreat-ment to enhance viral protein synthesis. J. Virol. 19:232-242.
11. Holtzman, E.,I.Smith, and S. Penman. 1966. Electron
microscopic studies of detergent treated HeLa cell
nu-clei. J. Mol. Biol. 17:131-135.
12. Ishibashi, M., W. W. Chin, and J. V. Maizel, Jr. 1977. Thepolypeptides of adenovirus. IX. Partial purification ofearly proteins and observationsonlate proteins of
type5adenovirus. Virology 83:1-15.
13. Ishibashi, M., and J. V. Maizel, Jr. 1974. The polypep-tides of adenovirus.VI. Early and late glycopolypep-tides.Virology 58:345-361.
14. Jeng, Y.-H., W. S.M.Wold, K. Sugawara, Z. Gilead, and M. Green. 1977. Adenovirustype 2 coded single-stranded DNA bindingprotein:in vivophosphorylation and modification. J.Virol.22:402-411.
15. Kornfeld, R., and S. Kornfeld. 1976.Comparative
as-pects ofglycoproteinstructure. Annu. Rev. Biochem. 45:217-237.
16. Levinson, A.D., and A. J. Levine. 1977. ThegroupC
adenovirus tumor antigens: identification in infected andtransformed cells andapeptidemapanalysis. Cell
11:871-879.
17. Levinson, A.D., E. H. Postel, and A. J. Levine. 1977. In titoandinvitrophosphorylationoftheadenovirus
type 5 single-stranded-specific DNA-binding protein. Virology 79:144-159.
18. Lewis, J. B., J. F.Atkins, P. R. Baum, R. Solem, R. F. Gesteland, andC.W.Anderson. 1976.Location and identificationofthegenesforadenovirustype2 early polypeptides. Cell 7:141-151.
19. Linne,T., H. Jornvall, and L. Philipson. 1977. Purifi-cation and characterization of the phosphorylated DNA-bindingprotein from adenovirus-type-2-infected cells.Eur. J. Biochem.76:481-490.
20. Poretz, R. D., andI.J.Goldstein. 1970. An examination
of the topography of the saccharide binding sites of concanavalin A andthe forces involved incomplexation. Biochemistry 9:2890-2896.
21. Rho, H. M., Y.-H. Jeng, W. S. M. Wold, and M. Green. 1977. Association ofadenovirus type 2 early proteins with a soluble complex that synthesizes adenovirus DNA in vitro. Biochem. Biophys. Res. Commun. 79:422-428.
22. Russell, W. C., and G. E. Blair. 1977. Polypeptide phosphorylation in adenovirus-infected cells. J. Gen. Virol. 34:19-35.
23. Saborio, J. L., and B. Oberg.1976.In vivoandin vitro synthesis of adenovirustype2early proteins. J.Virol. 17:865-875.
24. Shanmugam,G., S. Bhaduri, M. Arens, and M. Green. 1975. DNAbinding proteins in the cytoplasm and ina
nuclear membrane complex isolated from uninfected and adenovirus 2 infected cells. Biochemistry 14:332-337.
25. Spiro, R. G. 1973. Glycoproteins, p. 349-467. InC. B. Anfinson, J. T. Edsall, and F. M. Richards (ed.),
Ad-vances in protein chemistry, vol. 27. Academic Press Inc., New York.
26. Stohlman, S.A.,0.R.Eylar, and C. L. Wissman,Jr. 1976. Isolation ofthedengue virus envelope glycopro-teinfrommembranesofinfected cells by concanavalin Aaffinity chromatography. J. Virol. 18:132-140. 27. Sugawara, K., Z. Gilead, and M. Green. 1977.
Purifi-cation and molecular characterization of adenovirus
on November 10, 2019 by guest
http://jvi.asm.org/
VOL. 28, 1978
type 2DNA-binding protein. J. Virol. 21:338-346.
28. Sugawara, K., Z. Gilead, W. S. M. Wold, and M.
Green. 1977. Immunofluorescencestudy of the adeno-virustype 2 single-stranded DNA binding protein in infected and transformed cells. J. Virol. 22:527-539. 29. van derVliet, P. C., and A. J.Levine. 1973. DNA
bindingproteinsspecific for cells infected by adenovi-rus.Nature(London)246:170-174.
30. vanderVliet, P.C.,A. J.Levine, M. J.Ensinger, and H. S.Ginsberg.1975.ThermolabileDNAbinding pro-teins from cellsinfected withatemperature-sensitive mutantof adenovirus defective in viral DNAsynthesis. J. Virol. 15:348-354.
31. vanderVliet, P.C.,J.Zandberg, and H.S. Jansz. 1977.Evidenceforafunction of the adenovirus
DNA-EARLY GLYCOPROTEIN
binding proteinin initiation of DNA synthesis aswell as elongation of nascent DNA chains. Virology 80:98-110.
32. Waechter,C. J., and W. J. Lennarz.1976. The role of polyprenol-linked sugars in glycoprotein synthesis. Annu.Rev. Biochem.45:95-112.
33. Wall, R., J.Weber, Z. Gage, and J. E. Darnell. 1973. Production ofviral mRNA in adenovirus-transformed cells by the post-transcriptional processing of hetero-geneous nuclear RNA containing viral and cell se-quences. J. Virol. 11:953-960.
34. Wold, W. S. M., M. Green, and W. Buttner. 1978. Adenoviruses, p. 673-768.In D. P.Nayak (ed.), The molecular biology of animal viruses, vol. 2. Marcel Dek-ker. Inc., New York.

![FIG. 3.(G)infected(B)versuscellvolumesantiserum Identification ofpolypeptides labeled in vivo with [3H]glucosamine and immunoprecipitated by against 72C4 cells](https://thumb-us.123doks.com/thumbv2/123dok_us/1525562.105130/6.500.99.422.57.331/infected-versuscellvolumesantiserum-identification-ofpolypeptides-labeled-glucosamine-immunoprecipitated-cells.webp)
![FIG. 4.ofimmunoprecipitated21K. Partial proteolysis (S. aureus V8protease)the 44K and 20/21K [35S]Met-labeledpolypeptides by T2C4 antiserum (A) 44K, (B) Both polypeptides were treated with 5 pLg ofprotease.](https://thumb-us.123doks.com/thumbv2/123dok_us/1525562.105130/7.500.116.396.347.591/ofimmunoprecipitated-partial-proteolysis-protease-labeledpolypeptides-antiserum-polypeptides-ofprotease.webp)
![FIG. 6.(D)withoutCH;Ad2-early [35S]Met-labeled polypeptides induced in infected monkey (CV-1) cells, with and CH (25 pg/ml) pretreatment](https://thumb-us.123doks.com/thumbv2/123dok_us/1525562.105130/8.500.38.221.78.661/withoutch-early-labeled-polypeptides-induced-infected-monkey-pretreatment.webp)