0022-538X/94/$04.00+0
Copyright © 1994,American SocietyforMicrobiology
European Brown Hare
Syndrome
Virus: Relationship to
Rabbit
Hemorrhagic Disease Virus and
Other Caliciviruses
CHRISTOPHWIRBLICH,' GREGORMEYERS,' VOLKER F. OHLINGER,' LORENZOCAPUCCI,2
ULRICHESKENS,3 BERNDHAAS,' ANDHEINZ-JURGEN
THIEL'*
Federal Research Centrefor Virus Diseases of Animals, 72001 Tuibingen,1 and StaatlichesMedizinal-, Lebensmittel- und Veterinaruntersuchungsamt Mittelhessen, 35396
Giessen,3
Germany,and IstitutoZooprofilattico Sperimentale della Lombardia edell'Emilia, 25124Brescia, Italy2
Received 18 February 1994/Accepted 13 May 1994
Monoclonalantibodies directedagainstthecapsid proteinof rabbithemorrhagicdisease virus (RHDV)were used toidentify fieldcasesofEuropeanbrown haresyndrome (EBHS)and todistinguishbetween RHDV and the virus responsible for EBHS. Western blot (immunoblot) analysis of liver extract of an EBHS virus
(EBHSV)-infected harerevealed a single major capsidprotein species ofapproximately 60 kDathat shared
epitopeswith thecapsidproteinofRHDV.RNA isolated from the liver ofanEBHSV-infectedhare contained twoviral RNAspeciesof 7.5 and2.2kb thatcomigratedwith thegenomicandsubgenomicRNAs of RHDV and wererecognized bylabeled RHDV cDNA inNorthern(RNA) hybridizations.The nucleotide sequence of the 3' 2.8 kb of theEBHSVgenome wasdetermined from fouroverlappingcDNA clones.Sequenceanalysisrevealed anopenreadingframe that contains part of theputativeRNApolymerasegeneand thecompletecapsid protein
gene. This particular genome organization is shared by RHDV but not by other known caliciviruses. The deduced amino acid sequence of thecapsid proteinofEBHSVwascomparedwith thecapsid proteinsequences ofRHDV and othercaliciviruses.The aminoacid sequencecomparisonsrevealed that EBHSV iscloselyrelated toRHDVand distantly related to other caliciviruses. On the basis of their genomeorganization,it issuggested
that caliciviruses be divided into three groups.
Since 1980 the occurrence of an apparently new disease affecting wild and farmed hares has been reported in many
European countries. The term European brown hare
syn-drome (EBHS) has been used by mostworkers to designate the disease. The etiology of EBHS remained unclear until it
was shown by animal experiments and electron microscopy analysis(12, 21) that EBHS is caused byanonenveloped virus, termedEBHSvirus(EBHSV), which appearedtobesimilarto
the rabbit hemorrhagicdisease virus(RHDV).
Numerous datapoint toward arelationship of EBHSV and
RHDV. EBHS and RHD are very similar with regard to
clinical signs as well as pathological and histopathological changes. Both diseases are characterized by high mortality, reaching 90to100% in adult animals(9, 15, 28). Degeneration ofhepatocytes and necrosis of the liverare the predominant lesions in diseased animals (28, 29). Morphologically, the virions ofRHDVandEBHSVareapparentlyindistinguishable
(3,9,21). Antigenic relatedness betweenRHDVand EBHSV has been demonstrated by Western blot (immunoblot) using hyperimmune serum against RHDV (9). Thus far, neither virus has been adapted to continuous growth in cell culture. The striking similarities between EBHS andRHD prompted
several investigators to perform cross-species infections.
Al-though transmission was reported to be successful in some
instances, most cross-species infections have failed to induce disease(5, 9, 31). Discrimination betweenRHDVandEBHSV was possible by immunoelectron microscopy (9), hemaggluti-nation (28, 29), and enzyme-linked immunosorbent assay
(ELISA) (5). These findings suggested that a virus related to,
*Correspondingauthor.Mailing address: Federal Research Centre
for Virus Diseases of Animals, P.O. Box 1149, 72001 Tubingen,
Germany.
but distinct from, RHDV is responsible for outbreaks of EBHS.
RHDVhasbeenidentifiedas amember of the Caliciviridae family (30, 33).Caliciviruses arenonenveloped positive-strand
RNAviruses with adiameter of about 35 to 39nm(37).The
virionsarecomposed ofasingle-strandedRNAabout 7.5to8 kb inlengthand asingle majorcapsid protein withamolecular massof60 to 71 kDa. Members oftheCaliciviridaefamilyare
the feline calicivirus (FCV), the San Miguel sea lion virus
(SMSV), and the prototype, vesicular exanthema of swine
virus. Sequence analyses of the genomes of FCV (7, 25) and
RHDV (24) revealed a large open reading frame that was
proposed to code for nonstructural proteins. This prediction
wasbased on thepresence of conservedamino acid motifs that
exhibit homology to picornavirus nonstructural proteins 2C, 3C,and 3D. The genescodingfor the nonstructural proteins are located in the 5' two-thirds of the genome. The capsid protein is encoded in the 3' third of the genomicRNA(8,24, 26).Asimilargenomeorganization has beenreported for the Norwalkvirus (17) and the Norwalk-like Southamptonvirus
(20). In terms of the number of functional open reading
frames, RHDV is different from other caliciviruses. The genomicRNAofRHDVcontainsalargeopenreadingframe of 7 kb that includesthegenesfor the nonstructuralproteins and thecapsid proteingene.FCV, SMSV, andNorwalkvirus
encodethecapsid proteininaseparate openreadingframe.In
addition, a small open reading frame encoding a protein of
unknownfunctionhasbeen describedfor all caliciviruses that
have been examined to date. This open reading frame is several hundred nucleotidesinlength and coversthe 3'-most
region of the genome. Like other caliciviruses, RHDV
pro-ducesasubgenomicRNAthat is 3'-coterminaltothegenomic
RNAandcoversthecapsid proteingene (23). Invitro
trans-lation of this 2.2-kb RNA gave rise to a protein with an apparentmolecular massof60 kDa thatcomigratedwith the
5164
on November 9, 2019 by guest
http://jvi.asm.org/
RELATIONSHIP OF EBHSV TO RHDV AND OTHER CALICIVIRUSES 5165 mature capsid protein (4). N-terminal sequence analysis of
peptide fragments obtained by CNBrcleavage of the capsid protein of RHDV (32) suggested that most of the capsid
protein starts at the first methionine encoded by the
sub-genomicRNA.
Inthe presentstudywereportanalyses of EBHSV primarily with respect to its relationship to RHDV and other
calicivi-ruses.Inparticular, cloningandsequencing of the 3'-terminal 2.8 kb of the EBHSV genome including the complete capsid protein gene is described. The deduced amino acid sequence of thecapsid proteinwascompared with published sequences ofRHDVandother caliciviruses.
MATERIALSAND METHODS
RNA isolation and Northern (RNA) blot hybridization.
RNA was isolated from frozen liver by using guanidinium isothiocyanate and centrifugation through a cesium chloride cushion(10) aspreviouslydescribed
(30).
Twomicrograms ofRNA wasglyoxylatedfor 40 minat56°Cinatotal volume of12
RI
ofglyoxylationmixture(10mMsodiumphosphate[pH 6.8],1.2 M glyoxale) and electrophoresed on a 1%
phosphate-buffered agarose gel containing 5.5% formaldehyde (6). The
RNA wastransferredtoNylon membranes,bakedat80°C for
2h,andhybridizedto
32P-labeled
cDNAprobesat52 and68°Cas described elsewhere
(22).
The RNA size standard forelectrophoresiswas obtained from Bethesda Research Labo-ratories.
cDNAcloning.Fivemicrogramsof total RNAisolated from the liver ofan EBHSV-infected hare was used to synthesize
double-stranded cDNA. First-strand cDNAsynthesis with 50
pmol of
oligo(dT)12_18
(Pharmacia) as a primer and size selection ofcDNAmoleculeslargerthan1 kbwerecarriedout as reported elsewhere (34). Cloning inLambdaZapll
(Strat-agene) and in vitro packaging using Gigapack Gold (Strat-agene) were performed as recommended by the supplier. Ligationof thecDNAinplasmid pBluescript (Stratagene)and transformation intocompetent Escherichia coli XL1-Bluewerecarriedoutaccordingtostandardprotocols
(35).
Screeningof the phage library with 32P-labeled cDNA was carried out asdescribedbyBenton andDavis (2). Screening of theplasmid library by colony hybridizationwasperformed accordingtothe
procedure of Grunstein and Wallis (13).
Sequence determination. Nucleotide sequence determina-tionwas
performed by
thedideoxy
chain termination method(36).
A nested set of deletions of the cDNA inserts wasconstructed with exonucleaseIIIand nucleaseS1(14). Alkali-denatured
plasmid
DNAwassequenced
with a modified T7DNApolymerase
according
tothe instructions of thesupplier (Pharmacia).Nucleotide sequenceswereanalyzedon aDigitalMicrovaxll with the GeneticsComputer Groupprogram
pack-age
(release 7.3)
(11).Construction of expression plasmids and purification of
fusionproteins.RHDV cDNA
fragments
wereexpressedinE.coliby using plasmid pEX34b, which is derived fromplasmid pEX31
(38).
Plasmid pEX34b codes for the amino-terminal partof theRNApolymerase ofbacteriophageMS2.This part is followedbyapolylinkerregion
whichallowsconstruction of in-frame gene fusions. Expression of the gene fusions is controlledbythe lambda leftward promoterand thecI repres-sor. Induction is achieved by a temperature shift from 28 to42°Cin anE. coli strain that harbors a temperature-sensitive
mutant of the cI repressor. The fusion
proteins
that areproducedupon induction are composed of 99 aminoacids of the MS2 RNA polymerase, a small number of amino acids encoded
by
thepolylinker,
thepolypeptide
which is encodedby
theinserted DNA, and a small number of amino acids at the C-terminalend which are encoded by vector sequences.
Standardcloning procedures were used to construct expres-sionplasmids pEX-H, pEX-G, andpEX-I (35). Numbering of the amino acid and nucleotide positions is according to the published sequence (24). Plasmid pEX-H containsa 1,397-bp HindIII-BamHI cDNAfragment (position 4001 to 5398) in-serted into the BamHIsite ofpEX34b. This fragment codes for amino acids 1332 to 1796 of the ORF1 polyprotein and contains theputativeRNApolymerase geneand afew codons of thecapsid protein gene. Plasmid pEX-Hwasdigested with XhoI (position 5182) and PstI (whichcutsin the 3' polylinker region of pEX34b) to remove a 239-bp fragment coding for amino acids1727 to 1796. Theshortenedplasmidwastreated with Klenow polymerase to convert sticky ends to blunt ends andreligated to obtain plasmid pEX-G, which codes for amino acids 1332 to 1727 of the RHDV polyprotein. To obtain plasmid pEX-I, a 2-kbBamHI-BamHI cDNAfragment that codes for amino acids 1796to 2343 was excised from cDNA clone pRHDml (23) and ligated into the BamHI site of pEX34b. This cDNA fragment starts at nucleotide position 5395 and extends beyond the stop codon ofORFi into the
multiple cloning site of plasmid pBluescriptSK-. Plasmid constructions were verified by restriction enzyme digestion, andcorrectplasmidswereused to transform E. coli537 cells
containing a temperature-sensitive mutant of the lambda
re-pressor genecI on akanamycin resistance plasmid.
Expression of the gene fusions, purification of inclusion
bodies, and extraction of fusion protein was carried out as
describedby Strebeletal.(38).Extracted fusionproteinswere
furtherpurifiedbypreparativesodiumdodecyl
sulfate-polyac-rylamide gel electrophoresis (SDS-PAGE). After
electro-phoresis, the gelswere stained for about 10 minwith Coom-assie blue (0.25% Coomassie blue, 50% methanol, and 10% aceticacid), andbandscontaining fusion proteinwereexcised after2 to 3hofdestaining(30% methanol, 10% acetic acid). Fusionproteinwaselectroeluted for24hat4V/cm byusinga
commercially available elution device (Biotrap; Schleicher &
Schull). The eluted proteinwas dialyzed against ammonium carbamate buffer (100 mM, pH 7.5), lyophilized, and
resus-pendedinphosphate-buffered saline (PBS).
Immunization of rabbits. Serum was collected from each rabbit before immunizationto serve as acontrol. Theanimals
wereprimedwith 20to200
jig
of fusionproteinemulsified incompleteFreundadjuvant. Injectionsweredelivered subcuta-neously over multiple sites in the back. The rabbits were
boostedat3-weekintervals with 20to200 ,ug of fusionprotein emulsified in incomplete Freund adjuvant, and blood was
collected 10days after each boost.
Immunoblotting. Extracts obtained from homogenized
liv-ersof infected and control animalswereclarifiedby
centrifu-gation. Samples containing50 ,ugof totalproteinwereboiled insamplebuffer(5%
3-mercaptoethanol,
2% SDS, 6 Murea, 62.5mMTris-HCl[pH6.8])andelectrophoresedonSDS-10%polyacrylamide gels (19). Transfer to nitrocellulose was per-formed inaHoefersemiphorapparatusfor 1 h at 100 mA as
describedbyTowbin et al. (39). The nitrocellulosewas
incu-bated for 1 h at room temperature in PBS containing 2.5% low-fat milkpowder toblock unspecific binding. Rabbit anti-serum wasaddedand allowed tobind for1 h. The membrane
waswashed three times with PBScontaining 0.1% Tween 20
(PBST),andalkalinephosphatase-conjugated rabbit anti-bodies(Dianova)wereaddedat a1:5,000dilution. After1 hof incubation with secondary antibody, the membrane was washed threetimes with PBST. The blotsweredevelopedwith nitroblue tetrazolium (Fluka) and BCIP
(5-bromo-4-chloro-3-VOL. 68,1994
on November 9, 2019 by guest
http://jvi.asm.org/
1 2 3 4 5
110
K-84
K-47
K-*_
33 K-
2416
K-FIG. 1. Cross-reactivity of anti-RHDV MAbs with EBHSV.
Viri-ons werepurified bysucrosedensity gradient centrifugation from livers
of EBHSV-infected hares according to the method previously de-scribed for the purification of RHD virions (30) and subjected to
SDS-PAGE. Western blotting was performed with the anti-RHDV MAbs3H6, 6D6, 1H8, 5D1, and 3H2atadilution of 1:500 (lanes 1to
5, respectively). Numbers indicate the molecular weights (in thousands [K]) of the protein standards (Bio-Rad).
indolylphosphate toluidinium) (Fluka) as substrates in AP buffer(100mMNaHCO3, 1 mM MgCl2 [pH 9.8]).
Immunoblotsto testthe reactivity of monoclonal antibodies (MAbs) to RHDV and EBHSV antigen were performed as described previously (30). Dilutions of 1:500 of the anti-RHDVMAbs3H6, 6D6, 1H8, 5D1, and 3H2were incubated
overnight. Specific binding was detected with
peroxidase-conjugated anti-mouse antibodies (Dianova) diluted 1:2,500 and2,2-chloronaphthol (Sigma) as asubstrate.
ELISA. RHDV andEBHSV antigen samples were diluted
1:3 in PBST and incubated at 37°C for 1 h in 96-well
flat-bottomplates (NuncI; Nunc) coated with rabbit anti-RHDV
hyperimmune serum as described previously (30). After
re-peated washings with PBST, the plateswere incubated first(1
hat37°C)withanti-RHDV MAbs diluted 1:500 inincubation
buffer (PBST, 2%rabbit normal serum)and then with
perox-idase-conjugated anti-mouse antibodies (Dianova) at a dilu-tion of 1:1,000 in PBST (30
min).
O-Phenylenediamine (Sig-ma) was used as a substrate. MAb 075, which is directedagainst foot-and-mouth disease virus, servedas acontrol.The cutoff values for negatives inan ELISAfor routine diagnosis weredeterminedasthreefold standard deviations derived from apanelof 60EBHS-negativeliversamplesfrom hares and 83
RHD-negative liver samples from rabbits. Incomparison with negative controls, the relative optical density readings for
MAbs 3H6 and 6D6rangedbetween 2 and 9(3H6)and 3 and 12(6D6).
Nucleotide sequenceaccession number. The nucleotide
se-quence datareported inthis article have been depositedwith the EMBL and GenBank data libraries under accession no. U09199.
RESULTS
Cross-reactivityof anti-RHDVMAbswithEBHSV.Apanel
of anti-RHDV MAbs (3H6, 6D6, 1H8, 5D1, and 3H2), all reactivewith theRHDV capsid proteinVP60 (5), wastested
for recognition of EBHSV-specific antigen by using ELISA andWesternblot(Fig. 1and Table1). It turnedoutthat three
of fiveanti-RHDV MAbsreacted with EBHSV. Interestingly, twocross-reacting MAbs (3H6 and 6D6)werepositive only by
ELISA, suggesting that they recognized discontinuous epitopes (4a). In contrast, MAbSD1 reacted exclusively with
[image:3.612.127.251.74.189.2]the denatured EBHSV capsid protein. These results confirm
TABLE 1. Recognition of EBHSV antigens inaWesternblot and ELISAby anti-RHDV MAbs
Recognitionof EBHSVantigens
MAba
ELISAb Westernblot'
3H6 +
6D6 +
1H8
5D1 +
3H2
aImmunoglobulin isotypeGIexceptfor5D1(G2b).
bRabbit anti-RHDVhyperimmuneserum wasused to trap viralantigens from
liver samples ofEBHSV-infectedhares.
'Highly purifiedvirions ofEBHSVwereused.
previous studies (5, 9) which demonstrated that EBHSV preparations contain a cross-reactive 60-kDa protein which
mostlikely represents thecapsid protein.
Inorder to investigate whether anti-RHDV MAbs can be used to identify field cases of EBHS, a blind study was
performed with liver material from diseased hares. MAb 3H6 was used for detection of EBHSVantigen in tissue samples
using ELISA. The non-cross-reactive MAb 1H8 served as a
control to excludeinfection with RHDV (Fig. 2). All EBHS
samplesscoredpositivefor MAb 3H6. Nosignificantreactions
weredetected with hares 5 and9, whichwerediagnosed with pseudotuberculosis. MAb 6D6 as well as a polyclonal
anti-serum to RHDVscored EBHSsamplesexactlylike MAb 3H6 (data not shown). Together with the results obtained by
pathology and histopathology (data not shown), it can be concluded that acombination of the anti-RHDV MAbs 3D6 and 1H8 is suited to reliably detect cases of EBHS and to
discriminate between both viruses.
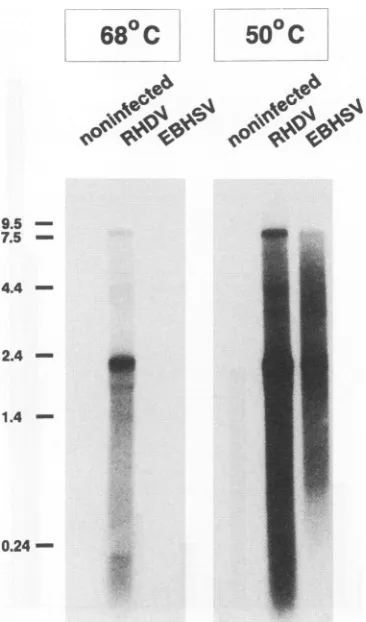
Demonstration of viral nucleic acid. To demonstrate the presence of viral RNA, Northern hybridizations were per-formed with RNAextracted from liver tissue ofan EBHSV-infected hare.Atlowstringency(50°C),twoRNAspeciesof 8 and 2.2 kbweredetectedbyacDNAprobecoveringthe 3' 5.5 kbof the RHDV genome(Fig. 3).ThelargerRNAcomigrated
with thegenomic RNAofRHDV.The smallerRNA species
was equal in length to the subgenomic RNA of RHDV. It should be notedthat,accordingtosequencedata,thegenomic
RNA of RHDV is about 7.5 kb in length notincluding the
poly(A)tail.Becauseof thepoly(A)sequenceof undetermined length and probably also the VPg, the RNA migrates more
slowly than the 7.5-kb marker. Athigh stringency(68°C),viral
RNAcould be detected inRNAextractedfrom the liver ofan
RHDV-infected rabbit butnotinRNApreparedfrom theliver ofanEBHSV-infected hare. Nosignalwasobtained withRNA
isolated from liver tissue of a control rabbit at either low
stringencyorhighstringency. Accordingly,EBHSV represents
avirus relatedtobutdistinct fromRHDV.
cDNA cloning and sequence analysis. Sequence
compari-sons should give more insight into the relationship between EBHSV and RHDV. Itwas,therefore,decided to clone and sequence the capsid protein gene of EBHSV. It has been shownfor differentcaliciviruses that theregion codingfor the
capsid protein is located within the 3' third of the genome(8,
16, 23, 27).Since the 3' end of the genomicRNAofRHDV
and other caliciviruses ispolyadenylated,oligo(dT)wasusedas aprimerforfirst-strand cDNAsynthesis.TotalRNAextracted from the liver ofan EBHSV-infected hare servedas starting
material for cDNA synthesis. A 2-kb large RHDV cDNA
fragment, which contains 95% of thecapsid proteingene and extends closetothe 3' end of the genome,wasusedas aprobe
on November 9, 2019 by guest
http://jvi.asm.org/
[image:3.612.324.564.95.177.2]RELATIONSHIP OF EBHSV TO RHDV AND OTHER CALICIVIRUSES 5167
OD
492 nm0
1 2 3 4
[image:4.612.95.535.84.415.2]samples
obtained
from hares
RHD
FIG. 2. Reactivity ofanti-RHDV MAbs with liversamples from hares. Liver homogenates from 14 hares were tested forreactivitywiththe anti-RHDV MAbs3H6 (left bar)and1H8(middle bar)in anELISA using a rabbit anti-RHDV hyperimmune serum to trap viral antigens. The liversamples were derived from hares infected with EBHSVexperimentally (samples 1 and 2) and naturally (samples 3, 4, 6 to 8, and 10 to 14) aswell asfrom haresdiagnosedwithpseudotuberculosis (samples 5and9). RHDV antigen and thefoot-and-mouth disease virus-specificMAb 075(right bar) servedascontrols.Theresultsrepresentthe meanopticaldensity(OD)values of fourreplicates obtainedat492 nm. Inthe field samplesobtained formhares,degradationof viral antigencannotbe ruled out and may have contributed to the low OD reading of sample 8.
forscreeningofacDNA libraryconstructedinpBluescript. A
positiveclone(pEB-1)witha1.4-kblargeinsertwasidentified.
Additional EBHSV-specific cDNAcloneswereobtained from asecond cDNA library constructed in LambdaZapll. Intotal, 2,806nucleotides from the 3' region of theEBHSV genome
weredeterminedfromfouroverlapping cDNAclones(Fig. 4). Nucleotidesequence comparisons revealed that thisregion is
homologous to nucleotides 4651 to 7437 of the RHDV
ge-nome. An alignment generated by the program Gap (gap
weight, 5; gap length weight, 0.3) showed that the two
se-quencesmatchat70.5% of thealigned positions.Theidentities withthenucleotidesequencesoftwoother caliciviruses,FCV (7)andNorwalk virus (17),are40.5 and42.1%, respectively.
Computeranalysis of thenucleotidesequence revealed two
open reading frames in theplus strand of thegenome. ORF1
extends from the 5' end ofthecDNAto nucleotide position
2378 and encodes a polypeptide of 794 amino acids with a calculated molecular massof84.9 kDa. ORF1 overlaps by8 nucleotides withan openreadingframe (ORF2) thatextends from nucleotide 2378 to 2719. ORF2 encodes a putative
polypeptide of 114 amino acids with a calculated molecular massof 12.4 kDa. The sequenceanalysisrevealedthree silent
nucleotide substitutions in ORF1, one silent variation in ORF2, andtwovariations in the noncodingregion.
Amino acid sequence comparisons showed that the 84.9-kDa protein exhibits 77% homology (identical amino acid
residues) to the corresponding region of the polyprotein
encoded in ORF1 ofRHDV.Thehomology isnotdistributed evenly along thepolypeptide (Fig. SAand B). There are two
regionsof veryhigh homology. The first region is locatedatthe
Nterminus of the 85-kDa protein (amino acids
[aa]
1to 110)and includes the amino acid motifs GLPGS, YGDD, and FKLR, which are conserved among RNA-dependent RNA
polymerases of
picornaviruses
(1). The second highlycon-served region (aa 285 to450) isseparatedfrom the firstby a morevariableregion and exhibitssimilaritytothe VP3capsid protein of picornaviruses (data not shown). Both conserved regions arevisible in adot matrixcomparison with FCV and Norwalk virus (Fig. SC and D). For these comparisons, the
polypeptidesencoded in
ORF1
and ORF2 of FCV and Nor-walk virus, respectively, werefused to result in a contiguous polypeptide. The dot matrix comparison with FCV showed that the conservedregions arenotonthesamediagonal.
The second conserved region is shifted along the horizontal axis.VOL.68,1994
on November 9, 2019 by guest
http://jvi.asm.org/
5168 WIRBLICH ET AL.
68°
C
50o
C
,
.,._.,,..._..
*CZNGO49i>4 4NX 4>
9.5 -7.5
-4.4
-2.4
-0.24
0,4:.,a
FIG. 3. Demonstration of viral RNAby Northernblot
hybridiza-tion.Twomicrogramseachof RNAs extracted from liver tissue ofan EBHSV-infectedhare, an RHDV-infectedrabbit, and a control rabbit werehybridizedtoradiolabeled RHDV cDNA at 68 and50°C.RNA ladder sizes(inkilobases) are indicated on the left.
Pol. Capsid
/
~~~~
aa.
> CL~FC
J.,~~~~~~~~~~~a
RHDV
IPal.
[ CapsidIn U~~~~~~~~~~~~~~~
z /~~~~~~~~~~~~~~IC
'U /
..I~~~~~~~~~~~~a
FCV
In I
m
lU
In I co
Pol. I Capsid ]
//'
Ia.
RHDV
Pol. Capsid
]
/
-NV
FIG. 5. Dot matrix comparison between the 85-kDaprotein en-coded in ORF1 of EBHSV and the corresponding polypeptides of RHDV(AandB),FCV(C),and Norwalkvirus(NV)(D).InpanelA awindow size of 20wasused ata stringency of 24. In panel B the stringency was increasedto29. For thecomparison with FCV (C) and NV (D), a window size of 20was used at a stringency of 13. The polypeptides encoded inORF1 and ORF2of FCV andNV, respec-tively, were fused to produce a contiguous amino acid sequence. Vertical and horizontal lines indicate the boundary between the polymerase(Pol.)gene(ORF1 in FCV andNV)and thecapsidgene (ORF2inFCV andNV).InpanelC thepositionof the N terminus of thematurecapsidproteinof FCV is also indicatedbyavertical line.
Thisshift is dueto anamino acid stretch in the FCV sequence that is not present in thepolyprotein of EBHSV. The addi-tional amino acids in the FCV sequence correspond to the N-terminalregion of thecapsid protein precursor of FCV; this region is cleaved off to generate the maturecapsid protein (8).
SacIl XhoI SmaISmaI
1 400 800 1200
4
SmaI Sacd
1600 2000 2400 2806
pEB-3
[image:5.612.323.555.69.325.2]pEB-4
FIG. 4. Thephysical mapof the analyzed region of theEBHSV
genome is shown at the top. Locations of EBHSV-specific cDNA
clonesare shownbelow. Solidbars, sequenced regions of thecDNA clones. Sequenceswereobtained from both strands of pEB-1,pEB-2,
andpEB-4 andfrom the antisense strand of pEB-3 as indicated by arrows.
When EBHSV was compared with Norwalk virus, the two
conservedregionswerealsoclearly detectable, but in thiscase
theyarelocated onthe samediagonal.
An open reading frame that corresponds to ORF2 of EBHSV is present in the genomes of allcaliciviruses that have been examinedtodate. This openreadingframeoverlapsthe
codingregion of thecapsid protein and extends closetothe 3' end of the genome. An amino acid sequence comparison revealed71% identity between the putative 12.4-kDa protein and the corresponding 117-aa polypeptide of RHDV. The homologytothecorresponding polypeptides of FCV, SMSV, and Norwalk virus is less than30%.
Amino acid comparisons and putative N terminus of the capsid protein of EBHSV. A recentanalysis suggested that at
leastamajor portion of thematurecapsid protein ofRHDV starts with the first methionine encoded by the subgenomic
RNA of RHDV (32). The amino acid sequence comparison shown in Fig. 6 reveals that the methionine residue is
con-served at the corresponding position in the polyprotein
en-coded in ORF1 of EBHSV(amino acid position219). Initia-tion at this methionine would give rise to a protein with a
calculated molecular mass of 60.05 kDa, which is in good
agreement with the apparent molecular weight of the capsid protein. Initiation at the following potential start codon lo-cated at
position
244(nucleotides730to732) wouldgive riseto a protein with a molecular mass of 57.6 kDa. Both start
codonsareflankedby purinesatthe -3 and +4positions and
5- I I I I I I I I II I I I
---30
11
J. VIROL.
I
on November 9, 2019 by guest
http://jvi.asm.org/
[image:5.612.91.274.70.381.2] [image:5.612.64.305.534.660.2]RELATIONSHIP OF EBHSV TO RHDV AND OTHER CALICIVIRUSES 5169
a.)
1000 1200 1400 1600 1800 2000 2200 2343
l l l l l l
+ I Capsid protein
Fusion protein H
1332 1796
Fusion protein G
1332 1727 179(16
Fusion protein I
Anti-I Anti-H Anti-G
!EBHSV RHDV|EBHSV RHDV+EBHSV RHDV
+I + -_ - + + i- + +
200
-
9771
-2343
b.)
151 DERGVQLEELQIHAAAHGEEFFELVKKELRRQQAFTRFSVFDYQTARKTL 200 1698 EERGVQLEELQVAAAAHGQEFFNFVCRELERQQAYTQFSVYSYDAARKIL 1747
end offusion protein G 0L
201 GDRKRIVSVVPDDSFVNVJEGKPRA....DAPGTATTASVPGTTTDGMDP 246
1748 ADRKRVVSVVPDDEFVNVKEGKARAAPQGEAAGTATTASVPGTTTDGMDP 1797
endof fusionprotein H-0-D
FIG. 6. Putative initiation site on the subgenomic RNA of EBHSV. (a) Location of the capsid protein in the RHDV polyprotein and regions ofthepolyprotein that are covered by fusion proteins H, G, andI. The location of theconservedGDD motif ofthe putative RNA polymerase (asterisk) and amino acid positions are indicated. (b) Amino acidalignment between EBHSV(upper sequence) and RHDV (lower sequence)that covers theregionaround the suggested amino terminusof the capsid protein of RHDV. Methionine residues are boldfaced. The putative amino terminus of the capsid protein of RHDVcorresponds to themethionineatposition1766. Fusion protein Hextendstotheaspartic acidatposition1796(arrow).Fusion protein G extendstothe leucineatposition 1727(arrow),andfusionprotein I covers aa 1796 to 2343. Amino acid numbering of the RHDV polypeptide refers to thepublishedsequence (24).
are, therefore, inafavorable context for translation initiation (18).InordertomaptheNterminusof the capsid protein of EBHSV,aWesternblot analysiswasperformedwith antisera
against fusion proteins containing different parts of the
polyproteinencoded in the 3' regionofORF1 of RHDV(Fig.
6and7).Antiserum against fusionproteinHthat includes the first 31 amino acids of the RHDV capsidprotein specifically
reacted withaprotein of about 60 kDa inaliverhomogenate of an EBHSV-infected hare. This protein migrated slightly
more slowly than the capsid protein of RHDV. The 60-kDa
proteinwasalsorecognized by antiseraagainstfusionprotein
I, which contains the C-terminal 58-kDa large part of the RHDV capsid protein but not by antisera against fusion protein G, which lacks the 70 C-terminal amino acids of the
RHDVpartoffusionproteinH. Nosignalwasobtained with liver extracts from control animals. Thus, the cross-reactive 60-kDaproteinrepresentsthecapsidproteinofEBHSV. The
RHDV part of fusion protein H extends from the putative
RNApolymerase into the capsid protein and ends after the second methionine encoded by the subgenomic RNA of
RHDV.Thefact that antiseraagainst fusion proteinH recog-nized the capsid protein of EBHSV shows that this fusion protein covers the N-terminal region of the mature capsid protein. Therefore, the AUG codonatposition 244cannotbe the initiation site at which capsid protein synthesis starts.
Moreover, the data show that the N terminus of the capsid proteinis located closetothemethionineatposition 219, since antiseraagainstfusionproteinG didnotrecognizethecapsid
4529
-
18-kDa
1 2 3 4 5 6 7 8 9 10 11 12
FIG. 7. Detection of the capsid protein of EBHSV with anti-RHDVantibodies. Clarifiedliverhomogenatesof a control hare(lanes 1, 5, and 9), and EBHS-positive hare (lanes 2, 6, and 10), an RHDV-infected rabbit(lanes 3,7,and11),and a controlrabbit (lanes 4, 8, and 12) were separated by SDS-PAGE and transferred to nitrocellulose. The membrane was then incubated with rabbit sera directed against fusionproteinG, H, or I(Fig. 6).
protein. There is no methionine between the C terminus of fusion protein G and the methionine at position 219. We,
therefore,conclude that this methioninemostlikelyrepresents the functional initiation codon at which translation of the
subgenomic RNAof EBHSVstarts.
Amultiple alignment between the capsidproteinsequences of RDHV, EBHSV, FCV, SMSV, Norwalk virus, and
Southamptonviruswasobtained with the programPileup(Fig.
8), and the percentage of identical amino acid residues was
calculated from this alignment for each pair of caliciviruses
(Table 2).The overall homology between the capsid proteins of RHDV and EBHSV is 76%. The homology to other caliciviruses isconsiderably lower,andtheNorwalk virus is the
most distantly related virus. Interestingly, the homology
be-tween RHDV and EBHSV is higher than the homology
between serotypes 1and4 ofSMSV(72%) and thehomology
between Norwalk virus and Southampton virus (72%) but
considerably lower than the homology between the F9 and
CFI/68strains of FCV (90%).Thereare atotal of 135 amino acid changes between the capsid proteins of EBHSV and
RHDVincludingadeletion of4 aa neartheNterminus of the EBHSV sequence andaninsertionof 1aa nearthe C terminus of the EBHSV sequence. Most of the changesare located in
theC-terminal half of thecapsid protein.TheN-terminalhalf
(aa219 to511) isconsiderablymoreconserved(Fig.SB). Only
44(32%) of the amino acidchangesarelocated in this part of the capsid protein, and 86% of the amino acid residuesare identical. The following region from position 512 to 648 exhibits the highest variability (64 amino acid changes; 55%
identity).Thisregionrepresents25%of thetotal sequence yet
accountsfor47% of the amino acid
changes.
Inthefollowing
C-terminal portionof the capsid protein (aa 645 to
794)
thehomology risesto 79%. VOL. 68,1994
on November 9, 2019 by guest
http://jvi.asm.org/
[image:6.612.340.526.71.299.2] [image:6.612.62.298.82.230.2]FCV-F9 NCSTCANVLK YY WD...D PHFKLVINP NNF...LS VGFCSNPMlC~CYPKLLPEFG TVHDCDRSPLEIYLESILGD DKXASTFDAV FCV-CFI/68 MCSTCANVLK YYDWD... PHIKLVINP NKF...LHVGFCDNIPLNC CYPKLLPKFG TNN1DCDQSPL QVYLKSILODDEWSSTHE.AI
SR4SV1 NATTHT.LLS FDDLEFLLHK KDLTDLYGER CGTLNLVINP YDLFLPDDDDDWCMDPFNC CFSDVYTSIG TEYSYIDPPD LIYEZHCATN OHNPDG.TPC SMSV4 MATTHT.LLS FDDLEFLLHIR KDLTDLYGER COTLNLVINP YELFLPDELD DDCCDDPFNC CFPDVYASIG TEYSYIDPPELIHZEHCATN GTwPNG.DPC SHV .. . . .. . . .. . . .. . . .. . . .
NV.. . . .. . . .. . . .. . . .. . . .
235 283
EBHSV... ...E"R... DJPGTATTLtS VPGTT`TDGWE PGVVAS.. .TDVVTRDNVAASVATAGICoGPP QQASPQESWR RHDV... KOKARAAPQG .ALAGTATTASVPGTTTLD0aM PGVVAT..TSVIThEESSA.SIATAIG1GPP QQVIDQQEITW
FCV-F9 DPVVPPMIIII AAKIFQPHP QVLWWHLIQKVA&OWDPDLP...LIRLEADDOS. ..ITAPEQGTMVQGVIAZPSAQKSTAADM& TGKBVDSEWI
FCV-CFI/68 DPVVPPNHND KAGKIFQPHP GVLKHNLCK VAZ0WDPNLP...LFRLZADDGS. ..ITTPEQGTMVOGVIAZPNAQMSTAADMA,TOKSVDSEWK SMSV1 EPILP?PFVII GTNIHYYATKP GEAVSGILSKLGSANDPDLQ STVDTKPDFVFRAZSDGPQGADIVTKZ&QGT VVQQQPVPAQSALTTLAAAS TGXTVDCZWT
SI4SV4 EPILPPFTIT GTIHYYATKP GZVVSGILSK LGSSWDPSLRSTIDNSNSFTFRAESDGPGS AEIVTKEQGT VVQQQPAPAPTAL&TL&TAS TGKSVEQENM SHV... ...1UM&4SKAPQ SADGASGAGQ LVPEVNTAkDP LPIMPV'AGPT TAVATAGQVN NV... ...DOE&MSKATS SlVDGA.SGAGQ LVPKVNASWDP LADPIVAGSS T&AVATAGQV Consensus --- ---D---2---GV---- ---A---T---W
Consensus - - - -- - -
-324 371
EBHSV VNFFY...NDVFTW SVTDAPOSIL YSVQHSPQNNPFTQVLSO8YAGWAGGMQFR FIVAGSGIFGORLVCAIIPP GIQIQPGLKVR....FPHVV
RHDV TNFYY...NDVFTW SVADAP0SIL YTVQHSPQNNPFTAVLSQ8Y A014AGGMQFRFIVAGSGVFG GRLVR&VIPPGIKIGPGLEVR....FPHVV
FC`V-F9 AFFSF...HTSVNW STSETQGKIL FKQSLGPLLNPYLEHLARLY VP.NS0SIKVR FSISGSGVFGGKIAAIVVPP GVDPVQSTSM L....YPHVL FCV-CFI/68 AFFSF...HTSVNW STSETQGKILFKQSLGPLLNPYLTHIAKLY VANSGSVDVRFSISGSGVFGGKLAAIVVPP GIDPVQSTSM L....YPHIVL
SMSV1 TFFSY...HTAVNW STTE.AQGKIL FSRALSPEI.N PYLRHIISSLY STNSOGIDVR FTVSGSGVFG GKLAALIVPP GIEPVESPTM L....YPHVL
SMSV4 TFFSY...HTSINW STVESQGKIL YSQ&LNPSINPYWDNIAKLY STNSGGIDVR FTVSGSGVFGGKLRALLVPP GV3PIKSVSM L....YPNVL SNV MIDPWIVNNF VQSPQOEFTI SPNNITPGDIL FDLQLGPHLN PFLSHLS(KYNGWVISNERVR ILLAGNAFSAGKIIVCCVPP GFTS.SSLTI AQ&TLFPRVI
NV PIDPVIINNF VQAPQGEFTI SPNNTPGDVLFDLSLGPHUNPFLIJHLSQKY NGVG&UMVRIMLAGNAFTAGKIIVSCIPP GFGS.HNLTI AQATLFPHVI Consensus --F---NS---G-IL ----L-P--N P-L-HL---Y --W-G---VR F---GSG-FOGKL----PP G---
-Q----PffV-Consensus ---S---G--L ----P--N
P---Y--W-0----R----G---0--G----PPG---Q----PNV-418 466
EBHSV IDARSLEPVT ITMPDLRPKM YHPTGDPGLVPTLVVSVYNN LINPFG...GTTSAIQVTVE TRPSZDFEFVLISAPSSKTV DSVNPSNLL. .TTPVLTGAG RHDV IDARSLZPVT ITHPDLRPNM YHPTGDPGLVPTLVLSVYNN LINPFG...G STSAIQVTVETRPSKDFEFV MIRAPSSKTVDSISPAGLL. .TTPVLTGVG
FCV-F9 FDARQVZPVI FCLPDLRSTL YHLUSDTD.TTSLVIMVYND LINPYAND.ANSSGCIVTVETKP0PDFKFH LLKPPGSMLTHGS!PSDLIP KTSSLWIG.. FCV-CFI/68 FDARQVEPVI FSIPDLRSTLYNLHSDTD.T TSLVIMVYND LINPYAND.SNSS0CIV-TV TKPGPDFKFHLLKPPGSMLTHI0SIPSDLIP KSSSUNIG.. SMSV1 FrDARQTEPVI FTIPDIRKTLYHNSDDTD.TTRLVIMVYNE LINPYEQS.ZPKSSCSITVE TRPSSDFPTFS LLKPPGSLLK HGSIPSDLIP RNSRHNMG..
SMSV4 FDASQTEPVI FTIPDIRKTLFHN8DKTD.T TKLV...INPYRNGVE NKTTCSITVETSPSADFTFALLKPPGSLIK H0SIPSDLIP RNSAHWflG..
SHV ADVRTLEPIZMPLKDVRNVLYHTND.NQPT MRLVCMLYTP LRT000SQNS DSFVVAGRVL TAPSSDFSFLFLVPPTIEQK TRAFTVPNIP....LQTL
NV ADVRTLDPIEVPLEDVRNVLFNNNDRNQQT MRLVCMLYTP LRTGGGTO.. DSFVVAGRVMTCPSPDFNFLFLVPPTVKQK TRPFTI.PNLP ....LSSL
Consensus -DAR--EPV- ---PD-R--LYH---T --LV---Y-- LINP---TVE T-PS-DF-F- -L-PP-S---P--L-P-- 0----Consensus -D-R---P----D-R--H---LV---V- T-P--DF-F- -P
---512 558
EBHSV SDNRW0APrV0GLQPVP.... GGFSTSNRHWNIQNGSTYO.WS SPRFDDIDHP SGNVSYPTGSATNTIETWYANAGTATTNPI SNIAPDGFPD,GAPF....
RHDV NDNRNN0QIV GLQPVP.... GGFSTCNRHN NLNGSTYGWS SPRFGDIDNR RGSASYSGSN ATNVLQFWYA N&GSAIDNPI SQVAPDGFPD 8SFVPF....
FCV-F9 ..NRYWSDTDrITR,,.... FVF.QANRNF DFNQKTAGWS TPRFRPISVT ITEQ...N0& KIGI.... 0.VATDYIVPGI.PDGWPD... FCV-CFI/68 ..NRFWSDIT DFVRP.... FVF.QANRNF DFNQKTAGWS TPRFRPITIT ISVK...ZSk KLGI.... O.VATDYIVPGI.PDOWPD...
SMSV1 ..NRNWSTID GFVQP.... RVF.QSNRNF DFDSTTTGNS TPYYIPIEVT LE.KWZRGGO YFKV.... .TDTEKSLVPGL.PD0WPD...
SMSV4 ..NRNNSTIS GFSVQP.... RVF.QSNRHF DFDSTTTGWS TPYYVPIEIK TQ0KV0SNNKWFHV.... T.DTDKALV PGI.PDGWPD...
SHV SNSRFPSLTQ GS4ILSPDASQ VVQFQN0RCL .IDOQLLG.TTPATSGQLFRVRGKINQGARTUNLTEV....D0KPFXAF DSPAPVGFPDFO;KCDWNHMRI
NV SNSRAPLPIS SIGISPDNVQ SVQFQNGRCT . DGRLVG.TTPVSLSHVAK IRGTSN..GTVTNLTZL....DGTPFHPF ZOGPAPIGFPD LOGCOWN..I
Consensus --NR----T---P--- VF-Q-NRHN----T-GWS TP----I---
--PDG-PD---Consensus ---R----I----P---R-0---G---
--P-G-PD---625
EBHSV SGTTIPTGAN VGFOQYNNAS NGTPYVGTVQ AYELG...FANG...A PSSTRPVTTT ...TG&QLVA KSIYGVAIAQ RHDV NGP0IPAAGW VGFGAIWNSN SGAPNVTTVQ AYELO...FATG...A PGNLQPTTNT ...SGAQTVAKSIYAVVTGT FCV-F9 ..TTIP .GEL IPAGDYAITN GTGNDITTATGYDTADTI.. ..KNNTNFRGMYICGSL...OAK WGD.KKISNTAFITTATW.GDNNNKINPOI
FCV-CFI/68 ..TTIP.GKLVPV0DYAITNGTNNDITTAAQYDARTZI.. ..RNNTNFRGMYICGSL...QRA NGD.KKTSNTAFITTGTVDGA....KLIPSN
Sl4SV1 ..TTIP.TAM...TA SNGNYDYTVA HYRTT...NNGTHFK0 FYIMGNLTT. KVKGSDNL.. .GKTQQTSRT LFASVGNYKD Q..NTTNPTH
SMSV4 ..TTTP.DZT...K TNGNFSYG.ESYRAGSTTIK PNKNSTHFKG TYICOTLSTV EIPENDKQQI KTFAZKKKSQT MYVVTADFKD ...TIVKPQHI SNV SKTPNNTOSG DPMRtSVSVQT NVQGFVPHLG STOF...DKVFNHPTGDYTGTIEW...ISQPS
NV NMTQFGHSSQ T....YDVDT TPDTFVPHL0 S10k...NOT...0S0 NYVG.VLSW..T..SPPS
Consensus --T-TP---Y---0---T
---Consensus -- -- - --
-668 716
EBHSV NQSSAGIIFL...SKQAVSTPGVAATTYT....TTPGTPVAAP 105..NTPTM FS&VVSRTGDVN&0PGSVNGTQYGVGSQPLSVTLGLSLTN
RHDV AQNPAGLFVM...ASGITSTPNAQRITYT....TTPGTPAAAP VGK..NTPTM FASVVRRTGD VN&TAGSANG TQYGTGSQPL PVTIGLSLNN FCV-F9 TIDQSKIVVFGONHN...VGK KAQTSDDTLALLL0YTGIGZQRISTLPETG. R0G. .NNPIF YIOISIKLOYVTRS.TDVFN...SQILHTSRQLSLNN
FCV-CFI/68 TTDQTXIAVFQ)TH...ANKHVQTSMOTLRLLL0YT0GIGZ RISVLPZR0A RAGO..NNPIF NKNSIKLGYVIRS.IDVIN...SOILHTSRQLSLNH SM4SVI KITSNSLVVYDANNVSAATAKTTTWHSTMS NLGYVLVDIS RIATLPKAFT HOG..NFPVF FTNKTOIGHFDRAHTKCFN...SQL MTSQKLABNH SMSV4 KISPQKLVVY....FDGPEKDLTMSATLS PLGYTLVDEQRIATLPZAFrTQGG..NYPIF YVNKTKVGYFDRATTNCYN...SOIL MTSQRALZGN
SNV TPPGTDINLW KTPDYGSSLS QAANLAPPVFPPGFGEA... FVSAFPGPNNRSAPNDVPCL LPQKY.ITTNFV... ... SK PTMGDAKLLH
NV NPSGSQVDLWKTPNY0SSIT KATNLAPSVY PPGFGZV... FMdSKNPGP.. ..GKYNLPCLLPOZY.ISHLA... ...0MKPTV0KAALLN Consensus --- -GY---P---- -GO---N-P---N---SQ-L -T---
L-L--Consensus.--- ---P---P.--- ----
---765 794
EBNS9V YSSALQPGQF FVWQL.NFASGFNEVI3OITD GYFYAGTGKY S0KMDLTDLI D.VRPVG. .V RPNTSTLVFNLKOVATTGYS YV
ANDV YSSALNP0QFFVWQL.TFAS GFMZXOLSVD GYFrYAGTGKS TTLIDLTELI D.VRPVG. .P RPSKSTLVFN LGGTA.NGFS YV FCV-F9 Y..LLPPDSFAVYRAIDSNG SWFDTGTDSD GFSFVGVSGFG.KLEFP..LS.ASYNG. .I QLRKIRLASNIRSP14TKL..
FCV-CFI/68 Y. .LLSPDSFAVYRIIDSNG SWFDIGIDND GFSFVGVSSI G.KLEFP. .LT.ASYMG..I QLRKIRLASNIRSVKTKL..
SMSV1 Y. .TLPPDSLLVYRITDAAS SWFDLGINHD GFSYVGISTI P.ZDFP..LT.FNLNG..VQLAKVKI.KSKVKTSKTTI..
SMSV4 Y. .NLPPDSL AVYRITDSSS QNFDIGINHD GFSYVGLSDL PNDLSFP. .L T.STIMG..VQIARVALASAVAKETITA..
SNV YVDPDTNRNLGZFKL. .YPGGYLTCVPN...OGVGAGPQQLPLNGVF LFVSNVSRFY QLKPVGTASTARGRLGVRRT
NV YVDPDTGRNLGOFAR..YPD GFLTCVPN...GASSGPOOLPINGVIVFVSWVSRFY QLKPVGTASSARGRLOLRR.
Consensus Y---L-P-O---G---D G---G---L---0---- QL----
LAS---Consensus
Y---0---G---5170
on November 9, 2019 by guest
http://jvi.asm.org/
RELATIONSHIP OF EBHSV TO RHDV AND OTHER CALICIVIRUSES 5171 TABLE 2. Aminoacididentities of EBHSVwith
othercalicivirusesa
%Identicalresiduese
Virus
RHDV FCV F9 FCVCFI/68 SMSV-1 SMSV-4 SHV NV
EBHSV 76 26 25 27 25 20 19
RHDV 25 25 26 24 18 18
FCV F9 90 47 47 21 21
FCVCFI/68 47 47 21 20
SMSV-1 72 20 21
SMSV-4 19 21
SHV 72
aThe sequences are aligned in Fig. 8. SHV, Southampton virus; NV, Norwalk virus.
bCalculated with the program Distances (Genetics Computer Group program package).
One main difference between the N-terminal part and the C-terminal part withregardtoamino acid composition is that the former is more highly charged and contains a lower percentageofsmall amino acids (A, G, S, andT). Astriking feature of themultiplealignment was that more than 25% (14 of49) of the strictly conserved residues are proline residues. The capsidprotein ofRHDV and EBHSV contains a single cysteine residue that is not conserved. Disulfide bonds are, thus,not involved in maintenance of the tertiary structure of thecapsid protein.
DISCUSSION
The datapresented in this article show that EBHSV exhibits properties characteristic of known caliciviruses. The typical size, structure, and composition of the virions have already been noted(5).TheWestern blotanalysis demonstrated that EBHSVforms amajorcapsid proteinspecies of about 60 kDa that shares epitopeswith thecapsid proteinofRHDV. Cross-reaction between EBHSV andRHDVhas also been shown in other studies (5, 9). On the other hand, it is well known that there are antigenic differences between both viruses as dem-onstrated hereby the RHDV-specific MAbs (1H8 and 3H2).
Amino acid sequencecomparisonsrevealed thatnearly half of the total amino acidchanges between the capsid proteins of
RHDVand EBHSVareconcentrated in the centralportionof thecapsid protein sequence (positions 512to 648). With the
exception of a short highly conserved amino acid stretch
(PDGFPD [aa 547 to552), this partexhibitsnostrict
conser-vation between different caliciviruses. Incontrast, the preced-ing part (aa 298 to 511) of the capsid protein exhibits high
conservation in themultiple alignment,andmorethan80% of the amino acid residues that are strictly conserved among calicivirus capsid proteins are located in this region. It is reasonabletoassumethat thehighlyvariable central stretch of the capsid protein sequence is located at the surface of the virions whereas the conserved N-terminal half constitutes the interior of thevirions. SurprisinglytheN-terminal part of the
capsid proteincarriesanetnegative charge (pl= 4.6).Abasic character may have beenexpectedbecausethe interior part of
the capsid is assumed to interact with the nucleic acid and a high content of basic amino acids is a characteristic feature of manynucleicacid-binding proteins. Nevertheless many of the basic residues in the N-terminal part are strictly conserved in the multiple alignment of the capsid protein sequences. In contrast, there is no conservation of basic residues in the C-terminal portion, suggesting that the positive charge of the basic residues is essential for the function of the N-terminal partbut not for the function of the C-terminal part.
The capsid protein of EBHSV is encoded in the 3' region of the genome and is preceded by the putative RNA polymerase gene.Both genes are part of the same open reading frame. The location of the polymerase gene in front of the capsid protein gene is the same for all caliciviruses, but only EBHSV and RHDV encode both genes in a continuous open reading frame. Other known caliciviruses encode the capsid protein and thepolymerase in separate openreading frames (8, 17, 20, 26,27). It is clear from the analysis of the EBHSV genome and from the data reported for RHDV (24, 32) that the presenceof
a continuous open reading frame that includes the capsid protein gene and the genes coding for nonstructural proteins is
not aunique property ofaparticularvirus isolate.
RNA isolated from the liver of an EBHSV-infected hare contains two viral RNA species of 7.5 and 2.2 kb that are similar in size to the genomic and subgenomic RNAs of other caliciviruses. ThesubgenomicRNAthat isproduced by calici-viruses covers thecapsid protein gene and probably serves as
mRNAtosynthesize the capsid protein oraprecursorthereof that may be posttranslationally modified to generate the
mature protein (8, 23, 27, 32). The subgenomic RNA of EBHSVis identicalinsizetothesubgenomicRNAofRHDV, and the sizes of the capsid proteins of both viruses are very
similar. It is, therefore, likely that the subgenomic RNA of EBHSVfunctions as mRNAforcapsidprotein synthesis.
Translation of the genomic RNA of RHDV and EBHSV initially will give rise to a polyprotein precursor that
encom-passes the polymerase and the capsid protein. Production of the functional polymerase probably requires cleavage of this
polyprotein.Anobviousquestionis whether thecapsidprotein species generatedbythis cleavage is different from the
trans-lation product of the subgenomic RNA. The results of an
N-terminal amino acid sequence analysisof CNBrpeptidesof the RHDV capsid protein suggested that at least a major
fraction of the capsid protein ofRHDV begins with the first methionine encoded by thesubgenomic RNA (32). The data further suggest thatmostof thecapsidproteinisderived from thesubgenomic RNA, buttheydo notexclude the possibility
that a minor fraction of the capsid protein is generated by
proteolytic cleavage of the polymerase capsid polyprotein. Dependingonthe location of thecleavagesite, thisproteolytic
product may be similar in sizetothe translationproductof the
subgenomic RNA, but it is also possible that the cleavage
product isconsiderablydifferent insize. TheWesternblot did
not show a capsidprotein species that migrated moreslowly
than the majorband. This finding suggests that the assumed cleavage site is located downstream of the first methionine encoded bythesubgenomicRNA.
FIG. 8. Multiple alignmentof thecapsidproteinsequencesofEBHSV,RHDV(24),FCVstrainF9(8),FCVstrainCFI/68 (27),serotypes 1 and 4of SMSV (26), Southamptonvirus (SHV) (20), and Norwalk virus (NV) (17). Thealignment wasgeneratedwith the program Pileup
(Genetics Computer Groupprogrampackage)withagapweight settingof 2 andagaplengthweight settingof 0.06.Upperconsensussequence, amino acid residues conserved inatleastsix of thealignedsequences;lowerconsensussequence,amino acid residues conserved in allsequences. Amino acid differences between thecapsid proteins of EBHSV andRHDV (asterisks),gapsinserted in the sequencestomaximize homology
(dots), andamino acidpositionsinthe EBHSVsequenceareindicated. VOL.68, 1994
on November 9, 2019 by guest
http://jvi.asm.org/
Initiationatthefirst AUGencoded by thesubgenomicRNA
of RHDV would give rise to a protein with a calculated
molecular mass of 60.4 kDa. Initiation at the corresponding AUGin theEBHSVsequencewouldgiverisetoaproteinwith
a calculated molecular mass of 60.05 kDa. Remarkably, the
apparent molecularweight of thecapsidprotein of EBHSVis higher than the molecular weight of the capsid protein of
RHDV (Fig. 8). Atpresent,wedo notknowifthisunexpected differenceis dueto(i) different aminoacidcompositions of the capsid proteins, (ii) different proteolytic processing of the primarytranslation product, or (iii) other different posttrans-lational modifications. N-terminal amino acid sequence
anal-ysis of the capsid protein of EBHSV could help
clarify
this question.According to their genome organization and the
mecha-nisms that may be used to express the capsid protein gene,
caliciviruses can be divided into three groups. One group,
comprising RHDV and EBHSV, ischaracterized by the pres-ence of a continuous open reading frame that contains the
capsid protein gene together with the genes of the nonstruc-tural proteins. These viruses may use the genomic RNA in
addition to the subgenomic RNA to synthesize the capsid protein. The members of the second group, which includes
FCV and SMSV, generate the mature capsid protein by proteolyticprocessingofaprecursorthat is translated from the
subgenomic RNA. The N-terminal cleavage product of the
capsid proteinprecursorrepresents anadditional viralprotein
of unknown function that is not encoded in the genomes of
RHDV, EBHSV, and Norwalk virus. In the case ofNorwalk
virus, proteolytic processing may not be required to produce
the mature capsid protein (17), and the Norwalk virus is,
therefore,considered amemberof the thirdgroup of calicivi-ruses.This division is consistentwith the relationship between these viruses as revealed by the multiple alignment of their
capsidprotein sequences.
ACKNOWLEDGMENTS
Thisstudywassupported byagrantfrom the Consiglio Nazionale delle Ricerche(BTBS,92.01280.PF70)andagrantfrom theDeutsche
Forschungsgemeinschaft (Th298/3-1).
REFERENCES
1. Argos, P.,G. Kamer, M. Nicklin, and E. Wimmer. 1984.Similarity ingene organization and homology between proteins of animal
picornaviruses andaplant comovirussuggestcommonancestryof
thesevirus families. Nucleic AcidsRes. 12:7251-7267.
2. Benton, W.,and R. Davis. 1977. Screeninglambda GT
recombi-nant clones by hybridization to single plaques in situ. Science
196:180-182.
3. Biermann,U., andH. Krauss. 1991.Detection of virus in
connec-tion with "European brown hare syndrome" in Hesse, FRG. J.
Vet. Med.B 38:21-24.
4. Boga, J.A., M. S.Marin,R.Casais,M. Prieto, and F.Parra.1992.
Invitro translation ofasubgenomicmRNAfrompurified virions
of theSpanish field isolate AST/89 of rabbit hemorrhagic disease
virus(RHDV). VirusRes. 26:33-40.
4a.Capucci,L.,etal. Unpublisheddata.
5. Capucci, L., M. T. Scicluna, and A. Lavazza. 1991. Diagnosis of
viral haemorrhagic disease of rabbits and the European brown
haresyndrome. Rev. Sci. Tech. Off. Int.Epizoot 10(2):347-370. 6. Carmichael, G. C., and G. K. McMaster. 1980. The analysis of
nucleic acids in gels using glyoxal and acridineorange. Methods Enzymol. 65:380-391.
7. Carter,M. J.,I.D. Milton,J.Meanger, M. Bennett, R. M. Gaskell,
and P. C. Turner. 1992. The complete nucleotide sequenceofa
feline calicivirus.Virology 190:443-448.
8. Carter, M. J.,I.D.Milton, P. C. Turner,J.Meanger,M.Bennett, and R. M. Gaskell. 1992. Identification and sequence determina-tion of thecapsid protein gene offeline calicivirus. Arch. Virol. 122:223-235.
9. Chasey, D., M. Lucas, D. Westcott, and M. Williams. 1992. European brown hare syndrome in the UK.; a calicivirus related to but distinct from that of viral haemorrhagic disease in rabbits. Arch. Virol. 124:363-370.
10. Chirgwin, J. M., A. E. Przybyla, R. J. MacDonald, and W. J. Rutter. 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 18:5294-5299. 11. Devereux, J., P. Haeberli, and0. Smithies. 1984. A comprehensive
set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:6587-6601.
12. Eskens, U., and K. Volmer. 1989. Untersuchungen zurAtiologie
der Leberdystrophie des
Feldhasen
(Lepus europaeus pallas). Dtsch.Tierarztl.
Wochenschr. 96:433-472.13. Grunstein, M., and J. Wallis. 1979. Colony hybridization. Methods Enzymol. 68:379-389.
14. Henikoff, S. 1984. Unidirectional digestion of exonuclease III in DNA sequence analysis. Methods Enzymol. 155:156-165. 15. Henriksen, P., D. Gavier, and F. Elling. 1989. Acute necrotising
hepatitis in Danish farmed hares. Vet. Rec. 125:486-487. 16. Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992.
Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532.
17. Jiang, X., M. Wang, K.Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. 18. Kozak, M. 1987. An analysis of 5'-noncoding sequences from 699
vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8131. 19. Laemmli, U. K. 1970. Cleavage of structural proteins during the
assembly of the head of bacteriophage T4. Nature (London)
227:680-685.
20. Lambden, P. R., E.0.Caul, C.R.Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519.
21. Lavazza, A., and G. Vecchi. 1989. Osservazioni su alcuni episodi di mortalita nella lepre evidenziazione al microscopio elettronico di una particella virale. Nota preliminare. Estr. Sel. Vet. 30:461-468. 22. Meyers, G., N. Tautz, E. J. Dubovi, and H.-J. Thiel. 1991. Viral cytopathogenicity correlated with integration of ubiquitin-coding sequences. Virology 180:602-616.
23. Meyers, G., C. Wirblich, and H.-J. Thiel. 1991. Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology 184:677-689. 24. Meyers, G., C. Wirblich, and H.-J. Thiel. 1991. Rabbit hemor-rhagic disease virus. Molecular cloning and nucleotide sequencing of a calicivirus genome. Virology 184:664-676.
25. Neill, J. D. 1990. Nucleotide sequence of a region of the feline calicivirus genome that encodes picornavirus-like RNA-depen-dent RNA polymerase, cysteine protease and 2C polypeptides. Virus Res. 17:145-160.
26. Neill, J. D. 1992. Nucleotide sequence of the capsid protein gene of two serotypes of San Miguel sea lion virus: identification of conserved and non-conserved amino acid sequences among cali-civirus capsid proteins. Virus Res. 24:211-222.
27. Neill, J. D.,I.M. Reardon, and R. L. Heinrikson. 1991. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J. Virol. 65:5440-5447.
28. Nowotny, N., A. Fuchs, F. Schilcher, and G. Loupal. 1990. Zum Auftreten der rabbit haemorrhagic disease (RHD) inOsterreich.
I. Pathomorphologische und virologische Untersuchungen. Wien.
Tierarztl. Monschr. 77:19-23.
29. Nowotny, N., T. Steineck, F. Tataruch, F. Schilcher, and H.
Weissenbock. 1991. European brown hare syndrome
(EBHS)-Experimentelle Untersuchungen. Wien.Tierarztl.
Monschr. 78: 370-378.30. Ohlinger, V. F., B. Haas, G. Meyers, F. Weiland, and H.-J. Thiel. 1990. Identification and characterization of the virus causing rabbit hemorrhagic disease. J. Virol. 64:3331-3336.
31. Ohlinger, V. F., andL. Ronsholt. Unpublished data.
32. Parra, F., A. J. Boga, M. S.Marin,and R. Casais. 1993. The amino terminal sequence ofVP60from rabbit hemorrhagic disease virus
on November 9, 2019 by guest
http://jvi.asm.org/
RELATIONSHIP OF EBHSV TO RHDV AND OTHER CALICIVIRUSES 5173
supports itsputative subgenomic origin. Virus Res. 27:219-228. 33. Parra, F., andM.Prieto. 1990. Purification and characterization of
acalicivirusasthe causativeagentofalethalhemorrhagic disease
inrabbits. J. Virol. 64:4013-4015.
34. Rumenapf,T., G. Meyers, R. Stark, and H.-J. Thiel. 1989. Hog choleravirus-characterization of specific antiserum and identifi-cation ofcDNA clones. Virology 171:18-27.
35. Sambrook, S., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning:alaboratory manual, 2nd ed. Cold Spring Harbor
Labo-ratoryPress,Cold Spring Harbor, N.Y.
36. Sanger, F., S. Nicklen, andA. R.Coulson. 1977.DNAsequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA
74:5463-5467.
37. Schaffer, F. L. 1991.Caliciviridae. Arch. Virol. 1991(Suppl. 2):300-302.
38. Strebel, K., E. Beck, K. Strohmaier, and H. Schaller. 1986. Characterization offoot-and-mouth disease virusgene products with antisera against bacterially synthesized fusion proteins. J. Virol. 57:983-991.
39. Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci.
USA76:4350-4354. VOL. 68, 1994