0022-538X/88/020472-07$02.00/0
Copyright C) 1988,American Societyfor Microbiology
Mouse Mammary Tumor
Proviruses
from
a
T-Cell
Lymphoma
Are
Associated with the
Retroposon
LlMd
JAQUELIN P. DUDLEY
Department ofMicrobiology, UniversityofTexasatAustin, Austin, Texas 78712-1095 Received17July 1987/Accepted 4 November 1987
Four Charon 4A clones containing mouse mammary tumor virus(MMTV) proviruses and their cellular flanking sequences were obtained from partial EcoRI libraries ofa C57BL/6 T-cell lymphoma with both endogenous and newlyacquired MMTV proviruses. The cellularflanking sequencesof three of four MMTV proviruses contained DNA homologousto the 3' endofthe longinterspersed retroposon LlMd. Two of the three proviruses werenewly acquiredinthelymphoma DNA,and these MMTVproviruses appearedtobe5 kilobases downstream and in thesametranscriptional orientationastheLi sequence. The thirdproviruswas endogenous Mtv-9 and waslocated less than 500 base pairs from the 3' end of Li. Seven additional clones containingMMTVproviruseswereisolated frompartial MboI librariesofaB6 T-celllymphoma. Five of the sevenclones contained Li elementsinthe cellularDNAflankingMMTVDNA. At leasttwoclones(including onewiththe Mtv-8provirus)hadmultipleLicopiesflankingthe MMTVprovirus, andoneclone containeda single MMTV long terminal repeatdirectly integratedintoatruncatedLi sequence.Althoughthefrequencies ofBi and Li in random library clones were similar, only one MMTV-containing clone hybridized to the abundantrepetitive elementBi. These datasuggesta nonrandomassociation betweenMMTVandLlMd. The mouse mammary tumor virus(MMTV)isinvolvedin
the induction of breast cancer in mice (19) although the mechanism of this event is not well understood. High mammary cancer incidence strains, such as GR and C3H, have a breast cancer incidence of greater than
90%,
and these strains transmit virus (also called exogenous or milk-borne MMTV) through the milk to theiroffspring
(19). Low-incidence strains, suchas C57BL/6 (B6) and BALB/c, lack the milk-transmitted virus but harbor the endogenous MMTV proviralcopies
common to most inbred strains of mice (15, 36, 37). Mammary tumors from high tumor inci-dence strainscontain additional integrated copies ofMMTV DNA (3, 17).The MMTV provirus, like those of other retroviruses, structurally resemblestransposable elements(22, 27) which canactivate expression ofgenesin orneartheirintegration sites(21,33,34).IntheC3Hstrain, transcriptionofaunique geneknownasint-I isdetectablein mammary tumors which contain anMMTVprovirus within 20 kilobases (kb) of this gene(32, 33).Twootherloci have beenidentifiedas common MMTVintegration sites in BR6 (C57BL x RIII)and Czech II mammarytumors (14, 34, 35).
Some murine T-cell lymphomas also contain additional copies ofMMTV DNA(4, 6, 30). Severalfactorsdistinguish these lymphomas from MMTV-induced mammary tumors. Acquired MMTV proviruses in B6 T-cell tumors originate from endogenous MMTV DNAand are transcribed at high levels, although MMTVproteins and virus are undetectable (6, 7). Molecular cloning of integrated MMTV copies from B6 lymphomas reveals extensive restriction enzyme poly-morphism in the acquired MMTV proviruses (7), and in both GR and B6lymphomas,proviruses have deletions within the U3 region ofthe MMTV long terminal repeat (LTR; 25, 31; L. Hsu and J. P. Dudley, unpublished data). Additional MMTV proviruses in B6
lymphomas
map to at least two different chromosomes which lack the endogenous MMTV loci of this strain (7). No common regions of MMTV integration have been identified inlymphomas.
Retroviralintegrationappears to occurwithout specificity
for host sequences(43). Toinvestigatethenatureof MMTV integration sites in T-cell lymphomas further, molecular clones of MMTVproviruses and their cellular flanking DNA were characterized. Restriction mapping and hybridization analysesindicatedcommonregionsof cellular DNAflanking theproviruses. One suchregion wasthe repetitive retropo-sonLlMd(38, 40, 47). Full-length copies of this repeat are approximately 7 kb long and have two overlapping open readingframesof1,137and3,900basepairs (bp),similarto those found in retroviruses (26, 29). Within the mouse genome, the 3' end of Li isalmost 10 timesmoreabundant (85,000copies)than the 5' end(10,000copies) (26), indicat-ingthat mostLi elementsaretruncatedwith respecttotheir 5' ends(38,40). Li elementshavea structuresimilartothat of the I elements which control hybrid dysgenesis in Dro-sophilamelanogaster (10).
This papercharacterizesthe Li repeatslinkedto MMTV proviruses in B6 T-cell lymphomas. The frequency of Li copies is approximately three times that expected in ran-domly selected clones containing murine DNA. This fre-quencyis mademore striking bythe presenceofasingle Bi repeat (23, 24; the AluI sequence equivalent [18]) in the cellular DNAs of these clones. The data suggest that inte-gration ofMMTVproviruses is notrandom with respect to mouse DNA.
MATERIALS ANDMETHODS
Restriction enzyme digestions, nick translations, Southern blotting, and hybridization analysis. Restriction enzyme digestions were performed as recommended by the manu-facturer (New England BioLabs or Bethesda Research Lab-oratories). Proceduresfor Southern blotting (42), nick trans-lation, and hybridization analysis have been described previously (6).
Molecular clones. Isolation of the phage clones AACI 1, AACI2,AACI 6, and AACI 7 containingMMTVproviruses has been previously described (7). Each of the clones was derived from a partial EcoRI library of the B6 lymphoma P3Cwhich contains additional MMTVproviruses.Subclone
472
on November 10, 2019 by guest
http://jvi.asm.org/
MMTV PROVIRUSES ASSOCIATED WITH LlMd 473
p2-12 (probe A) was obtained by transferring the 1.5-kb EcoRI-HindIII fragment 5' to the AACI 1 provirus into plasmid vector pUC9 (44). Subclones of the Li repeat in M13mp phages were generously provided by M.Comer, C. Hutchison, and M. Edgell and transferred into pUC9 before DNA purification on CsCl gradients containing ethidium bromide. Clones containing the mouse repetitive sequence Bi (38) were obtained from D. Endean and 0. Smithies, University of Wisconsin,Madison. Mm3l contains a 1,050-bp HindIlI insert in pBR322, and Mm35 contains a 2,500-bp EcoRI-HindIIIinsert in pBR322 (23, 24). The pVC217 probe (a 1.35-kb EcoRI-to-BglII fragment immediately 3' to the Mtv-8 provirus provided by G. Peters) (36) and the GR-40 clone (20) (from N. Hynes) were used to confirm the presenceof Mtv-8 in AACI 14.
The phage clones AACI 8, AACI 9, AACI 10, AACI 11, AACI12, AACI 13, and AACI 14 were obtained by using the lambda vector EMBL3 (12). High-molecular-weight DNA from the cell line E102P4C(16) wasextracted as described previously(6),partially digested withMboI, and treatedwith calf intestinal alkaline phosphatase(25U/ml) before gradient fractionation. Approximately 15- to 20-kb DNA fragments wereprecipitated with ethanol for ligation to EMBL3 DNA (PromegaBiotech)doubly digested withEcoRI and BamHI. Ligated DNA was packaged by using Packagene extracts (PromegaBiotech). Packaged phage were plated at approx-imately 104 phage per 150-mm (diameter) petri dish and screened with an MMTV LTRprobe(28).
Restriction enzyme mapping. Mapping of the EMBL clones was accomplished by using partial digestions and Southern blotting before hybridization with probes for the endsof linearDNAfragments. This is amodification ofthe method ofSmith and Birnstiel (41). Maps obtained by this method were confirmed by subcloning and/or single and doubledigestions of recombinant phage DNA.
Dideoxynucleotide sequencing analysis. Nucleotide se-quencing analysiswasperformed by the method of Sangeret al. (39). Reactions were analyzed on 8% acrylamide buffer gradient gels containing7 M urea as described byBiggin et al. (1). Plasmid DNA (1 to 2 ,ug) purified by ethidium bromide-CsClgradient centrifugationwasincubated in0.2 N NaOH-0.2 mM EDTA at 22°C for 5 min, neutralized by addition of0.2 M ammonium acetate, pH 4.5, and precip-itated with ethanol. The plasmid was added to 25 ng of 17-merprimers from nucleotides1273to 1289ofthe MMTV LTR (22) or from nucleotides 4848 to 4864 ofthe LlMd sequenceof Loebetal. (26). Sampleswereheated for 3 min at
100°C,
cooled in icewaterfor10min,and thenincubated at37°C for5minbeforedideoxynucleotide sequencing.
With theseprimers,the sequencewasobtainedfrom both strands. Primerswere synthesized bytheOligonucleotide
Synthesis
Facilityatthe University ofTexasatAustin.
Determination of LI and B1 frequencies and statistical analysis.
Approximately
1,000 to1,500
phage
wereplated
onto 100-mm
(diameter)
dishes and transferred to nitrocel-lulose intriplicate. Eachset washybridized
to anLi, Bi,
or lambdaprobe. Onlyplaques whichhybridizedtothe lambda probewerescored. Nitrocellulose transfersweremarked to facilitatealignment of plaques whichhybridized
toboth theLi
andBi
probes.Thestatisticalanalysis
wasperformed
by
usingthe binomial expansion.
RESULTS
Acommon repetitiveelementin theflankingDNAsof three Charon4AclonescontainingMMTVproviruses.Anumber of
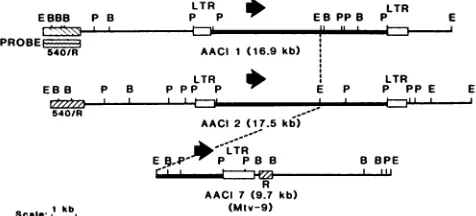
mouse T-cell lymphomas contain additional MMTV provi-ruses (4, 6, 30). Several proviruses and their adjacent flank-ing DNA sequences from a B6 T-cell lymphoma were cloned and characterized previously (7). Two of these proviruses from AACI 1 and AACI 2 were acquired in the lymphomas, whereas AACI 6 and AACI 7 contained the 3' ends of the endogenous Mtv-17 and Mtv-9 proviruses, respectively (7; unpublished data). To detect common regions within the AACI 1 and AACI 2 proviral integration sites, the cellular DNA flanking the MMTV proviruses was subcloned. The DNAimmediatelyadjacent to the provirus in clonesAACI1 and AACI 2 was found to be unique and did not cross-hybridize to the other clones (7; unpublished data). How-ever, a 1.5-kb EcoRI-HindIII fragment upstream of the AACI 1 provirus (Fig. 1; probe A) hybridized to two of the three other Charon 4A clones, AACI 2 and AACI 7 (con-tainingMtv-9; Fig. 2). The cross-reactive region inAACI 2 was localized to a 2.1-kbPstI-EcoRI fragment and, inAACI 7, to a PstI fragment of5.9 kb(Fig. 2, lanes 5 and 11). The AACI 6 clone (containing the 3' portion of the Mtv-17 provirus and approximately 13kb of flanking cellular DNA) didnothybridize toprobe A.
Hybridization analysis revealed that probe A contained repetitive sequences. Total B6 liver DNA was labeled by nicktranslationandhybridizedto Southern blotsofprobe A, AACI 1, and AACI 2 DNAs digested with HaeIII, EcoRI, BamHI,PstI, andHindlll. Results of this experiment (data notshown)indicatedthatprobe Acontainshighly repeated sequences. Moreover, the pattern ofhybridizationon AACI 1 andAACI 2 DNAs with probe A (Fig. 2) was identicalto thatobserved for totalcellular DNAprobes. Thisindicated that theonly repeatedsequences inAACI1andAACI2were homologous to the 1.5-kb probe Asubclone.
Therepetitive elementinAACI 1, AACI 2, and AACI 7 is part of the retroposon LlMd. The Charon 4A clones were tested for homology to a variety of cellular repetitive se-quences. Only probes specific for the 3' portion of the retroposon element LlMd(Bam540and R;2, 8, 28, 48;Fig. 3)hybridizedtoAACI 1,AACI2, andAACI7DNAs(Fig. 1). DNAsfromthethreeCharon4Aclones weredigestedwith theenzymesBamHI, EcoRI, and PstI, subjectedto South-ern analysis, and hybridized to a probe for the Bam540
LTR
E BBB P B P P
PROBE
540/R AACI 1(16.1
LTR
EB B P B P PP P
CtztA 540/R
LTR
EB PP B P E
_1 _1
.9k b)
Et P PLTRPPE E
AACI2(17.5kb)
,,-* LTR
E...P' P PB B B BPE R
AACI7(9.7kb)
Scale:.1kb, MtV-9)
FIG. 1. Restrictionmapsof Charon4AclonescontainingMMTV proviruses. The maps of these clones have been reported previ-ously;the sizeofthecloned insert is shown inparenthesesnext to
the clone number. The boxcontainingawavy line underAAC1 1 represents the1.5-kb EcoRI-HindIIIfragmentsubcloned intopUC9 (probe A). Open boxes represent the viral LTRs. Hatched boxes represent sequences which have homology to probe A. Large
arrowsindicate thetranscriptionalorientation of eachprovirus.The central EcoRl sitein eachprovirusisconnectedby adashed line. Restrictionenzyme sitesareabbreviatedasfollows: E, EcoRI;B, BamHI;P,Pstl.
VOL. 62, 1988
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.326.564.519.627.2]474 DUDLEY
12
3
4 5
6
7
8
9
10
11
12
kb kb
23.1
94-E
u:__
~~~94
-23.16.7 - 6.7
4.4 - 4.4
2.3-
20--W 4.*
- 2.3
- 2.0
FIG. 2. Southern blots of DNAs from clones AACI 1, AACI 2, andAACI7. Cloned DNA (1 ,ug)wasdigested singlyordoublywith
restriction enzymes andsubjected toelectrophoresis on0.8%
aga-rosegels before Southern transfer. BlotswerehybridizedtoprobeA
(Fig. 1). Lanes1, 4, 7, and 10 contain DNAsdigestedwithEcoRI, lanes 2, 5, 8, and 11 contain EcoRI-PstI-digested DNAs,and lanes
3, 6, 9, and 12 contain PstI-digested DNAs. Lane 1 contains two
bands, oneof which representsthe 5'EcoRI fragmentof AACI 1.
The smaller bandatapproximately 9 kbrepresentsadeletionwhich
accumulates inlarge-scale preparations ofthis clone. Similar faint
bands, whichrepresentdeletions ofAACI2,canbeseenin lanes 4 to6.
fragments in AACl 2. These results place LlMd approxi-mately 4.5 to 5.0 kb upstream of the MMTV proviruses in clones AACI 1 and AACI 2 (Fig. 1). If the positions of Bam540 and R correspondtothose observed in thesequence
of Loeb et al. (26), these results suggest that LlMd and MMTV arein the same transcriptional orientation.
Homology of the R probetothe AACI7 insert(containing Mtv-9) was localized to a 0.5-kb BamHI fragment within a
5.5-kb PstI fragment. Southern blotting experiments with ad-ditionalenzymesindicated that the Rregion is located just 3' to the Mtv-9provirus (datanot shown). If the Mtv-9 LTR is 1.3 kb, the Li R regionis withinafew hundred bases of theLTR. Isolation of additional MMTV clones from partial MboI libraries. Retroposon LlMd maps approximately 5 kb
up-stream and in the same orientation to two newly acquired MMTV proviruses locatedonseparatemousechromosomes
(7). The right-hand LTR of the endogenous provirus Mtv-9 also is flankedby the Li Rregion. Moreover, Fanningetal. have reported thatendogenous provirus Mtv-8is integrated into the 5' portion of the LlMd repeat (9). To confirm the high frequency of Li repeats in the cellular DNA flanking MMTV proviruses, seven additional clones were isolated from partial MboI libraries of B6 lymphoma DNA. Restric-tion maps of clones AACI 8, AACI 9, AACI 10, AACI 11,
AACI 12, AACI 13, and AACI 14 were constructed by
digestions with KpnI, SalI, and EcoRI, followed by
South-8 LTR K O E
a a
LTR a
region of LlMd (8, 26). This region includes the 3' portion of the3,900-bpopenreading frame within the7-kb repeat(Fig. 3). In both AACI 1 andAACI 2, Bam540 hybridization was
localizedtoaBamHIfragment of approximately 600 bp (Fig.
1). This BamHI fragment is located withina2.1-kb
EcoRI-PstIfragment in clones 1 and 2 and confirms hybridization dataobtained withprobe A (Fig. 2) and total cellular DNA probes. The Charon 4A clone AACI 7 (containingMtv-9)did nothybridize with the Bam540 probe.
Similar hybridizations were performed with an LlMd R
(26) probe. This region lies 3'totheBam540regionanddoes
notincludeopenreading framesequences(26; Fig. 3). These
experiments indicated that Li DNAwaslocalizedtoa1.4-kb
PstI-BamHI fragment in both clones AACI 1 and AACI 2. This homology was within 2.2-kb BamHI and 10.5-kb PstI
fragments in AACI 1 and 2.6-kb BamHI and 10.5-kb PstI
AACI (20.6kb)
kb-LTR KU K KU
8 LTR
AACI (17.0kb)
81 LTR K K a
AACI10(12.6 kb)
rkb
8LTRE E K SK E
AACI11(1I5.5 kb)
8 K 1 LTR E KE
.~~~~-(3
. O ' AACI 12 (16.3 kb)8 E EK E K K E E E LTRE8
180
AACI 13(13kb)
8LTR K l E
' I
-3
kb-LTR E E K
AACI 14(19.4 kb)
B B 840 B 4000
PROBE'-Bam Bam
200 840
Scale: kb
6540B B
RI Bam R
[image:3.612.67.283.75.244.2]1350 540
FIG. 3. Organization andconsensus map ofretroposon LlMd.
The open boxes above the restriction map represent the open
reading frames (orf) of LlMd described by Loeb et al. (26). The sizesofthese orfregions and the distances between restrictionsites
are given in base pairs. The regions of Li used as probes are
delineatedas thinlines under the restriction map. The 3' ends of
theseelementsoften contain A-rich regions. The restrictionenzyme
sites shown(EcoRI [E] and BamHI [B])aretheconsensusof several
different Li elements.
[image:3.612.314.553.355.609.2]l: kb
FIG. 4. Restriction enzyme maps of EMBL clones containing
MMTV proviruses. The openboxes represent MMTV LTRs, and the boldface arrows show the transcriptional orientation of the
proviruses. The smallerarrows indicate the approximate distance
between the viral LTRs and the closest Li elements. The size of
eachinsert isgivennext tothe clone designation and is flanked by Sallsites whicharecontributedby the EMBL3vector.The 540 and
Rregions of Li areshown ashatchedboxes, the 1350 regions are
filled boxes, the 840 region isadouble-hatched box, and the 200 region is a stippled box. A slash between Bam540 and R regions
(i.e.,540/R)indicates thatthe orderof theseelements isnotknown. Restrictionenzymesitesaregivenasfollows: E, EcoRI; K,KpnI;S, Sall.
1137 orf
I
E 2900
E E KE
I I a ..
3900orf
E 1350 E 3'
E E ES
_
,z-~
_.. . I
J. VIROL.
9 K KE A I.
5'
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.57.292.571.649.2]MMTV PROVIRUSES ASSOCIATED WITH LlMd 475
ernblotting and hybridizations with MMTV-specific probes (Fig. 4).
Four of the seven clones hybridized to probes for the 3' end of the Li repeat. In AACI 9, homology to the Li R regionwaslocalized toa2.2-kb SalI-KpnIfragment,
where-as hybridizations with AACI 11 DNA showed that the R
region is locatedon a1.2-kb EcoRI fragment withina2.1-kb
KpnIfragment (Fig. 4). Another clone, AACI 14,contained Li R sequences on 1.0- and 2.1-kb EcoRI fragments; the
homology was limited to a 10.5-kb KpnI fragment and a 6.4-kb SalI-KpnI fragment (Fig. 4). BamS40 and R region
probes gave identical hybridization patterns to Southern transfers ofeither AACI 9orAACI 11 DNA, suggesting that
Bam540 andRare linked within these clones (Fig. 4).
The EcoRI 1350 region (2, 26; Fig. 3) also was used in
Southern hybridization experiments. In AACI 11, the 1350
sequences werelocalizedto2.3- and 2.8-kbEcoRIfragments which did not contain BamS40 or R regions. Therefore, it
appearsthatthese regions areunlinkedtothe 1350sequence
in AACI 11 DNA and that they represent two distinct truncated copies of Li. In AACI 14 DNA, hybridizations with the 1350 probe indicate linkage of the 1350, Bam540, and R regions within a6.4-kb SaIl-KpnI fragment (Fig. 4).
Because the Rprobe also detects a 10.5-kb KpnI fragment
which contains the 3' half ofthe MMTVprovirus, the data suggest that there areat leasttwo truncated Li elements 3' totheprovirus in AACI 14. Restriction mapping and hybrid-ization analysis indicate that AACI 14 contains the
endoge-nous,Mtv-8provirus (20, 37; data not shown).
AAC1 13 was unusual, since the MMTV LTR and 1350
probes hybridized toan identical EcoRI fragment of 1.9 kb
(Fig. 4). No other MMTVorLlMd probes tested hybridized
tothisclone. Sequencinganalysis ofa1.9-kb subclone (Fig.
5) revealed that the MMTV LTR is inserted between nucle-otides 5033 and 5034 of the 3.9-kb Li open reading frame
described by Loeb et al. (26; Fig. 3). The data also indicate that theorientation of the 1350 element is oppositetothatof the LTR. Preliminary experiments indicate that the 1350
sequence continues uninterrupted on the other side of the
LTR(data notshown), suggesting that viral integration was
not accompanied by deletion of cellular DNA.
The MMTV provirus in AACI 10islinkedtoanLl 5' end.
All recombinant phage clones were hybridized to probes which detect the 5'ends of the LlMd element. Only AACI 10 DNAdemonstrated homologytothese probes. Homology of AACI 10 DNA to Bam840 was localized to a 0.8-kb
Sall-KpnI fragment withina2.3-kb KpnI fragment (Fig. 6A, lanes
4and 5), indicating that the Bam840sequences wereatone
endof the phage insert. TheAACI10 insert alsohybridized
to a 200-bp probe representing Li DNA immediately 5' to
Bam840 sequences (Fig. 3 and 6B). Since Li elements
usuallyaretruncatedattheir5'ends, these datasuggestthat the MMTV provirus in AACI 10 is in the same
transcrip-tional orientation and approximately 5 kb upstream of a
full-length LlMd element.
Restriction maps oftwo clones, AACI 8 and AACI 12, which contain no LlMd elements in their flanking DNAs
also are shown (Fig. 4). The MMTV provirus in AACI 12
lacks the virtually ubiquitous internal EcoRI site (15, 17). Representation ofother repetitivesequences in the MMTV proviralclones.BecausetheLi copynumberapproaches 105 (38, 40), the Charon 4A and EMBL clones were screened
with probes specific for other sequences ofequivalent or
greater copy number. One such element, the mouse Bi
sequence (38), has homology to and abUindance similar to
that of the humanAluIelements (18).DNAfrom each of the
LTR
5' CCCGGTGACCCTCAGGTCGGCGCACTGCGACATCTGGTGGAAT ---G
---A----*4- Li 1350
TTTTAGGGTCACTTATATATACTATCATATCATCTGCAAAAAG
____________________---G---TGATATTTTGACTTCTTCCTTTCCAATTTGTATCCCCTTGATC
---C--T---TCCTTTTGTTGTCTAATTGCTCTGGCTAGGACTTCAAGTACTA 3' ---G---AT---FIG. 5. Dideoxynucleotide sequencing analysis of the MMTV
LTR-LlMdjunction inAACI 13. Thetranscriptionalorientations of the MMTVLTRandLi sequences aregiven byarrows.Sequences included within the boxdelineate the 3' end of the MMTV LTR. Nucleotides different from the endogenous Mtv-8 sequence of Kennedy et al. (22) and the Li sequence of Loebet al. (26) are indicated byletters, whereas identical nucleotidesareindicatedby dashes.
clones was digested with EcoRI, subjected to Southern blotting, and hybridized to a Bi probe. Only AACI 6 (containingMtv-17) showed homology totheBi probe(Fig. 7, lane 3). Approximately 27% of random MboI clones hybridizedtotheBi probe,whereastheLiRprobedetected 24% of the total number ofclones. This is in good agreement with results reported by others for partial EcoRI libraries (26, 45). Thefrequency oflibraryclonescontaining bothLi andBi wasapproximately6%, suggesting arandom associ-ation
between
Li andBi.Statisticalanalysis. Theprobability ofobtaining linkageof Li and MMTV proviruses by chance was calculated. As-sumingthat thefrequency ofLi Ris24%inrandom clones (26, 45;seeabove;M. Edgell, personalcommunication),the
probability
of isolating6 of 11 MMTV-positiveclones withLi
Ris 0.028.DISCUSSION
The nature and location of LlMd sequences flanking severalMMTVproviruseshavebeen determinedby restric-tion mapping and hybridization analysis. Such analyses indicate that retroposon LlMdislocatedwithin7 kbofthe MMTV LTR in 8 of the 11 clones obtained from partial EcoRI or MboI libraries ofa B6 T-cell lymphoma. Ofthe eight clones containing LlMd, AACI 1 and AACI 2 con-tained
Li
approximately 5 kb upstream and in the same transcriptional orientationasthe MMTVprovirus, AACI 10 hadLi
approximately 5 kb downstream and in the same transcriptional orientation, and AACI 13 had a solitary MMTV LTR inserted directly into Li 1350in theopposite
transcriptional orientation.Threeotherclones contained Li flanking the provirus, but their orientation was not deter-mined. In two of these cases, AACI 11 and AACI 14 (containing Mtv-8),atleasttwoseparateLi elementsflanked MMTV DNA. Inviewofthelargenumberof these elements (about 85,000[26])inthemousegenome,itcanbecalculated (on thebasis ofa mousegenomesizeof3 x 109
bp [28])
thatLi
DNA is presentapproximately
every 35 kb.Excluding
MMTV
proviral
sequences,approximately
112 kb ofcellular DNAhas beencloned,and thusonewouldexpect3.2copies
VOL.62, 1988
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.324.550.76.137.2]of Li. The observed valie of 10 elements is higher than expectedin random mouse DNA. MMTVproviruses are ins,rted in nonrandoms DNA also is supported by the presence of element in cloned flanking DNA (23, 24). frequency of Li capture in random library clI higher (because of its larger size) thanthat of ] 490-bp region ofLi(R)also is representedin6 containing MMTV proviruses, and 1 clone (A two distinct R elements. The probability of 0.028, a value which does not account forthe multiple R elements in AACI 14, other porti MMTV flanking DNA, the distance of R fron provirus, or the presence ofa high
percentag
DNAin cloned inserts. This finding is inconsis idea that retroviruses integrate without specif DNA (43).
Nonrandom distribution of Li sequences ported in the genome ofrats (13). Several fa favorable for Li integration, including highA the presenceofthehexanucleotide TACTCA, ofalternating purine-pyrimidine residues (13). tion ofMMTV proviruses with Li, butnotB] consistent with the nbotion thatLielemnentsare dispersed in mammalian genomes.
Thetranscriptional activity of integration siti the distribution of MMTV and Li. LlMdistra discrete cytoplasmic polyadenylated
transcrip
detectable in B6 lymphomas with or with MMTV proviruses (5). In contrast to
cytor
scripts, a wide range of Li transcripts is pi nuclear RNAs ofthese and other murine cell probably representing readthrough transcriptic cent cellular promoters (47). Transcriptiona thoughttobe essential forLi dispersal (46, 47 oftranscriptionaredetectable fromacquired b
A
kb 1 2 3 4 5
31.0-* 12.5-_
10.0-B
kb 1 2 3
26.0-
.7
0
-0.8FIG. 6. Hybridization of AACI 10 DNAtoLi 5'l
DNA (1 Kg) was digested with restriction enzyme,
electrophoresis inagarose, and transferredtonitroc
wasdigested with EcoRI (lane1), EcoRIandSall(lar
3),SallandKpnI (lane 4),orKpnI (lane 5). DNAwa aprobefor the Li Bam840 region (panel A)orthe
(panel B).
considerably The factthat ites inmouse a single B1 Although the [ones may be Bi capture, a 5of 11 clones kACI 14) has this event is presence of ons of Li in
n the MMTV
ge
of MMTV itentwith the icity for hosthas been re-tctors appear + Tcontent, andastretch The associa-l, elements is nonrandomly
esmay affect .nscribed into
1 2 3 4 5 6 7 8 9 10 11 12
kb _ -23.1
- 9.4
_~ _-6.7
_4A
-2.3
- 2.0
FIG. 7. Hybridization of Charon 4A and EMBL clones to the mouse Bi probe. Cloned DNA (1 ,ug) was digested with EcoRI (lanes 1 to 4) or Sall (lanes 5 to 11) and subjected to Southern blotting afteragarosegelelectrophoresis.Thesourcesof DNAwere
asfollows(lanes): 1, AACI 1; 2,AACI 2; 3,AACI6;4,AACI 7; 5, AACI 8;6,AACI9; 7,AACI 10; 8, AACI 11; 9, AACI12; 10, AACI 13; 11,AACI 14; 12,50 ngofuncutMm31DNA.
)ts which are enous MMTV proviruses in B6 lymphomas, and no read-out acquired through transcripts between MMTV and LlMd have been
plasmic
tran- observed (7). MMTVtranscription is affected bytheproviral resent in the integration site(11).
types (5, 8), Both MMTV and
LlMd
are RNA polymerase II tran-rn fromadja-
scripts (38, 43, 47), whereas mouseBi
elements are tran-ilactivity
is scribed by RNA polymerase III (18, 47). In view of other ).High
levels datapresented here, retrotransposons, suchasMMTV, and ut notendog- retrotransposonlike elements(LlMd)
may integrate prefer-entially into regions of active RNApolymerase II transcrip-tion. This would explain the apparent nonrandom distribu-tion of LlMd elements and MMTV proviruses and the relativepaucity of murine Bisequences.Italsosuggeststhat 4 5 the regions of MMTV integration in T cells are actively transcribed in T cells. Other locicontaining active polymer-ase II transcription units contain multiple LlMd elements (46, 48). For example, eight LlMd elements have been described within thebeta-globin
locus (ca. 65 kb), yetonly
[image:5.612.359.516.80.269.2]one Bi element resides in the same region(45, 46). The endogenous provirus Mtv-8 (unit II) reportedly is integrated into the Bam840 region ofLlMd (9). Thisresult was not confirmed by hybridizations with Li 5' probes for Bam840 or the A type Li 200 sequence (Fig. 6). This -4.0
discrepancy
may indicate thepresence of several truncated Li copiesattheMtv-8integration site(Fig.4) andexplains why unique cellular flanking probes have been obtained from this region (36). Two other proviruses, Mtv-9 (in AACI 7) and an LTR-only provirus in AACI 13, also have Li se-quences adjacent tothe MMTV LTR. Since LTR-only and endogenous virus transcripts are not detectable in most T-celllymphoma
lines (7), it is possible, as suggested by Fanning et al. (9), thatLi
has a negative or damping effect probes.Cloned upon the transcription of MMTV proviruses within a-few s, subjected to hundred bases.ellulose. DNA ACKNOWLEDGMENTS
ie2),SaIl(lane
ishybridizedto This work was supported by Public Health Service grant CA
Li
200region 34780 from theNational Cancer Institute and by grant F-956 from theRobert Welch Foundation.on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.612.61.299.466.665.2]MMTV PROVIRUSES ASSOCIATED WITH LlMd 477 Iacknowledge AnnGlasgow andCharmaineFabritiusfor
techni-cal assistance and Henry Bose and BillFolk for helpful suggestions on the manuscript. I also thank Lilian Hsu for the sequencing analysis.
LITERATURECITED
1. Biggin, M. D., T. J. Gibson, and G. F. Hong. 1983. Buffer gradient gels and 3"S label as an aid to rapid DNA sequence determination. Proc. Natl. Acad. Sci. USA80:3963-3965. 2. Brown, S. D. M., and G. Dover. 1981. Organizationand
evolu-tionaryprogress of a dispersed repetitive family of sequences in widely separated rodentgenomes. J. Mol. Biol. 150:441-466. 3. Cohen, J. C., P.R. Shank, V. L. Morris, R. Cardiff, and H. E.
Varmus. 1979. Integration of the DNA of mouse mammary tumor virus invirus-infectednormal andneoplastictissue of the mouse. Cell 16:333-345.
4. Dekaban, G. A., and J. K. Ball. 1984. Integration of type B retroviral DNA in virus-induced primary murine thymic lym-phomas. J. Virol. 52:784-792.
5. Dudley, J. 1987. Discrete high molecular weight RNA tran-scribed from the long interspersed repetitive element LlMd. Nucleic Acids Res. 15:2581-2592.
6. Dudley, J., and R. Risser. 1984. Amplification and novel loca-tions of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J. Virol. 49:92-101.
7. Dudley, J. P., A. Arfsten, C.-L. L.Hsu,C.Kozak, and R. Risser. 1986. Molecular cloning and characterization of mouse mam-mary tumor proviruses from a T-cell lymphoma. J. Virol. 57: 385-388.
8. Fanning, T. G. 1982. Characterization of a highly repetitive family of DNA sequences in the mouse. Nucleic Acids Res. 10:5003-5013.
9. Fanning, T. G., D. W. Morris, R. D. Cardiff, and H. D. Bradshaw,Jr. 1985. Characterizationof anendogenous retrovi-rus-repetitive DNA chimera in the mouse genome. J. Virol. 53: 998-1000.
10. Fawcett, D. H., C. K. Lister, E. Kellett, and D. J. Finnegan. 1986. Transposable elements controlling I-Rhybrid dysgenesis in D. melanogasterare similartomammalian LINEs. Cell 47: 1007-1015.
11. Feinstein, S. C., S. R. Ross, and K. R. Yamamoto. 1982. Chromosomal positioneffects determinetranscriptional poten-tial of integrated mammary tumor virus DNA. J. Mol. Biol. 156:549-566.
12. Frischauf, A.-M., H.Lehrach,A.Poustka, and N.Murray.1983. Lambdareplacementvectors carrying polylinkersequences. J. Mol. Biol. 170:827-842.
13. Furano, A. V., C. C.Somerville, P. N. Tsichlis, and E. D'Am-brosio. 1986. Target sites for the transposition of rat long interspersed repeatedDNAelements(LINES)are notrandom. Nucleic Acids Res. 14:3717-3727.
14. Gallahan, D., and R. Callahan. 1987. Mammary tumorigenesis in feral mice: identification ofa new int locus in mouse mam-mary tumor virus (Czech II)-induced mammary tumors. J. Virol. 61:66-74.
15. Gray, D. A., E. C. M. L. Chan, J. I. Maclnnes, and V. L.
Morris. 1986. Restriction endonuclease map of endogenous mousemammarytumorvirus lociinGR, DBA, andNFSmice. Virology 148:237-242.
16. Green, P. L., W. W. Lamph, J. Dudley, A. Arfsten, R. Risser, L.L. Lanier, N. L.Warner, J.-S. Tung, and M. P. Scheid. 1985. Phenotypicvariation in clonal Abelson virus lymphomacells. J. Immunol. 134:1268-1275.
17. Groner, B., E. Buetti, H. Diggelmann, and N. E.Hynes. 1980. Characterization of endogenous and exogenous mouse
mam-mary tumor virus proviral DNA with site-specific molecular clones. J. Virol. 36:734-745.
18. Haynes, S. R., and W. R. Jelinek. 1981. Low molecularweight RNAs transcribed in vitro by RNA polymerase III from Alu-typedispersed repeats in Chinese hamster DNA arealso found in vivo. Proc. Natl. Acad. Sci. USA78:6130-6134.
19. Hilgers, J., and P. Bentvelzen. 1978. Interaction between viral
andgenetic factors in murine mammarycancer. Adv. Cancer Res. 26:143-195.
20. Hynes,N.E.,N.Kennedy,U.Rahmsdorf,and B.Groner. 1981. Hormoneresponsive expression ofanendogenous proviralgene of mouse mammary tumor virus aftermolecular cloning and gene transfer into cultured cells. Proc. Natl. Acad. Sci. USA 78:2038-2042.
21. Jenkins, N. A., N. G. Copeland,B. A. Taylor, and B. K. Lee. 1981.Dilute (d)coatcolour mutationof DBA/2J mice is associ-atedwith thesite ofintegration ofanecotropicMuLV genome. Nature(London)293:370-374.
22. Kennedy, N., G. Knedlitschek, B. Groner, N. E. Hynes, P. Herrlich, R. Michalides, and A. J. J. van Ooyen. 1982. Long terminalrepeatsofendogenousmouse mammary tumourvirus containalongopenreading frame whichextendsintoadjacent
sequences. Nature(London) 295:622-624.
23. Kramerov, D. A., A. A. Grigoryan, A. P. Ryskov, and G. P. Georgiev. 1979. Long double-strandedsequences(dsRNA-B) of nuclear pre-mRNA consist ofafew highlyabundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 6:697-713.
24. Krayev, A. S., D. A. Kramerov, K. G. Skryabin, A. P. Ryskov, A. A.Bayev, and G. P. Georgiev.1980. The nucleotide sequence ofthe ubiquitous repetitive DNA sequence Bicomplementary to the mostabundant class ofmouse fold-back RNA. Nucleic Acids Res. 8:1201-1215.
25. Kwon, B. S., and S. M. Weissman. 1984. Mouse mammary tumorvirus-relatedsequencesin mouse lymphocytes are induc-ible by 12-O-tetradecanoyl phorbol-13-acetate. J. Virol. 52: 1000-1004.
26. Loeb,D.D., R. W. Padgett, S. C. Hardies, W. R. Shehee, M. B. Comer, M. H. Edgell, and C. A. Hutchison III. 1986. The sequenceofalarge LlMd elementrevealsatandemlyrepeated 5' end and several features found in retrotransposons. Mol. Cell. Biol. 6:168-182.
27. Majors, J., and H. E. Varmus. 1981. Nucleotide sequencesat
host-proviral junctions for mouse mammary tumourvirus. Na-ture(London) 289:253-258.
28. Maniatis,T., E. F. Fritsch, and J. Sambrook. 1982. InMolecular cloning: alaboratory manual. Cold Spring HarborLaboratory, Cold Spring Harbor,N.Y.
29. Martin, S. L., C. F. Voliva, F. H. Burton, M. H. Edgell, and C. A.Hutchison III. 1984. A large interspersed repeat found in mouseDNA contains alongopenreading frame that evolvesas
if it encodes a protein. Proc. Natl. Acad. Sci. USA 81:2308-2312.
30. Michalides, R., E. Wagenaar, J. Hilkens, J. Hilgers, B.Groner, and N.E. Hynes. 1982. Acquisitionofproviral DNA ofmouse mammary tumor virusin thymic leukemiacells from GR mice. J.Virol. 43:819-829.
31. Michalides, R., E. Wagenaar, and P. Weijers. 1985. Rearrange-ments in the long terminal repeat ofextra mouse mammary tumorproviruses in T-cell leukemias ofmouse strain GR result in a novel enhancer-like structure. Mol. Cell. Biol. 5:823-830.
32. Nusse, R., A. van Ooyen, D. Cos, Y. K.Fung, and H. E. Varmus. 1984. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature (London) 307:131-136.
33. Nusse, R., and H. E. Varmus. 1982.Manytumorsinducedby the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31:99-109.
34. Peters, G., S.Brookes, R. Smith, and C. Dickson. 1983. Tumor-igenesis by mouse mammary tumor virus: evidence for a com-mon region for provirus integration in mammary tumors. Cell 33:369-377.
35. Peters, G., A. L. Lee, and C. Dickson. 1986. Concerted activa-tion of twopotential proto-oncogenes in carcinomas inducedby mouse mammary tumor virus. Nature (London) 320:628-631. 36. Peters, G., M. Placzek, S.Brookes,C. Kozak,R.Smith,and C.
Dickson. 1986. Characterization, chromosomeassignment, and segregation analysis of endogenous proviral units of mouse mammary tumor virus. J. Virol. 59:535-544.
VOL. 62, 1988
on November 10, 2019 by guest
http://jvi.asm.org/
37. Peterson, D. O., K.G. Kriz, J. E. March, and M. G. Toohey. 1985. Sequence organization and molecular cloning ofmouse
mammarytumor virus DNA endogenousto C57BL/6 mice. J.
Virol. 54:525-531.
38. Rogers, J. H.1985. The origin andevolution ofretroposons.Int. Rev. Cytol. 93:187-279.
39. Sanger, F., S. Nicklen,andA. R.Coulson. 1977. DNA
sequenc-ing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467.
40. Singer, M. F., and J. Skowronski. 1985. Making sense out of LINES: longinterspersedrepeat sequences inmammalian
ge-nomes.Trends Biochem. Sci. 10:119-122.
41. Smith,Ij. O., and M. L. Birnstiel. 1976. A simple method for
DNArestriction site mapping. Nucleic Acids Res. 3:2387-2398. 42. Southern, E. M. 1975. Detection of specific sequences among
DNAfragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517.
43. Varmus, H., and R. Swanstrom. 1985. Replication of
retrovi-ruses,p.83-93. In R. Weiss, N. Teich, H. Varmus, and J.Coffin
(ed.), Molecular biology oftumorviruses: RNAtumorviruses,
2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
44. Vieira, J., and J. Messing. 1982. The pUC plasmids, an
M13mp7-derivedsystemforinsertion mutagenesis and
sequenc-ing withsynthetic universal primers. Gene 19:259-268. 45. Voliva, C.F.,C. L. Jahn, M. B. Comer, C.A.HutchisonIII,and
M. H.Edgell. 1983.The LlMd longinterspersedrepeatfamily in
the mouse: almost all examples are truncated at one end.
Nucleic Acids Res. 11:8847-8859.
46. Voliva, C. F., S. L. Martin, C. A. Hutchison III, and M. H.
Edgell. 1984. The dispersal process associated with the Li family ofinterspersed repetitivesequences. J. Mol. Biol. 178:
795-813.
47. Weiner, A. M., P. L. Deininger, and A. Efstratiadis. 1986. Nonviral retroposons: genes, pseudogenes, and transposable elementsgenerated by thereverseflowofgenetic information.
Annu. Rev. Biochem. 55:631-661.
48. Wilson, R., and U. Storb. 1983. Association oftwo different repetitive DNA elements near immunoglobulin light chain
genes. Nucleic Acids Res. 11:1803-1817.