Copyright © 1976 American Society forMicrobiology PrintedinU.SA.
Reovirus-Specific
Polypeptides: Analysis
Using
Discontinuous
Gel
Eelectrophoresis
RISE K. CROSS' AND BERNARD N. FIELDS2*
DepartmentsofCell Biology and Medicine,Albert Einstein College of Medicine, Bronx,New York 10461
Received for publication3February 1976
The electrophoretic analysis of reovirus-specific polypeptides in infected cells usingadiscontinuous gelsystemhas allowed the resolution ofadditional viral-specific polypeptides, includingonelarge-sized (A3) andtwo (orpossibly three)
medium-sized [,u3, u4, ,5(?)] species. The proteins designated ,uO, o-1, and o2 based on electrophoretic mobility in gel systems containing phosphate-urea correspondto,4,o-2,ando-1,respectively, when analyzed insystemscontaining
Tris-glycine. Itislikely that protein modifications (phosphorylation and glyco-sylation)are responsible foratleastsomeof thesedifferences.
Reovirus provides an excellent system for studying several aspects of gene expression in eukaryotic cells. The genome of thisvirus con-sists of 10 segments of double-stranded RNA that are functionally as well as structurally discrete molecules (reviewed in reference 12). Each segment of genome RNAistranscribedin its entiretyby avirion-associated transcriptase (1, 3, 11, 18, 23, 26, 27). All 10single-stranded RNAtranscripts can bedetected onpolysomes from infected cells andserve asmRNAspecies for the synthesis of unique viruspolypeptides
(5, 19, 31).
In addition, several temperature-sensitive (ts) mutants have been isolated and placed in seven groups on the basis of recombination tests(6, 8; R. K. Cross andB. N. Fields, in H. Fraenkel-Conrat and R. R. Wagner(ed.), Com-prehensive Virology, in press). Each group of
mutantspresumably carriesa ts mutation on a different genome segment. Although the spe-cific segmentresponsiblefor eachtsmutation is not known with certainty, it should shortly become possible to correlate each mutation with a unique RNA segment and polypeptide
product (CrossandFields,inpress). The corre-lation of each mutant gene productwith a de-fect in the infectious cycle should provide in-sightintothe functionof eachreovirus
polypep-tide.
The following presents studies that were done to maximize resolution of reovirus-speci-fiedpolypeptides from infectedcellsbyutilizing
a high-resolution, discontinuous
polyacryl-'Present address: The Rockefeller University, New York, N.Y. 10021.
2Present address:DepartmentofMicrobiologyand
Mo-lecular Genetics, Harvard MedicalSchool, Boston, Mass. 02115.
amide gel electrophoresis system. A prelimi-nary report of most of these results has been presented (R. Cross and B. N. Fields, Abstr. Annu. Meet. Am. Soc. Microbiol. 1974, V21, p. 204). Thetechniques used in these studies were thenappliedto ananalysisofcytoplasmic poly-peptidesintsmutant-infectedcells (7). A simi-lar analysis of reovirus cytoplasmic polypep-tides has been recently reported (5).
MATERIALS AND METHODS
Cells and virus. Mouse L cells were grown in
suspensionculture inEagle minimal essential
me-dium (Joklik's modification, Grand Island
Biologi-calCo., Grand Island, N.Y.) supplemented with 5%
fetalcalf serum. The Dearing strain of reovirus type
3wasused.
Mode of infection and virus purification. Cells
wereinfected and virus was purified as previously
described (8, 28).
Labeling of infected cells. After virus adsorption, cells were diluted to 106 cells/ml in growth medium. At appropriate times, the infected cells were
concen-trated in medium containing 5% of the normal
amountof methionine (0.05x methionine) and
high-specific-activity [35S]methionine (>200 Ci/mmol,
New England Nuclear Corp. or Amersham/Searle). The specific labeling conditions of each experiment
aredescribed in detail in the figure legends.
In pulse-chase experiments, infected cells were
concentratedto107cells/ml ingrowthmedium
con-taining 0.05x methionine and 50 ,uCi of
[35S]methionine per ml (final specific activity of
about 10 Ci/mmol) and pulsed as described in the
figure legends. Chases were subsequently carried
out by dilutingtheinfected cells 10-fold ingrowth
medium containing four times the normal concen-trationof methionine(final specific activityofabout
10Ci/mmol). These chase conditionsprevented
fur-ther incorporation of label into acid-precipitable
counts.
Harvesting of infected cells and preparation of
162
on November 10, 2019 by guest
http://jvi.asm.org/
cytoplasmic extracts. Infected cells in suspension
cultures wereharvested by centrifugation for 5 min
at1,500 rpm inanInternational (PR-J) centrifuge at
4C and then washed twice with cold Earle saline.
The cell pellet was resuspended at a concentration of
1 X 107 cells/ml in an isotonic saltsolution (0.14 M
NaCl-0.01 M Tris-hydrochloride (pH 7.4)-0.0015 M MgCl2) containing 0.5% Nonidet P-40 (Shell
Chemi-calCo.) and placed on ice for 15 to 30 min until the
cellsweredisrupted (4). The extract wasthen cen-trifuged at 2,000 rpm for 2 min at 4 C in an Interna-tional (PR-J) centrifuge to remove nuclei, and the
supernatant(cytoplasmic)fraction was saved.
Processing of cytoplasmic extracts for polyacryl-amide gel analysis of radiolabeled polypeptides. (i) Acetone precipitation. Four to 8 volumes of cold
acetone wereadded to radiolabeled cytoplasmic
ex-tracts, and the mixture was kept at -20 C
over-night. The precipitate was collected by
centrifuga-tionat10,000rpmfor 20 min in aSorvall RC-5
cen-trifuge, washed one to three times with cold
ace-tone, airdried, and dissolved in gel sample buffer.
(ii) Immune precipitation. The procedure
de-scribed by Fields et al. (9) was used for immune
precipitation. A 10- to30-,ul amount of reovirus type
3 hemagglutination inhibition chicken antiserum
(prepared by hyperimmunizing chickens with a
sus-pension ofmonkey kidney cells infected with the
Dearing strain of reovirus type 3, Grand Island
Bio-logical Co.) was added to 0.5 ml of a cytoplasmic
extractand allowed to incubate at4C for 30 min. To
this mixture 0.1 to 0.2 ml of rabbit anti-chicken
globulin (Miles Laboratories Inc.) was added and
wasfollowed by incubation for 18 to24h at4C.The
immuneprecipitate wasthencollected by
centrifu-gation at 2,000rpm for 10min at4 Cinan
Interna-tional (PR-J) centrifuge, washed three times with
cold phosphate-buffered saline, pH 7.0, and
dis-solved ingelsamplebuffer.
Polyacrylamide gel electrophoresis. (i) Ten
per-cent SDS-polyacrylamide gels in a discontinuous
Tris-glycine buffer system. This procedure is
de-scribed byMaizel (20). The resolving gel was
pre-pared to a final concentration of 10% acrylamide,
0.267%bisacrylamide, 0.05%TEMED
(N,N,N',NT"-tetramethylethylenediamine), and 0.1%sodium
do-decyl sulfate (SDS; PierceChemical Co.; sequanol
grade)inabuffer containing0.19M Tris-hydrochlo-ride, pH 8.8. Polymerization was catalyzed with
0.06% ammonium persulfate. The electrophoresis
buffer consisted of0.05MTris-glycine (Fisher
Scien-tificCo., G-46),pH 8.8, and0.1%SDS.Sampleswere
resuspendedin a solution containing 0.05 M
Tris-hydrochloride, pH 8, 1% SDS, 1% 2-mercaptoetha-nol, 10% glycerol, and 0.001% phenol red (Grand Island Biological Co.) and boiled for2 minpriorto
electrophoresis. Aliquots (10to20,ul)wereapplied
tosample wells,andelectrophoresiswascarriedout
asdescribedinthefigure legendson aslabgel
appa-ratus.After electrophoresis, gelslabswerefixed and
stained for 1hin a 50%methanol solution
contain-ing0.2%Coomassie blue(Schwarz/Mann) towhich a
final concentration of7% aceticacidwasadded
be-foreuse. Gelsweredestainedelectrophoreticallyin
afreshly prepared solution of7%aceticacid and5%
methanol for 15 to 30 min in aCanalco quick gel
destainer. Gels were then dried and
autoradiogra-phy was performed. Dried gels wereexposed to
Ko-dak BB-54 blue brand medical X-ray film.
Microden-sitometer tracings of autoradiograms were
per-formed on aJoyce, Loebl and Co. double-beam
re-cording densitometer.
When gels with acrylamide concentrations other than 10% were prepared, the concentrations of
acrylamide andbisacrylamidewereadjusted
accord-ingly, and the same proportions of the other
re-agents wereutilized. Gradient gels were prepared as
describedpreviously (2).
(ii) Ten percent SDS-polyacrylamide gels
con-taining urea. The composition of the 10% SDS-poly-acrylamide gels containing urea is identical to that
described by Zweerink et al. (34). Electrophoresis
wascarried out onslab gels at 60Vfor 14to 16 h.
Afterelectrophoresis, slabs were fixed, stained,
de-stained, and prepared for autoradiography as
de-scribed above.
RESULTS
Electrophoresis of viral polypeptides on
SDS-polyacrylamide gels in a phosphate buffersystemcontainingurea.Zweerinketal. (34) reported that nine cytoplasmic viral poly-peptide species could be resolved from cells in-fectedat31 or37 C by electrophoresison poly-acrylamide gels containing SDS, phosphate buffer, and urea. As shown in Fig. 1, when polypeptides from cells infected at 39 C with wild type wereprocessed byelectrophoresis on
slabgelsinthismanner, identical resultswere
obtained; two A polypeptides, three ,t polypep-tides, and four o polypeptides were resolved. Sincepolypeptide,u2is acleavageproduct of,1 (33, 34), thissystemis capable ofresolving only 8of thepresumed 10geneproductsof the dou-ble-stranded RNAgenome.
Electrophoresis of viral polypeptides on
SDS-polyacrylamide gels in a discontinuous Tris-glycine buffer system. Electrophoresis of reovirus polypeptides from infected cells was
nextcarried out onpolyacrylamideslabgelsin adiscontinuousTris-glycinebuffersystem
con-tainingSDS. This gelsystemis knownto
per-mit a higher degree of resolution of polypep-tides than the phosphate buffer system (20). Reovirus polypeptides are clustered in three sizegroups (A, ,u, andvT)andrangein molecu-lar weights from about 155,000 to 34,000 (28). Thus, several concentrations of acrylamide were tested to obtain the best conditions for maximallyresolvingall size classes simultane-ously.Some concentrations ofacrylamide (12to
15%) that wereparticularly effective in resolv-ing the small-sized (Cr) polypeptides were less able toresolvethe ,t speciesandvirtually
una-ble to resolve the A species, whereas linear
on November 10, 2019 by guest
http://jvi.asm.org/
)U2
X2
ILO LI
a2A
O1I
FIG. 1. Analysis of viral-specific polypeptides from infected cells by electrophoresis on a 10%
polyacryl-amide gel containing urea. A totalof107infected cells wasconcentrated in 1 ml ofgrowthmedium containing
0.05x methionine andpulsedwith50 XCiof[35S]methionine from5 to5.5hpostinfectionat39 C. Viral
polypeptides wereprecipitatedfroma cytoplasmicextract byimmune seraandresuspendeddirectly ingel
sample buffer for gelelectrophoresisasdescribed in the text.Electrophoresiswascarriedout on a 10%
SDS-polyacrylamide slab gel containing6M ureafor14h at 60 V.The directionofelectrophoresisisfromleft to
right onthis and subsequent gels. A microdensitometer tracing of a gelautoradiogram is shown.
gradientgels withconcentrationsranging from 5 or 7.5 to 20% had the reverse effect. Two concentrations werefound thatallowed the best resolution of all species: 10%gels and7 to 12% linear gradient gels.
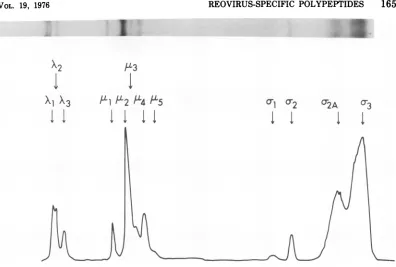
When viral polypeptides from infected cells labeled withP[5S]methionineduring the time of maximal viral protein synthesis at 39C were immunoprecipitated from cytoplasmic extracts and electrophoresed on 10%Tris-glycine gels,
additional reovirus-specific polypeptides were detected (Fig.2).Threelarge-sized (X), five me-dium-sized (,), and four small-sized
(cr)
viral polypeptides were resolved. The polypeptides within a sizeclass have beennumbered accord-ingto their decreasing molecular weight. The two largest of the polypeptides, Xl and X2, mi-grate as a tightly coupled doublet, whereas X3, which runsslightly ahead, isusually theleast abundant ofthelarge polypeptides. Ofthe five mediumspecies, two are thesame size ascapsid polypeptides ,ul andA,2,andthree,g3,
u4, andA,5,
whichmigrateahead of thesepolypeptides,
are not found in purified virions (see below). Polypeptide A,5 is a minor polypeptide that is often difficult to resolve clearly from
AL4,
the mostabundant of thesefaster-migrating ,u spe-cies. Polypeptides o-1,o-2,
ando-3arethe samesize as the- corresponding polypeptides
ex-tracted fromwhole virions. A degreeof
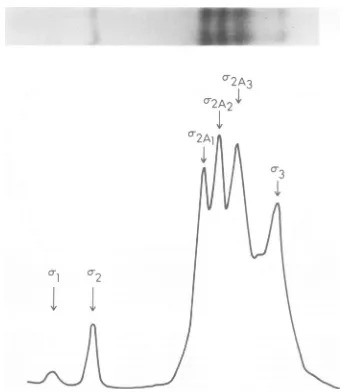
hetero-geneity has been observed in the polypeptide designated o2A, which can be resolved into three distinct species designated o-2A1, oc2Aa, and o-2A3 (Fig. 3). It isprobable, however,that this heterogeneity reflects lability of the o-2A polypeptide ordegradation of otherlarge poly-peptides since these species are more consist-ently observed inolder preparations.
All of these species were observed when in-fected cellular extracts were treated with im-mune sera, as described above. Since it had been shown previously that thisimmune sera
selectively precipitatesreovirus-specified poly-peptides (9),this methodwasutilizedtoensure that any new polypeptides detected on Tris-glycine gels were, in fact, reovirus specific. None of these polypeptides wasobserved when uninfectedcells were similarlyprocessed.
To verify that artifacts were not introduced by immunological precipitation, cytoplasmic extractsof infected cellswerealso examinedby analternate method.Acetoneprecipitationwas carried out on cytoplasmic extracts of cells
in-fected at 31 Cinthe presence ofactinomycin D andlabeledlate in the infection (at which times normal cellular proteins are sufficiently re-duced to allow clear distinction between viral and cellular polypeptides). The results shown in Fig. 4 clearly demonstrate that the same polypeptides observed when immunological
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.505.118.427.56.285.2]165
I
U
113
P-1
/1L2 kiL4
kL5
I I 1 I
Cr1]
2
52A
I
I
I
FIG. 2. Electrophoresis of immunologicallyprecipitatedviralpolypeptidesonTris-glycine gels.Atotalof
107infected cellswasconcentratedin1 mlofgrowthmediumcontaining0.05xmethionine andpulsedwith
50 pgCi of[35S]methionine from5 to5.5 hpostinfection at39 C. Viralpolypeptides wereprecipitated from
cytoplasmicextractsbyimmuneseraandprocessed for gel electrophoresisasdescribed in thetext.
Electropho-resiswascarriedouton a10%polyacrylamideslab gel for13hat50V.Theautoradiogramofthisgel andits
microdensitometertracingareshown.
precipitation is employed can be detected by this alternative approach. Furthermore, it is apparentthatthesepolypeptidesarepresent in reovirus-infected cellsincubatedateither 39or 31 C. Acetone precipitation, however, is less effective at 39C, where there remains an
ap-preciable background of hostcell protein
syn-thesis (9). A cellular protein (not shown) that migrates slightly faster than o-2A is often de-tected, however, in acetone precipitates of in-fected and uninin-fected cell extracts prepared from actinomycin D-treated cells incubated at
31 C.
Comparison of the phosphate-urea and
Tris-glycine gelsystems. (i)Evidence for dif-ferentmigrationof the, polypeptides. Using thephosphate-urea gelsystem,Zweerinketal. (34)resolved three ,L polypeptides, the largest ofwhich, uO, wasfound only in infected cell
extracts(Fig. 1). Nopolypeptides migratingin
the position of the ,O polypeptide can be
de-tected on Tris-glycine gels. However, three nonstructural polypeptides, ,3, ,u4, and ,u5, which migratefaster thanpolypeptides ,p1and
,u2,areresolvedonTris-glycine gels.This find-ingsuggestedthatoneor moreof thesespecies mightbeidentical tothe ,O polypeptide.
To test this hypothesis, thefollowing obser-vation was utilized. After treatment of reovi-rus-infected cellswith cycloheximide at a con-centrationsufficient to allow synthesis of only the four"early" species ofreovirus mRNA (32), followedbyremoval ofthedrugtopermit
trans-lation, only four viral polypeptides, X2, ,O, o2A, and o-3, are synthesized (15). Thus, since
,uOis the onlymedium-sized polypeptide
syn-thesizedunder theseconditions, its positionon
Tris-glycine gels couldbe directly determined. Accordingly, the technique of cycloheximide
treatmentand releasewascarriedout,and the
polypeptidesweresynthesizedafterremoval of the drug analyzed on Tris-glycine gels. As shown in Fig. 5, polypeptide u4 was the only medium-sized polypeptide synthesized under these conditions. The presence ofX2 and o3 is consistent with previous findings. Polypeptide a2A wasnotresolved from a-3 onthisgel, but other experiments have clearly confirmed its synthesis after release from cycloheximide. On phosphate-urea (not shown), the X2, ,LO, and o(3 polypeptides could be discerned as previ-ously reported. Thisfindingconfirms the iden-tity of ,uO with the ,u4polypeptideseenon Tris-glycine gels. No conclusioncould be drawnon
X1
X3
I
I
cr3
I
VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.505.53.449.55.322.2]. |
_F;-2
Fr3
Is_M
1,2A
_.:cr2A1
T.3
[image:5.505.101.450.64.456.2]O0i
4,
FIG. 3. Autoradiogram and microdensitometer tracing of the small-sized (o-) polypeptide region. A total of
107cells infected at 31 C was pulsed from 12 to 14 h postinfection with 50 ,uCi of [35S]methionine. A
cytoplasmic extract was then prepared, treated with immune sera, and processed for gel electrophoresis as described in the text. Electrophoresis was carried out on a 13% polyacrylamide gel for 24 h at 60 V.
the identity of,u3 and ,u5 since these polypep-tides were not detected underthe condition of infection and labeling used in these experi-ments. These experimentscanalsonotexclude the possibility that ,u3 and p5 might also mi-gratewith the uO peakinthephosphate-urea system under conditions in which cyclohexi-mide isnot presentand all viral polypeptides aresynthesized.
(ii) Evidence for different migrationof the c- polypeptides. A comparison ofpolypeptides
extracted frompurifiedvirions and
electropho-resed onTris-glycine gels withthose analyzed
onphosphate-urea gels revealed differences in
thepatternof thea-regionpolypeptides. Figure
6B shows the a- region as resolved on phos-phate-ureagels,inwhich thepolypeptideshave beenpreviously designated as a-1 to a-3 on the basis of their migration in this system (28). Polypeptide a-i, which has the slowest migra-tion of the a-polypeptidesinthisgelsystem, is aminorcomponent. OnTris-glycine gels (Fig. 6A), this minor component migrates between the other two structural a- polypeptides and, accordingly, it has been designated a-2,
whereas the otherremainingpolypeptideshave been designated al and a-3.
To further confirm this finding, advantage
on November 10, 2019 by guest
http://jvi.asm.org/
s1~~~~~~1
/A2 [image:6.505.52.447.54.305.2]ii
£
FIG. 4. Electrophoresis of acetone-precipitated polypeptides fromacytoplasmicextractofinfectedcells. A
total of5 x 106cells infectedfor16 h at31 C in thepresence of0.25 pgof actinomycin Dperml was
concentratedto5 x106cellslml andpulsedwith 100 Xtiof[3'5S]methioninepermlforIh ingrowthmedium
containing0.05xmethionine. Viralpolypeptideswereprecipitated fromacytoplasmicextractbyacetoneand
processedforelectrophoresisasdescribed in thetext.Electrophoresiswas on a10%polyacrylamideslabgel for
15hat45V.Theautoradiogramandmicrodensitometertracing ofthisgelareshown.
A. >12
l~~~~~~~O O
FIG. 5. Viral polypeptidessynthesized aftercycloheximide release. A totalofas 6cellslmlwasinfectedwith
100PFUlcellinthe presence of 20 0gofcycloheximidepermlat31 C.Cycloheximidewas washedout18h
postinfection, and105the cells were concentrated to cellslml and pulsed for 2 h with 200mei of o5S]methionineperml.Cytoplasmicextractswereprepared,treated with immunesera,andprocessed forgel
electrophoresisasdescribed inthetext.Electrophoresiswas on a10%polyacrylamideslabgelfor16hat45V.
Microdensitometertracingsof gelautoradiogramsareshown. (A) Controlpatternofreovirus-specific
cyto-plasmic polypeptides electrophoresed inparallel with (B); (B) viral polypeptide species synthesized after
releasefrom cycloheximide.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.505.109.392.392.584.2]168
A. 3 B.
FIG. 6. Microdensitometer tracings of
autoradi-ogramsof the small-sized(a-) polypeptides from
puri-fied virions. (A) Electrophoresison 10%Tris-glycine
gel for 16 h at 45 V; (B) electrophoresis on 10%
phosphate-urea gels for 16 h at 60 V.a-3istruncated
toallow greater sensitivity of theal- and a-2
polypep-tides.
wastaken ofthe fact thatonlyone o- polypep-tide (designateda-2onphosphategels)isfound in viralcoresprepared by chymotrypsin diges-tion. Coreswerepreparedandthepolypeptides found in cores were compared with those of wholevirions and the top component (particles lacking RNA) onTris-glycine gels.
As previously reported, the polypeptides in the top component are identical with those of virions (28). Eightpolypeptides, Xl,X2, X3, ,ul, ,u2,
a-1,
o-2, anda-3,
are found in thesestruc-tures (Fig. 7A andB). PolypeptideX3 was
pre-viously unresolved from the other two large
species (28). Thepolypeptide migrating ahead of ,u2 inthe virus preparation appearstobe a
degradationproduct that increases in amount
during storage. The origin of the additional species between theX and upolypeptidesseen
inthistop componentpreparationisunknown butmostlikelyrepresentsbreakdownproducts
of the X species. Cores contained
polypeptides
XA,X2, X3, andacl (Fig. 7C).Thesmallamount
ofut2representsaminoramountofundigested
material. The finding of
a-1
in viral cores con-firms theidentityof thepolypeptidedesignated a-i inthe Tris-glycine system with that desig-nated a-2 on phosphate gels. It also demon-strates the presence of all three X species incores. When identical preparations of these
polypeptides were electrophoresed on
phos-phate-ureagels (notshown),cores were, as
ex-pected,foundtocontainXi, X2,X3,ando-2. The
appearance of X3 inboth cores aswell asvirus
top components was also observed on these
gels.
Pulse-chaseexperiments. (i) Comparisonof phosphate-urea and Tris-glycine gels. Zweer-ink et al. (34) reported that polypeptide ,u2 is derived from ,ulby cleavage. Since this analy-sis wascarried out on phosphate-urea gels, it was ofinteresttoexamine this relationshipon
A2
A]
XIE
C
3 2I
A_3
'
FIG. 7. Resolution of virus, top component, and
corepolypeptidesonTris-glycine gels.Atotalof109
cells was infected and labeled for 24 h in growth
medium containing1
gCi
of [35S]methionine per ml.Cells wereharvested, and virus(a- = 1.36) and the
topcomponent(a- =1.29)werepurified as described
in the text. Cores were prepared by the treatment of
whole virus in1 xSSC (0.15 MNaCl plus 0.015 M
sodium citrate) for 60 min at37C with 100 Mgof
chymotrypsin perml.Thereaction mixture was then
centrifugedon apreformedCsCl gradient (a=1.3to
1.5g/cm3)in anSW41 rotor at 24,000 rpm for 16h. A
visible bandof cores (a = 1.42) was collected.An
aliquot ofeachof these preparations was pelleted by
centrifugation in an SW41 rotor for 1 h at24,000
rpm, resuspended in gelbuffer, and boiled for1min.
Electrophoresis was carriedout at 45 Vfor17.5h.
Microdensitometer tracings of gel autoradiograms
areshown. (A) Virionpolypeptides; (B) top
compo-nentpolypeptides; (C)corepolypeptides.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.505.68.261.62.301.2] [image:7.505.274.460.215.476.2]Tris-glycine gels where polypeptide ,u is clearly separated from the nonstructural ,i spe-cies (,u4). In anticipationof studying these poly-peptides in ts mutant-infected cells at 39 C, immunological precipitation was employed. Cells infected with the wild-type strain were labeled for 15 min with [35S]methionine at 5 h postinfection and then chased. Cytoplasmic ex-tractsof samples taken at the beginning of the chase and 120 min thereafter were treated with immune sera, and precipitated polypeptides were subjected to gel electrophoresis (Fig. 8).
When pulse-chase experiments were carried outusing immunological precipitation, the rel-ative amounts of all of the polypeptides
changed during the chase, although the total incorporation of radioisotope into acid-precipi-table material remained constant. Examina-tion of the viral polypeptides on Tris-glycine
gelsrevealed an increaseintheXl, ,u2, cr1, and a3 polypeptides during the chase, whereas
polypeptides ,ul, ,u3 to ,u5, o-2, and or2A
de-creased. Likewise, when identical aliquots were electrophoresed on phosphate-urea gels, the Xl,
u2,
cr2, and or3polypeptides increased,whereas polypeptides ,uO,
,tl,
cr1, andoa2A de-creased. The parallel reduction in polypeptides ,uO and ,u4aswellasthereversebehavior ofcr1
andor2onthe two gel systems is consistent with the differences in migration of these species already noted.
These results suggested that, since no fur-therincorporation ofradiolabel occurred during the chase, the changesobserved must have re-flected a differentialselection by immune sera of viralpolypeptides from the total pool of la-beled polypeptides. Particularly striking was the finding of a substantial decrease in the precipitation of nonstructural viral polypep-tides 3top5(MO) and cr2Abyantiseraduring the chase, such that after 120 min the polypep-tide pattern more closelyresembled that found in mature virions. Although a decrease in
Al
and an increasein,2 wereobserved,no conclu-sion regarding the relationship of these twopolypeptidescould be drawn from these experi-ments.
(ii)Comparison of precipitation by immune sera or acetone. To insure that there was, in fact, no changeinthetotalpoolofradiolabeled
A
FIG. 8. Pulse-chase experiment. Infected cells were concentrated to107cellslmlingrowth medium
contain-ing 0.05x methionineandpulsed with[35S]methioninefrom 5 to 5.25 h postinfectionat 39C. The cellswere
thendilutedto a concentrationoflO6cellslml in growth medium containing4xmethionine (see text).Aliquots
oflO7cellsweretakenout at0(A andC) and120 min(B andD) during the chase.Cytoplasmicextracts were prepared, treated with immune sera, and processed for gel electrophoresis as described in the text. Electropho-resis wascarriedout in 10%polyacrylamide gels in a Tris-glycine buffer for 13 hat50V(A and B)or in a
phosphate-urea buffer for14hat 60 V(C and D). Microdensitometer tracings ofautoradiograms of these gels
areshown.
19,
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.505.58.447.336.581.2]170
polypeptides during the chase, similar experi-ments were carried out in whichidentical sam-ples were precipitated either immunologically orby acetone. These experiments were carried outat31 C to ensurethat host protein synthe-sis wasadequately reduced. When cytoplasmic extracts wereprecipitated by immune sera and electrophoresed on Tris-glycine gels, a striking decrease in the nonstructural polypeptides
1A
and oa2Aagain was observedduring the chase (Fig. 9A and B). In addition, adecrease ingl
and an increase in ,u2 were also seen. The additional changesinamountsof viral polypep-tide observed during the chase at 39 C were not observed.
In contrast to the findings with immunologi-cal precipitation, when aliquots of identical samples were precipitated by acetone, there was nochangeinthe amounts of any
polypep-A.
B.
tides during the chase, except for a decreasein
,uland an increase in
pL2,
confirming the pre-cursor-product relationship of these polypep-tides (Fig. 9C and D). It is unclear at thistime why there was a greater decrease in the ul polypeptide than the corresponding increase in the ,u2 species. Whether this reflects inefficient cleavage of the ,ul polypeptide, resulting in smaller degradation products, turnover of the ,u3 polypeptide that isunresolved from the ,u2 speciesinthis experiment, or achangein the specific activity of this polypeptide after cleav-age hasnotbeendetermined.The results when immunological precipita-tion was employed were quite unexpected. Three possible explanations appear most likely. (i) The hyperimmune serum utilized in these experiments containsantibodies with different affinities for each of the viralpolypeptides. The
C.
M14
X1/X2
D.
FL.
lI
"II
II
II
II II
O'2A
II
K
FIG. 9. Comparison ofacetone and immunological precipitates during pulse-chase experiments. Cells
infectedat31 Cinthepresenceof0.25 ,ugof actinomycinDpermlwereconcentratedto107cellsingrowth
medium containing 0.05x methionine andpulsed with [35S]methionine (50 pCiIml) for 5 min at18 h
postinfection. Cellswerediluted10-fold ingrowthmediumcontaining4x methionine,andaliquotsof107
cellsweretakenat0(A andC)and60min(BandD) duringthechase.Cytoplasmicextractsofthesecellswere
dividedintoequal aliquots, andpolypeptides wereprecipitatedbyimmunesera(AandB)orbyacetone(C
andD) asdescribedin thetext.Electrophoresis wascarriedoutin10%Tris-glycine gels for16hat45 V.
Microdensitometer tracingsof gelautoradiogramsareshown.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.505.149.383.280.579.2]immunereactionfor certainpolypeptides (such
as ,u4and a-2A) maythus be weaker andmore sensitivetochangesinthe totalpool of polypep-tides during the chase. This possibility is sup-ported by the fact that the precipitation of ,u4 and o-2A (both nonstructural polypeptides) is greatlydiminished by thepresenceof 0.5% de-oxycholate (an ionicdetergent) (Cross, unpub-lished data).Immunologicalprecipitationof the otherviralpolypeptidesisvirtuallyunchanged by thistreatment. (ii) During the chase there may be a selective and rapid increase in the pools of the ,4Aand oa2Apolypeptides. The con-ditions utilized, thus,may nolongerbeoptimal forimmuneprecipitation. At these suboptimal
conditions, less precipitation would occur and the relative amounts ofprecipitated polypep-tides wouldnot represent theiractual amount in the cytoplasm. (iii) It is also possible that during thechase the antigenicity of the u4or a-2Apolypeptides is selectively altered through configurational changes or interactions with other proteins such that these species are no longerprecipitated immunologically.
In viewof these apparent difficulties, pulse-chase experiments with ts mutants at 39C (which requires this technique) were not pur-sued. However, immune precipitationwas uti-lized to study the nature of the polypeptides synthesized by these mutants at the nonper-missive temperature,bearinginmindthat cau-tion mustbe taken in quantitating individual species (7).
DISCUSSION
Zweerink et al. (34) detected nine reovirus
polypeptidespeciesininfected cells using
poly-acrylamide gel electrophoresis with 10%
SDS-polyacrylamide gelsinaphosphate-ureabuffer system. Since one of these species (,ul) is a
precursor ofanother (,u2), only 8 of the pre-sumptive 10 primary gene products were
re-solved. Accordingly, an attempt was madeto
resolve additional gene products by utilizing
electrophoresis of radiolabeled virus-specific
polypeptides on SDS-polyacrylamide gels in a
discontinuous Tris-glycine buffer, asystem re-puted to allow a higher degree of resolution (20).
Using this gel system, several virus-specific
polypeptides have been demonstrated in cells infected at 31 and 39 C. Three large-sized spe-cies (Xl, X2, and X3) can be resolved; the two largest polypeptides (Xl and X2) migrate as a tight doublet, whereas the leastabundant spe-cies, X3, migrates slightly ahead. It seems rea-sonable that these polypeptides correspond to
thegeneproductsof the threelarge (L)
double-stranded RNA segments. Five medium-sized polypeptides (,ul, ,u2, ,u3, u4,and ,u5)have also been detected. Polypeptide ,u2 is a cleavage product of,ul,confirming thedata of Zweerink et al. (34) in the phosphate-urea gel system (28). The ,u3 polypeptide is often difficult to clearly resolve from ,t2. Further studies are necessary todefinitivelyprovethat this previ-ously unresolved and rather elusive polypep-tide (as well as the ul and ,L4 species) is, in fact, a primary gene product of medium-sized (M) double-stranded RNA segments. The ,u5 polypeptideis a minor componentthatis possi-bly identicaltothe ,t3polypeptide (seebelow). Four small-sized polypeptides (al, a2, o-2A, ando-3) thataregeneproducts of the small (S) genome segments (5, 10) were detected in in-fected cells as previously reported (34). Poly-peptides Xl, X2, ul, ,u2, a-, a-2, and a-3 were also foundinpurifiedvirionsand the top com-ponent (28) as was a small quantity of the X3 polypeptide. Polypeptides
tL4
and a-2A were foundonly incytoplasmic extracts. The detec-tionof the p3polypeptide onlyininfected cells could conceivably be due to the fact that it is obscured by largequantitiesof themajor struc-tural ,u2polypeptideinthevirion.The electrophoretic behaviorof some of these polypeptide speciesdepends onthe gelsystem utilized for their resolution. Polypeptide,u4 mi-grates morerapidly thanpu2onpolyacrylamide gels in aTris-glycine buffersystembut slower than pu2 in the phosphate-urea buffer system whereit was designated,u0.Based on the elec-trophoretic behavior ofthe,uOand ,u1 polypep-tidesinthe lattersystem, thesegene products were assigned to the Ml and M2 segments of genome RNA, respectively (34). On the other hand, themigration of the ,u4(,uO)polypeptide intheTris-glycinesystemwouldsuggest that it iscoded by theM3genomeRNAsegment. This assignmentis moreconsistentwiththefindings from Graham's laboratory (15, 21) (and con-firmedabove)that, after release from cyclohex-imide, whenonlythe transcript of thesmallest of these medium-sized double-stranded RNA segments(M3)ispresent(34),the ,u4(,uO) poly-peptide is translated. Ultimately, translation invitro witheach of theisolated mRNAspecies will allowaccurategene assignment.
In addition to differences in the electropho-retic behavior of the ,u polypeptides, the order ofmigration of the two larger a- polypeptides
(a-ianda2)on Tris-glycine gels is thereverse of that on phosphate-urea; i.e., the polypeptides designated as a-1 and a-2 on Tris-glycine gels correspond to the a-2 and al- polypeptides, re-spectively, on phosphate-urea gels. Consistent withthis observation is thefinding that viral
on November 10, 2019 by guest
http://jvi.asm.org/
172
corescontainpolypeptides Xl, X2, X3,andc1 by analysis on Tris-glycine gels (whereas the lat-terpolypeptide has been referred to aso-2after analysis of core polypeptides on phosphate-urea gels). Differences have also been detected in the pattern of reovirus polypeptides resolvedinthe Tris-glycine system as described by Maizel (20) from that seen on Tris-glycine gels as described by Laemmli (14) (Ramig, unpublished data). Forexample, the order of migration of,t3 and ,4 as described in the Maizel system may be reversed inthe Laemmli system. This finding hassuggested that the ,u5 component
occasion-allyseen intheMaizel system may be an alter-nateform of the ,u3 polypeptide.
These migrational differences may reflect posttranslational modifications such as phos-phorylationorglycosylation that areknownto alter the electrophoretic behavior of polypep-tides onSDS-polyacrylamide gels (Maizel, per-sonal communication). It has been found, for example, thata phosphorylated NS protein of
vesicularstomatitis virus (22) migratesslightly
faster than the N protein in the phosphate system but more slowly than this protein on Tris-glycine gels (29).
Krystal et al. (13) have, in fact, presented evidence that the polypeptide ,u2isa phospho-protein and that the phosphorylation process mightbe highly selective, since only one or less serineresidues (outofapossible total of59per
,u2 protein [24]) is phosphorylated. Thus, itis
possiblethat the difference inmigration of the
,2polypeptide (and its precursor ,ul) with re-specttothe nonstructural,A
(itO)
polypeptideintwogelsystems is the result of this phospho-rylation. The anomalous migration of the o-1 and o-2 polypeptide species inthe two gel sys-temssuggeststhat they may also be modified. However, since they are present in such small amounts in whole virions and cytoplasmic ex-tracts, modification would be extremely diffi-culttodetect.
Evidence for the presence of carbohydrate in reovirus type 3 has also been indirect until recently (reviewedinreference 25). Reovirions havebeen demonstratedtoagglutinatehuman type 0 erythrocytes, and thisproperty can be destroyed bypretreatment with periodate (16, 17, 30). Krystal et al. (G. Krystal, J. Perrault, and A. F. Graham, Virology, in press) have recently identified a sugar moiety, probably a trimerortetramer,linkedO-glycosidicallyto a serine or threonine residueof the ,2 polypep-tide. The carbohydrate contains aterminal N-acetyl neuraminic acid as well as N-acetylga-lactosamine and galactose. The prior difficul-ties in identifying this component are most likelydue to thefact that only 10 to 20 of the 550
J.
,2polypeptides per virion are glycosylated. It is currently unknown whether this minor de-gree ofglycosylation has an effect on the migra-tion of the ,u2 polypeptidesin thedifferent gel systemsdescribed inthisreport.
Both etal. (5) havedetected10primary gene products in cytoplasmic extracts of infected cells. An in vitro protein-synthesizing system derived from wheat germ was used to identify these species. The large polypeptides (Xl to X3) have been clearly identified as primary gene products. The migration pattern of these spe-cies inthe Laemmli discontinuoussystem (14) used by these authors is identical to that ob-served in the Maizel system, although more reproducible separation of these species (partic-ularly Al and X2) and other high-molecular-weightpolypeptides is, in fact, possiblein the Laemmli system (Ramig,unpublisheddata;5). On the other hand,it isdifficulttoresolve the
o-polypeptides in the Laemmli system (Ramig, unpublished data), and Both et al. (5), there-fore,utilized the Maizel systemtoresolve these four small primary gene products. Their pat-tern of , polypeptides is somewhat complex, and itis difficultto reconcile the pattern they obtained for these polypeptides with the find-ingsreported here.Inaddition, the assignment of primarygenefunction of the,uspeciesseems somewhat tentative, sinceoneof these polypep-tides (which they designate
it2)
isbarely detect-able in vivo and in vitro, and another, which has been classed as a cleavage product (P69),cannotbewellresolved againstabackgroundof
uncompleted productsintheir in vitro system. In light of the variability in the
electropho-retic migration of the reovirus polypeptides
(andespeciallythe ,species)presentedinour
study, it will be critical to examine the gene products synthesized in vitro and in vivo by
individual mRNA speciesinpolyacrylamide gel
systemscontainingdifferentbuffers.
ACKNOWLEDGMENT
This research wassupportedby Public Health Service research grant AI-10326 from theNationalInstitute of Al-lergy andInfectiousDiseases.
LITERATURE CITED
1. Banerjee,A.K., and A. J. Shatkin. 1970. Transcription
in vitro by reovirus-associated ribonucleic acid-de-pendent polymerase.J.Virol.6:1-11.
2. Baum, S.G., M. Horwitz, and J. V. Maizel, Jr. 1972. Studiesonthe mechanism of enhancement ofhuman adenovirusinfection inmonkey cells by simian virus
40.J. Virol. 10:211-219.
3. Borsa, J., and A. F. Graham. 1968. Reovirus: RNA polymerase activityinpurifiedvirions.Biochem. Bio-phys.Res.Commun. 33:885-901.
4. Borun, T.W., M. D. Scharff, and E. Robbins. 1967. Preparation of mammalian polyribosomes with the
on November 10, 2019 by guest
http://jvi.asm.org/
detergent Nonidet P-40. Biochim. Biophys. Acta 149:302-304.
5. Both, G. W.,S. Lavi,and A. J. Shatkin. 1975. Synthesis ofallof the geneproductsof the reovirus genome in vivo and in vitro. Cell4:173-180.
6. Cross, R. K., and B. N. Fields. 1972. Temperature-sensitive mutantsof reovirus type 3: studies on the synthesis of viral RNA.Virology 50:799-809. 7. Cross, R. K., and B. N. Fields. 1976.
Temperature-sensitive mutants of reovirus type 3: evidence for aberrant p1 and ;2 polypeptide species. J. Virol. 19:174-179.
8. Fields, B. N., and W. K.Joklik. 1969. Isolation and preliminary genetic and biochemical characterization oftemperature-sensitive mutants of reovirus. Virol-ogy 37:335-342.
9. Fields, B. N., R. Laskov, and M. D. Scharff. 1972. Temperature-sensitive mutants of reovirus type 3: studies on the synthesis of viralpeptides. Virology 50:209-215.
10. Graziadei, W. D., and P. Lengyel. 1972. Translation of in vitrosynthesized reovirus messenger RNAs into proteins of the size of reoviruscapsid proteins in a mouse Lcell extract. Biochem. Biophys. Res. Com-mun.46:1816-1822.
11. Hay, A. J., and W. K. Joklik. 1971.Demonstration that the same strand of reovirus genome RNA is tran-scribed in vitro and in vivo.Virology 44:450-453. 12. Joklik, W. K. 1974. Reproduction of reoviridae, p.
231-320.In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehesive virology. Plenum Press, New York. 13. Krystal, G., P. Winn, S. Millward, and S. Sakuma.
1975.Evidence for phosphoproteins in reovirus. Virol-ogy 64:505-512.
14. Laemmli, V. K. 1970. Cleavage of structural proteins duringtheassembly of the head of bacteriophage T4. Nature (London) 227:680-685.
15. Lau, R. Y., D. Van Alstyne, R. Berckmans, and A. F. Graham.1975.Synthesisof reovirus-specific polypep-tidesincells pretreatedwithcycloheximide.J.Virol. 16:470-478.
16. Lerner, A. M., E. J. Bailey, and J. R. Tillotson. 1966. Enterovirushemagglutination: inhibitionby several enzymesand sugars. J. Immunol. 95:1111-1115.
17. Lerner, A. M., L. D. Gibb, J. R. Tillotson, M. M. Carruthers, and E. J.Bailey. 1966. Enterovirus he-magglutination: inhibition by aldoses and a possible mechanism. J. Immunol. 96:629-636.
18. Levin, D. H., N.Mendelsohn,M.Schonberg, H. Klett, S.C. Silverstein, A. M. Kapuler,and G.Acs. 1970.
Properties ofRNA transcriptase in reovirussubviral particles.Proc.Natl. Acad.Sci. U.S.A. 66:890-897. 19. McDowell, M. J., W. K. Joklik, L.Villa-Komaroff, and
H. F.Lodish. 1972. Translation of reovirus
messen-gerRNAs synthesized in vitro into reovirus polypep-tidesby several mammalian cell-free extracts. Proc. Natl. Acad. Sci. U.S.A. 69-.2649-2653.
20. Maizel,J. V., Jr. 1971.Polyacrylamide gel electropho-resisof viralproteins, p. 177-246.In K.Maramorosch and H. Koprowski (ed.), Methods in virology, vol.5.
Academic Press Inc., New York.
21. Millward, S.,and A. F. Graham. 1974. Reovirus: early events (in theinfected cells) andthe structure of the double-strandedRNA genome, p. 651-676. In E. Kur-stakandK. Maramorosch (ed.), Viruses,evolution, andcancer. Academic PressInc., NewYork. 22. Moyer, S. A.,and D. F. Summers. 1974.
Phosphoryla-tionofvesicular stomatitis virus in vivoandin vitro. J. Virol.13:455-465.
23. Nonoyama,M., S. Millward,andA. F.Graham.1974. Control of transcription of the reovirus genome. Nu-cleicAcids Res. 1:378-385.
24. Pett, D. M., T. C.Vanaman, and W. K. Joklik. 1973. Studies on the amino and carboxyl terminal amino acid sequences of reoviruscapsidpolypeptides. Virol-ogy52:174-186.
25. Shatkin, A. J. 1968. Viruses containingdouble-stranded RNA, p. 351-392. In H. Fraenkel-Conrat (ed.), Molecular basis of virology. Reinhold,NewYork. 26. Shatkin, A. J., and J. D. Sipe. 1968. RNApolymerase
activityinpurifiedreoviruses.Proc. Natl. Acad. Sci. U.S.A. 61:1462-1468.
27. Skehel, J. J., and W. K. Joklik. 1969. Studies on the in vitro transcription of reovirus RNAcatalyzed by reo-virus cores. Virology 39:822-831.
28. Smith, R. E., H. J. Zweerink,and W. K.Joklik.1969. Polypeptide components of virions, top component, andcoresof reovirus type 3.Virology39:791-810. 29. Stampfer, M., and D. Baltimore. 1973. Identification of
the vesicular stomatitis large protein as a unique viral protein. J. Virol. 11:520-526.
30. Tillotson, J. R., and A. M. Lerner. 1966. Effect of perio-date oxidation on hemagglutinating and antibody-producing capacities of certain enterovirusesand reo-viruses. Proc.Natl. Acad. Sci. U.S.A. 56:1143-1150. 31. Ward, R. L., A. K.Banerjee,A.LaFiandra, andA.J. Shatkin.1972.Reovirus-specificribonucleicacidfrom polysomesofinfected Lcells.J.Virol. 9:61-69. 32. Watanabe, Y., S.Millward, and A. F.Graham. 1968.
Regulationoftranscriptionofthe reovirus genome.J. Mol.Biol.36:107-123.
33. Zweerink,H.J., and W. K.Joklik. 1970. Studies on the intracellular synthesis ofreovirus-specifiedproteins. Virology 41:501-508.
34. Zweerink, H. J., M. J. McDowell, andW. K.Joklik. 1971. Essential and nonessentialnoncapsidreovirus proteins. Virology 45:716-723.
on November 10, 2019 by guest
http://jvi.asm.org/

![FIG.5.plasmicpostinfection,Microdensitometerrelease100electrophoresiso5S]methionine Viral polypeptides synthesized after cycloheximide release](https://thumb-us.123doks.com/thumbv2/123dok_us/1557165.108314/6.505.52.447.54.305/plasmicpostinfection-microdensitometerrelease-electrophoresiso-methionine-polypeptides-synthesized-cycloheximide-release.webp)


