0022-538X/81/040173-11$02.00/0
Selective Inhibition of Avian Sarcoma Virus Protein
Synthesis
in
3-Deazaadenosine-Treated
Infected Chicken Embryo
Fibroblasts
C. MARTIN STOLTZFUS'* ANDJOHN A.MONTGOMERY'
Departmentof Microbiology, University of Iowa, Iowa City, Iowa 52242,1 and The Southern Research
Institute,Birmingham,Alabama
352052
Received10October 1980/Accepted5January 1981
Theproduction ofB77aviansarcomavirions wasinhibitedmore than 90%in
infected chicken embryo fibroblasts thatweretreatedwith 100
,uM
3-deazaaden-osine, an inhibitor ofadenosylhomocysteine hydrolase and, for this reason, aninhibitorofmethylation reactions. This nucleoside analog at aconcentrationof 100
,uM
inhibited theratesof overall cellular protein synthesisandpolyadenylated RNAsynthesis by40 to50%. Rates of viral proteinsynthesis werecompared,andthe results indicated that in infected cells treated with 3-deazaadenosine syntheses of both the precursor of the gag proteins
(pr765¶g)
and the precursor of the reverse transcriptase(pr180'`9 P"')
were inhibited. Synthesisof theprecursor ofthe viral envelope glycoproteins (pr92env) appeared to be affected less by the
analogtreatment.Mostof the hostpolypeptides also continuedto besynthesized in3-deazaadenosine-treatedcells. The fraction ofthe total RNArepresentedby
virus-specific RNA in the 3-deazaadenosine-treated cellswasapproximately40%
of the fraction of the total RNA represented byviral RNA in control cells, as
determined byhybridization kinetics. Therefore,there was aselective inhibition of viral RNAaccumulation in the presence of 3-deazaadenosine. Theamountsof
genome-sized 35S and 38S RNAswerereduced compared withthe amountsof 28S and 21S viral mRNA's. These resultssuggestthatselectiveinhibitionof the
synthesis of viral proteins is due to selective decreases in the amounts of the
mRNA's for these polypeptides.
Bader etal. have shown that the nucleoside analog 3-deazaadenosine,aninhibitor of S-ade-nosylhomocysteine hydrolase, reversibly
in-hibits the reproduction of the Bryan strain of
Rous sarcomavirus and the transformation of chicken embryocells (2). These authors
postu-latedthat this effect is due tothe inhibitionof methylation reactions required for virus repli-cation. They also proposed that inhibition of virus replication and inhibition of oncogenic
transformationarecausedbyaninhibition of 5'
capmethylations (2).However, the biochemical basis of inhibitionwas notinvestigated.Wehave shownpreviouslythat2'-O-methylcap methyl-ations and internal N6-methyladenosine meth-ylations of cellular mRNA and viral genome RNAare inhibitedmore than 90% in the pres-enceofcycloleucine,yet virusproductionunder these conditions is inhibited only slightly (7). Thissuggestedthatthemethylationsblockedin the presence of cycloleucine (i.e., the
2'-O-methyl cap and internal N6-methyladenosine methylations) are not essentialfor virus repli-cation. We have investigated the basis for the
inhibition of virusproduction by 3-deazaadeno-sine and the reasonsfor the differences in the effects of thetwoin vivomethylationinhibitors 3-deazaadenosine and cycloleucineon virus
re-production. In this work we confirmed for
an-other strain of aviansarcomavirus (ASV), B77
ASV, the observation of Baderetal. thatvirus
production is inhibited drastically in the pres-ence of 3-deazaadenosine (2). We then
deter-mined themetaboliceffects of 3-deazaadenosine
onbulk cellular mRNA and protein syntheses and on virus-specific RNA and
protein
syntheses. We found that thesyntheses of the
internal non-glycosylated virion
polypeptides
(gagproteins) are inhibited
selectively
in the presence of 3-deazaadenosine. This inhibition appears toresult,atleast inpart, from selective decreases in theamountsof viral mRNA's.MATERIALS AND METHODS
Virus and cells. Chicken embryocells were pre-pared from10-day-old embryosandweresubcultured
at3-day intervals. Cellswereinfectedwith B77ASV
at amultiplicityofinfection of2 inthepresenceof2 173
on November 10, 2019 by guest
http://jvi.asm.org/
,ugofPolybrene perml.The infected cellswere pas-saged one ortwotimesin ordertoinsure that allof the cells were infected.Cellsweremaintainedin me-dium 199containing 1%(vol/vol) dimethylsulfoxide. Forexperimentsinwhichthe effect of 3-deazaadeno-sine was tested, Earle minimal essential medium (MEM) wasused, and tryptosephosphatebrothwas notadded. The media containedafinal concentration of 5%(vol/vol)calfserum.
Purification of virus. Viruswaspurifiedfromthe medium of infected cellsbypreviouslydescribed tech-niques (21).Briefly, the virus waspelletedfrom the medium through a 20%(wt/vol) sucroseshelfontoa 70%(wt/vol)sucrosepad.Thepelletedviruswasthen banded to equilibrium in a continuous 20 to 70% sucrosegradient.
Isolation of RNA. Total RNAwasisolatedfrom cells by phenol-chloroform extraction, as described previously (20).Cellswerefractionated into nuclei and cytoplasmby lysiswith 1%(vol/vol)Nonidet P40-1% (wt/vol) deoxycholate,andthe polysomes were iso-latedby discontinuoussucrosegradientsaspreviously
described (8). The fraction containing polyadenylic
acid[poly(A)+]wasisolatedbyoligodeoxythymidylic
acid-cellulosechromatography (20).
Determination of hybridization kinetics of viral cDNAtocellular RNA. Varyingamountsof RNAfrom3-deazaadenosine-treated and control cells werehybridized to3P-labeledcomplementaryDNA
(cDNA) prepared by the endogenous reverse
tran-scriptase reaction of detergent-disrupted B77 ASV (20).Hybridizationwascarriedoutinasolution con-taining0.3 MNaCl, 0.01 M Tris-hydrochloride (pH
7.5), and0.001MEDTAat680Cfor 18 h. The resist-anceof the 32P-labeled cDNAtoS1 nucleasewasused as a measureof theextentofhybridization.
GelelectrophoresisofRNA,transferto
diazo-benzoxymethyl paper,andhybridizationto 32P-labeledpSal"'DNA. Gelelectrophoresisofpoly(A)+
RNAon 1% agarosegelscontaining 10mM
methyl-mercuryhydroxidewascarriedoutessentiallyby the method ofBaileyand Davidson(3). The RNA in the gelwastransferredto diazobenzoxymethyl paperby theprocedureof Alwineetal.(1).The paper was then
prehybridizedfor24h,andthiswasfollowedby hy-bridization with a 32P-labeled nick-translated DNA probe (5.7 x 107cpm/pig), asdescribedbyAlwineet al.(1). The3P-labeled probe used in the experiments described belowwas pSal'01,arecombinant plasmid DNAwhich contains the entire ASV Prague A strain genome insertedintothe SallsiteofpBR322. Astock culture of this recombinantplasmid wasgenerously provided by J. Thomas Parsons, University of Vir-ginia, Charlottesville. The location of theradioactivity onthepaperwasdeterminedbyautoradiography on Kodak BB-5X-rayfilm,using a Dupont Cronex Light-ning-Plusintensifyingscreen.
Preparation of
[8Hlleucine-labeled
cytoplas-mic extracts. Chicken embryo fibroblasts infected with B77 ASV in100-mmpetri dishes were incubated for17hwitheither10mlof MEM or 10mlofMEMcontaining 100 uM 3-deazaadenosine. The medium wasthen changed to either 5 ml of MEM without leucine or 5mlof MEM without leucine containing 100MiM3-deazaadenosine. Aftera1-h incubation, 250
,Ciof
[3H]leucine
wasaddedtoeach plate, andthecells were labeled forvarying intervals, as described below. The cellswerethen washed once with 4 ml of asolution containing 140 mMNaCl, 5 mM KCI, 0.6 mMNaHPO4, 5.5 mM glucose, and 25 mM Tris-hy-drochloride(pH 7.2) (TBS) at 4°C, scraped into 2 ml of a solution containing 10 mM Tris-hydrochloride (pH 7.4), 1 mM NaCl, and 1.5 mM MgCl2, and swelled onicefor 10min. After this the cell suspensions were eachbrought to a final concentration of 1% (vol/vol) Nonidet P-40, homogenized by five strokes of a Douncehomogenizer, and centrifuged for 30 min at 10,000 x g to remove nuclei and cell debris. The supernatant was removed and stored in
100-pl
portions at-700C.Immunoprecipitation of virus-specific pro-teins. Formalin-fixed StaphylococcusaureusCowan Istrainwaspreparedby the method of Kessler (11). Fixedbacteria(100
pl)
at aconcentration of 10%(vol/vol)weresedimented and then suspended in 400,l of asolutioncontaining 150 mMNaCl, 5 mMEDTA, 50 mMTris-hydrochloride (pH 7.4), and 0.5% (vol/vol) Nonidet P-40 (buffer A). To this suspension
approxi-mately 100
id
of[3H]leucine-labeled cell extract was added, and the suspension was incubated for 30mi at roomtemperature. The bacteria were then pelleted, and we added to thesupernatant an amount of anti-serum determined to be sufficient toquantitatively immunoprecipitate protein from 10 ug of detergent-disrupted virions. The samples were incubated for 30 minatroomtemperature, followed by the addition of 100p1of a 10%(vol/vol) suspension of Formalin-fixed bacteria. In some cases, varying amounts of disrupted nonradioactive viruswerepresentduringthe incuba-tion (seebelow). The bacteriacontaining the immu-noprecipitated proteins were pelleted, the superna-tants were discarded, and the pellets were washed three times with buffer A.Thepellets were suspended in a solution containing 6 M urea and 4% (wt/vol) sodium dodecyl sulfate and heated for 3 min at 1000C. The releasedproteinwasrecoveredafter centrifuga-tion of the fixed bacteria and eitherdirectly counted forradioactivityorappliedtopolyacrylamidegels.Polyacrylamide gel electrophoresis of pro-teins. Discontinuous sodium dodecyl sulfate-10%
polyacrylamide slab gelswerepreparedessentially by the method of Laemmli (12). The samples were elec-trophoresed at 80 V until thetrackingdye reached the bottomedgeofthegel.Radioactivelylabeled proteins werelocatedbyfluorographyessentially as described by BonnerandLaskey (4). Kodak BB-5 X-ray film was exposed to the dried gels at -70°C and then
developed to visualize the bands produced by the radioactiveproteins.
Materials. Embryonated gs chf chicken eggs wereobtained from SPAFAS, Inc., Norwich, Conn. 3-Deazaadenosine was synthesized by a previously de-scribed procedure (15). L-[methyl-3H]methionine (9
Ci/mmol), [5,6-3H]uridine (41Ci/mmol),
[2-3H]aden-osine(22Ci/mmol),L-[4,5-3H]leucine (149Ci/mmol), and deoxycytidine 5'[a-3P]triphosphate (2,000 Ci/ mmol)wereobtained fromAmersham Corp., Arling-tonHeights, Ill.S1 nuclease was obtained from Miles Laboratories, Inc.,Elkhart,Ind.Aminobenzoxymethyl paperwaspurchased fromSchleicher & Scheull Co., J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
INHIBITION OF ASV PROTEIN SYNTHESIS 175 Keene, N.H. Goat antisera directed against
detergent-disrupted avian myeloblastosisvirus (AMV) and p27
wereobtained from the Division of Cancer Cause and
Prevention, National Cancer Institute. Anti-p19
anti-serum wasgenerously supplied by Volker Vogt,
Cor-nellUniversity, Ithaca, N.Y.
RESULTS
Effect of 3-deazaadenosine on the
pro-duction of B77 ASV virions by infected
chickenembryofibroblasts. Totestthe effect
of3-deazaadenosine onthe production of B77 virions,plates of confluent infected chicken em-bryo fibroblastsweretreatedwitheither MEM or MEM containing 100 LM 3-deazaadenosine for14h. The radioactiveprecursors[3H]uridine
and[3H]methionine (toassayfor RNA and
pro-tein,respectively)werethen addedtothe media. Themediawerecollected after12h oflabeling, the viruswaspurified, and theamountsof
radio-activity in the virus bands fromanequilibrium sucrose gradient were determined. Table 1
shows thatboth theincorporation of [3H]aden-osine and the incorporation of[3H]methionine
into virionswere reducedbymorethan 90% in
thepresenceof3-deazaadenosine. These results
indicated that theproductionof B77virionswas
inhibited in the presence of 3-deazaadenosine
and confirmedthe results of Baderetal. (2)for
adifferent ASV strain. Theproductionof
trans-formation-defective virionswas alsoinhibited in
the presence of3-deazaadenosine (Table 1).In agreementwith Baderetal.,wealsofound that
the inhibition of virusproductionwasreversible.
Uponremoval of3-deazaadenosine, therateof virus production, as determined by
incorpora-tion ofradioactivity orbyreversetranscriptase activity,wasrestoredtonormal levelswithin 12 h(datanotshown).
Effect of3-deazaadenosne on host and
viralpolypeptide synthesis.Onepossible ex-planationfor the inhibition of virusproduction by3-deazaadenosine would beageneral
block-ade of host protein synthesis. Therefore, we
tested the effect of thisdrugontheoverallrate
ofprotein synthesis.Cellsweretreated for
vary-ingtimes in the absence and presence of 100
ILM
3-deazaadenosine. Cell cultures were then pulse-labeledwith [3HJleucine for 1 h, the cells were harvested, and the incorporation of [3H]leucine into hot trichloroacetic acid-insoluble material wasmeasured (Table 2). A 35 to 50% decrease inincorporation was obtained in the presence of 3-deazaadenosine. These results agree reasona-bly well with those obtained by Bader et al., who reported aninhibition of approximately 20% in the rate of
[3H]leucine
incorporation after treat-ment of chicken embryo cells with 100ItM
3-deazaadenosine for24h(2).To test for a specific effect of 3-deazaadeno-sine on viral protein synthesis, ASV-infected cells were labeled with [3H]leucine for 1 h, cy-toplasmic extracts were prepared, and the virus-specific proteins were detected by incubation with appropriate antisera to avian retrovirus proteins, followed by adsorption of the
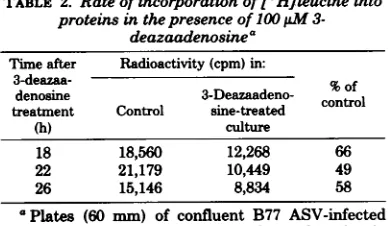
antigen-TABLE 2. Rateofincorporation of[3HJleucineinto
proteinsinthepresence of100X jM 3-deazaadenosinea
Time after Radioactivity(cpm)in:
3-deazaa-denosine 3-Deazaadeno- %of treatment Control sine-treated control
(h) culture
18 18,560 12,268 66
22 21,179 10,449 49
26 15,146 8,834 58
Plates (60 mm) of confluent B77 ASV-infected cells were treated with 100jiM3-deazaadenosine in MEM forvarying times. The mediumwas then
re-placedwith theappropriatemediumlackingleucine. After 1h 50juCiof[3H]leucine was added, and the cells were incubated for an additional1h. After this, thecellswerescrapedfrom each dish and suspended in1ml ofabuffercontaining0.1MNaCl,0.01MTris
(pH7.5), 0.001 MEDTA, 1%sodiumdodecylsulfate, and 1% 8-mercaptoethanol. From thesesamples, 0.1
ml-portionswerewithdrawn into 1-mlportionsof 10%
[image:3.494.251.445.296.409.2]trichloroacetic acid,and thesampleswereheated for 15minat900C.Thesampleswere then filtered and counted forradioactivity.
TABLE 1. InhibitionofradioactiveB77virionproductioninthepresence of100 M3-deazaadenosinea
Radioactivity (cpm)in:
Virus8lwtopCo3-Deazaadenosine- %Inhibition
Control treatedculture
B77 [3H]methionine 106,000 9,400 91
B77 [3H]adenosine 17,000 800 95
tdB77 [3H]uridine 4,400 500 88
'Plates(100 mm) of confluent B77 ASV-infected cellswerelabeled with800jiCiof[methyl-3H]methionine
per mlor20,iCiof[3H]adenosineper ml for12haftera14-htreatmentwith3-deazaadenosine. Cells infected withtransformation-defective B77 ASV(tdB77)werelabeled with20jiCiof[3H]uridineper ml for12h. The viruswaspurifiedfrom the mediaasdescribedpreviously (21),and theamountofradioactivityinthepeakat adensityof1.15to1.16g/mlwasdetermined.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.494.47.445.546.610.2]176 STOLTZFUS AND MONTGOMERY
antibodycomplexestoFormalin-fixed S.aureus (11). The materials in the immunoprecipitates
werethenanalyzed by sodium dodecyl sulfate-polyacrylamidegelelectrophoresis,andthe
rel-ative intensities of the bandscorresponding to
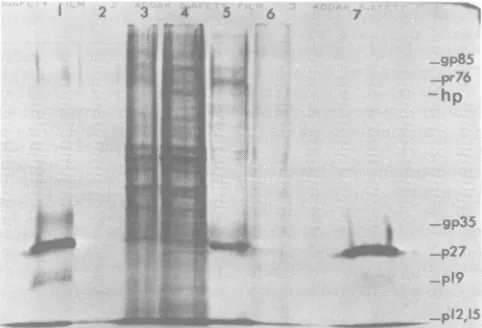
the viral proteinswerecompared (Fig. 1).Inthe control extracts, the major bands immunopre-cipitated by the AMV antiserum and detectable
onthegelscorrespondedtop27 and the precur-sorof thegagproteins, pr769ag (Fig. 1, lane 5). Figure1also shows thatuponaddition ofexcess
competing unlabeled viral protein during the immunoprecipitation (lane 6), the relative inten-sities of thepr76Wg andp27bandswere dimin-ished, indicating that the immunoprecipitation of these proteins was specific. Much smaller
amountsofpr769ag and p27were immunoprecip-itated fromanequivalentamountof [3H]leucine-labeledproteinextractobtained from 3-deazaa-denosine-treated cells (Fig. 1, lane 2). Similar results were obtained whenanti-p27 antiserum
wasused(datanotshown).
Theprofiles of the total labeled proteins from 3-deazaadenosine-treated and untreated cells
areshown inFig. 1, lanes3and4, andthe two
profileswerequite similar. Since themajorityof thelabeled proteins are cellular, thisresult
in-dicates that thesynthesis ofmostof thecellular proteinswas notinhibitedselectively by the
3-deazaadenosine treatment. An exception ap-pears to be apolypeptide withanapparent
mo-lecular weight of approximately 70,000 (desig-nated hp), which was present in the control
extract.Theposition of thispolypeptideonthe gel didnotcorrespondtotheposition ofanyof the virion proteins. This suggested that 3-dea-zaadenosinemayspecifically inhibit the synthe-sis of some host cell proteins, as well as the synthesis of viral proteins.
It was possible that the viral proteins were
synthesized in thesamerelativeamountsin the 3-deazaadenosine-treated cells but that these proteinsweredegradedrapidlyunderthese
con-ditions. If thiswere the case, wewould expect
thatlabeling forashortertime with [3H]leucine would result in the production of relatively largeramountsof viralproteinsin 3-deazaaden-osine-treatedcells.Therefore, theproductsofa
20-minpulse-labelingwith[3H]leucinewere
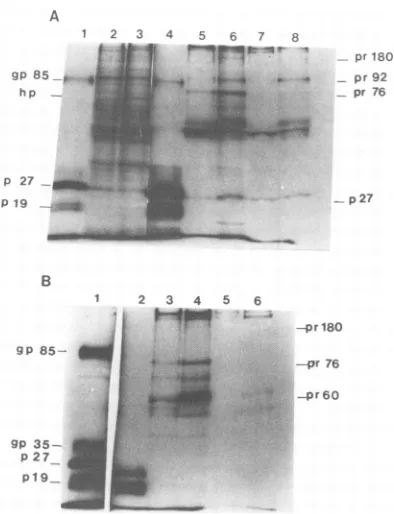
an-alyzed (Fig. 2). In these experiments we used
twoantisera of differentspecificities,anti-AMV and anti-p19. It was clear that the anti-AMV antiserum immunoprecipitated both the gag
proteinsand,to alesser extent,theenvproteins gp85and gp35 (compare Fig. 2A,lane 1 [whole disrupted virions] with Fig. 2A, lane 4 [immu-noprecipitate of whole disrupted virions]).The immunoprecipitation of gp85 and gp35 from
vi-FIG. 1. Polyacrylamide gel electrophoresis of viral proteins pulse-labeled for 1 hwith
[3HJleucine
and immunoprecipitated from extracts of3-deazaadenosine-treated and control infected cells.[3H]leucine-labeledcell extractswereprepared, andimmunoprecipitationwithanti-AMVantiserumto theFormalin-fixed Cowan Istrainof S. aureus and elution from the bacteria were carried out as described in the text. Equal input amountsofradioactivity from the two extracts were used. Polyacrylamide gel electrophoresis was carried out on theelutedsamples in discontinuous sodiumdodecylsulfate-polyacrylamide gels for 4.5 h at 80 V. The gel wasprepared forfluorography by using previously described procedures (4). Lane 1, Disrupted
[3H]leucine-labeled B77virions; lane 2,immunoprecipitation ofproteins from 3-deazaadenosine-treatedcell extract; lane 3,totalproteinsfrom3-deazaadenosine-treatedcellextract; lane 4, total proteins from control cellextract;
lane5, immunoprecipitation of proteins from control cellextract; lane 6, immunoprecipitation of proteins from control cell extract carried out in the presence of 100ug ofnonradioactive disrupted B77 sarcoma virions; lane 7, immunoprecipitation of disrupted[3H]leucine-labeledB77 virions.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.494.138.379.377.541.2]A
1 2 3 4 5 6 7
gp85_
hp
P 27_ Pl19
B
-pr180
gp
85-gp 35-J
P27_ p19
-Pr76
_pr60
FIG. 2. Polyacrylamide gelelectrophoresis ofviral proteins pulse-labeled for20 min with[3H]leucine
andimmunoprecipitated fromextractsof 3-deazaa-denosine-treated and control infected cells by anti-AMVantiserum(A)andanti-p19antiserum(B). The experimentalconditions were identicaltothose de-scribed in thelegendtoFig. 1,exceptthatthelabeling time was 20 min rather than 1 h. (A) Anti-AMV
antiserum. Lane 1, Disrupted B77 virions; lane 2, total proteins from 3-deazaadenosine-treated
ex-tract;lane3,totalproteins fromcontrolextract;lane 4, immunoprecipitation of virionproteins; lane 6, immunoprecipitation ofcontrolextract;lane5,same aslane 6, but in thepresenceof80pgof nonradio-activedisruptedB77sarcomavirions;lane8,
immu-noprecipitation of 3-deazaadenosine-treatedextract;
lane7,same aslane8,but inthepresenceof80pgof nonradioactive disrupted B77sarcoma virions. (B)
Anti-p19antiserum. Lane 1, DisruptedB77virions;
lane2, immunoprecipitation ofvirionproteins; lane
4, immunoprecipitation of proteins fromcontrolcell
extract;lane3,same aslane4,butcarriedoutin the
presence of80pg ofnonradioactive disruptedB77
sarcomavirions;lane6,immunoprecipitation of
pro-teins from 3-deazaadenosine-treated cell
extract-lane5,same aslane6,but carried out inthepresence
of80 pg ofnonradioactivedisrupted B77sarcoma
virions.
rionswasnotobvious in the 1-hpulse shownin
Fig. 2because theexposure time ofthe
fluoro-gramwas not sufficiently long to detect
radio-activity in these peaks. On the otherhand, the
anti-p19 antiserum precipitated primarily the
p19polypeptides, but it alsoprecipitated p27to
some extent; i.e., the antiserum was
contami-nated slightly with antibodies directed toward p27 (Fig. 2B, lanes 1 and 2). There was no
activity directed toward theenvproteingp85. Figure 2 also shows that in the control infected extract anumber ofbands werespecifically
pre-cipitated with the anti-AMV antiserum (Fig.2A, lanes5and6). Asexpected, the antiserum
pre-cipitatedpr769ag and aless intense band
corre-sponding top27. We also detected a
virus-spe-cific polypeptide migrating at the position of
pr180O9"9I)(, the presumptive precursor of the reverse transcriptase (15). In addition, a poly-peptide with a molecular weight of
approxi-mately90,000wasimmunoprecipitated, and this immunoprecipitation wasinhibitedinthe pres-enceofexcessunlabeleddisrupted virions.
Whentheanti-p19 antiserumwas used,both
pr7W6Ma and a band correspondingtopr609'9, a
previously described intermediate (23) in the processing of
pr769"¶,
werespecifically immuno-precipitatedfrom the extract. (Fig. 2B, lanes3and 4). Although not visible in Fig. 2, a band correspondingto pr1809'9P"wasalso detected inlongerexposures.However,the90,000-dalton polypeptide was not immunoprecipitated. We believe that thelatterpolypeptide corresponds
to the envelope protein precursor (pr92en") for the following reasons: (i) the polypeptide was
immunoprecipitated by the anti-AMV
antise-rum, which contained antibodies directed againstenvproteins,butwas not immunoprecip-itated bytheanti-p19antiserum,whichdidnot;
(ii) thepolypeptide had theapproximate appar-ent molecular weight of the env precursor, as
reported byotherinvestigators (14); and (iii)the
immunoprecipitation of this protein was in-hibited in the presence ofdisrupted virions,
in-dicating that it was related structurally to a
virionprotein.
Inthe extracts prepared from 3-deazaadeno-sine-treatedcells,wedetected littleor nopr769'9
ortheputative
pr1809h
P)o polypeptideineither the anti-AMV immunoprecipitates (Fig. 2A, lanes 7 and8) or the anti-p19immunoprecipi-tates (Fig. 2B, lanes 5 and 6). This result was
similar to the resultobtained inthe 1-h pulse-labeling experiment (Fig. 1) andsuggestedthat
thesyntheses ofthese proteinswereindeed
in-hibited in the presence of 3-deazaadenosine
rather thantheproteinsbeingrapidly degraded.
Incontrast, thepresumptive
pr92env
polypeptide appearedtobe present in the anti-AMVimmu-noprecipitates (Fig. 2A, lane 8). These results
suggested that detectable amounts of
pr92env
were synthesized in 3-deazaadenosine-treated cells, whereas comparable amounts of pr769`9
werenot.
Wecomparedtheprofilesof the totallabeled
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.494.51.248.66.323.2]178 STOLTZFUS AND MONTGOMERY
proteins from 3-deazaadenosine-treated and control extracts (Fig. 2A, lanes 2 and 3). As
observedwith the 1-h labeled extracts, the
pro-files appeared to be identical, except for the presence inthe control extract ofa heavily la-beled band having a molecular weight of
ap-proximately 70,000 (band hp), which appeared
tobemissingintheextractsfrom the 3-deazaa-denosine-treated cells.
Effect of 3-deazaadenosine on host and viral RNAsynthesis.Wenexttestedwhether the selectiveinhibition of viralprotein synthesis
might be due to a selective inhibition of viral
RNAsynthesis.Ithasbeenreported previously
that a 24-h treatmentwith100-,iM
3-deazaaden-osine results in an inhibition ofapproximately 30%intherateof[3H]uridineincorporationinto
the totalRNA of chicken embryo cells (2). To
test the effect of the analog specifically on
mRNA synthesis, cells were treated for 15 h
with3-deazaadenosine and labeled with [3H]ur-idine for12h,and the totalpoly(A)+RNAwas
isolated fromthe cells. Theamountof radioac-tive RNA per cell was determined. There was
an inhibition in the accumulation of both poly(A)+ RNA andpoly(A)-RNA (34and 22% inhibition, respectively) in the presence of
3-deazaadenosine.
Todeterminewhetherthepoly(A)+ RNA
syn-thesized in thepresenceof3-deazaadenosinewas
associated with polysomes, analog-treated and controlcellswerelabeledforvaryingtimeswith
[3H]uridine.
Afterthe labeling period,the cellswerelysedandseparated into nucleiand
cyto-plasm. The cytoplasmic fractions were further
separated into polysomal and nonpolysomal fractions bycentrifugationondiscontinuous su-crose gradients. The amount ofpoly(A)+ RNA
in eachfractionwasthen determined. Figure 3
shows that there was a decrease of
approxi-mately 50% in the rate of
[3H]uridine
incorpo-rationintonuclearRNA (Fig.3A).Therewas acorresponding decrease in the rate of incorpo-ration of
[3H]uridine
intopolysomalRNA (Fig.3B). The size distributions of the polysomes
were not significantly different in treated and controlcells (datanot shown). The decrease in the observed rate ofprotein synthesis shown in Table 2 was similar in magnitude to the de-creased rate at which mRNA associated with
polysomes. This result suggested that the inhi-bition ofprotein synthesis in
3-deazaadenosine-treatedcellswas due to lowered levels of mRNA in polyribosomes. Intheexperimentsshown in
Fig.3 andTable 2, the results were not corrected forpossible effects of3-deazaadenosine on cell growth rate.However,weobservedthatthe cell
densities under the two conditions remained comparable over the time periods of the
experi-_x0. B
C3
2-1 2 3 4
HR
FIG. 3. Kineticsofpoly(A) RNA synthesis in 3-deazaadenosine-treated and control infected cells. Confluentmonolayersin100-mmplasticpetridishes ofB77ASV-infectedchickenembryofibroblastswere incubatedfor15h with either MEMorMEM
con-taining100,uM3-deazaadenosine. At the endofthis time, the medium wasremoved, andfresh medium withorwithout100
liM
3-deazaadenosinecontaining20,uCiof[3HJuridineper mlwasadded. The cells wereharvestedafter varyinglabeling periods by first washingtheplateswith5mlofTBS and then resus-pending the cells in 5 ml ofTBS. The cells were collected by centrifugation, disrupted by detergent lysis, and separated into nuclear and cytoplasmic fractions. The cytoplasmwasseparated into polyso-mal andnonpolysomal fractions by discontinuous
sucrosegradientsas describedpreviously (8). Poly-somal andnuclear RNAs wereisolated by phenol-chloroform extraction, and thepoly(A)+ RNA was recovered by oligodeoxythymidylic acid-cellulose
chromatography. Symbols: 0, 3-deazaadenosine-treatedcells;0,control cells. (A)Nuclear RNA.(B)
Polysomal RNA.
ments.
We next tested whether all or part of the action of3-deazaadenosine onvirusproduction
was due to selective inhibition of viral RNA
synthesis.Thefractions ofvirus-specificRNA in
analog-treatedandcontrol cellswerecompared by determining the kinetics of annealing of a
viral cDNAprobetoRNA isolated frominfected cells (Fig. 4). The
Crtl/2
values for the RNAsJ. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.494.288.434.54.357.2]0 ~ ~ 0 0
C,t
FIG. 4. Kinetics of RNA from 3-deazaadenosine-treated and control infected cells hybridized to viral cDNA. Confluent monolayer cultures of B77-infected cells were incubated witheither MEM or MEM con-taining1(0) jtM 3-deazaadenosine for 18 h. After this, [8HJadenosine (final concentration, 20yCi/ml) was added, and the incubation was continued for an additional 12 h. Total RNA wasisolated from the cells by phenol-chloroform extraction, DNase diges-tion, LiCIprecipitation, and oligodeoxythymidylic acid-cellulose chromatography, according to previ-ously described techniques (21). Hybridization of RNA at varying concentrations to approximately
1,000)cpm of 32P-labeledcDNA was carried out in a solutioncontaining 0.3 MNaCI,0.01 M Tris-hydro-chloride (pH 7.5), and 0.001 M EDTA for 18 at
68°C. The extent ofhybridization was assayed by the sensitivity of the 32P-labeled cDNA to Si nuclease digestion. Symbols: 0, 3-deazaadenosine-treated cells; 0, control cells.
from the control and 3-deazaadenosine-treated cells were 4.7 and 11.5 mol.s/liter, respectively. This result indicated that the fraction of the total RNA represented by virus-specific RNA in 3-deazaadenosine-treated cells was 40% of the fraction of virus-specific RNA present in control cells. Therefore, there appeared to be a selective decrease in the amount of viral RNA in the analog-treated cells. Since the overall poly(A)-RNA synthesis was inhibited somewhat less than the overall poly(A)+ RNA synthesis, we also directly tested the poly(A)+ RNA from 3-deazaadenosine-treated cells for the fraction of virus-specific RNA byhybridization kinetics. In this case we obtained a value of 50% of the control value (data not shown). We concluded that there was a selective decrease in the steady-state levels of viral RNA in the 3-deazaadeno-sine-treated cells.
We then analyzed the RNA in polyribosomes for individual species of virus-specific mRNA. Chicken embryofibroblasts infected with ASVs reportedly contain subgenomic 285 and 21S mRNA's, as well as 38S genomic RNA (10, 24). The 285 and 21SmRNA's are thought to serve as messengers for theprecursor of the envelope
proteinsgp85 andgp35 and for the protein
re-quired for transformation
(pp6e)'c),
respectively(17-19). Onthe otherhand, at least part of the 38SgenomicRNA serves asthemRNA for the gag protein presursor (pr769"9) and the
pre-sumptive reverse transcriptase precursor
(prl80V'gPO') (16). The 28S and 21S mRNA's
probably arisefrom the 38S RNA by a process
in whichsequencesin the 3' halfof the genome
RNAaresplicedtoa5'-terminal leadersequence
(6, 13,20).Totestfor thepresence and relative amountsof the various mRNA species,we
elec-trophoresed poly(A)+RNAsfromthe polysomal
fractions of 3-deazaadenosine-treated and
un-treated cells on agarosegels containing 10 mM
methylmercury hydroxide. The separatedRNA
speciesweretransferredtodiazobenzoxymethyl
paperby the procedure of Alwineetal. (1), and
the virus-specific species were detected by
hy-bridization of theblotto aplasmidpreparation
containing the entire ASV Prague A genome
(pSal'01)
which had been labeled with 32P bynick translation (Fig. 5). Qualitatively, it
ap-peared from the gels that the intensities ofthe
38S and35SgenomeRNA bands inthe
polyso-mal RNA from 3-deazaadenosine-treated cells
werereducedrelativetotheamounts of28Sand
21S mRNA's when compared with equal amounts of polysomal RNAs from the control
cells.(The 35S RNAwasderived from the pres-ence of transformation-defective deletion mu-tants which contaminated the B77 ASV stock used in this experiment). To approximate the
relative amounts of the various virus-specific RNA species presentin the 3-deazaadenosine-treated and control infectedcells,the
autoradi-ograms werescanned with adensitometer, and
the areasunder thepeakswerecompared. The
sizes ofareasunderthepeaks correspondingto
the 21S, 28S, and 35S-38S RNAs were 47, 40,
and12%of the sizes of theareasunder thesame
peakswhenequalamountsof control RNAwere
used. These differenceswereobserved when the gelswereexposed for several different intervals. Therefore, there appeared to be a relatively larger decrease intheamountof 35S-38S RNA than in theamountsof thesubgenomic 28Sand
21S mRNA species in the 3-deazaadenosine-treatedcells.
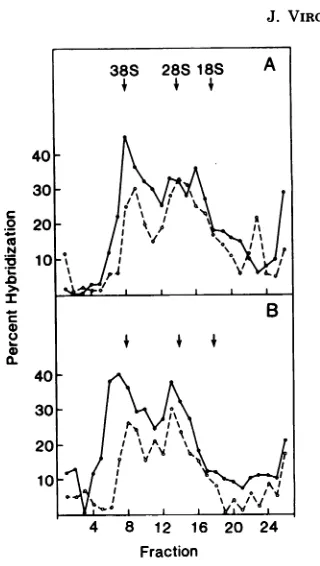
Further evidence for a decrease in the relative amount of the 35S-38S RNA in
3-deazaadeno-sine-treated cells compared with control cells
was obtained by hybridization of nuclear and
polysomal RNAs from glycerol gradient frac-tionstoradioactiveviral cDNA(Fig. 6).Clearly,
theresolutionof RNAspeciesinthesegradients
wasnotnearlyasgoodasin themethylmercury hydroxide gels shown in Fig. 5. However, the
data obtained from this hybridization of the
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.494.49.246.63.218.2].Y,0
N
0
0
.0
0.
I
1216
I I \\I \
[image:8.494.280.440.62.345.2]4 8 12 16 20 24 Fraction
FIG. 5. Electrophoresis ofviral RNAon 1%
aga-rosegels containing10 mMmethylmercury hydrox-ide. Thefractionation of cytoplasmic extracts into
polysomesandpostpolysomal cytoplasmwascarried outasdescribedin the text.Approximately 15 pgof RNAfromeachofthepolysome fractionswas
elec-trophoresed on 1% agarosegels containing 10 mM
methylmercury hydroxide for6 hat50 V. The loca-tions ofthe virus-specific RNA bands were deter-mined by procedures described in the text. The ex-posuretimewas24 h. LaneA, PolysomalRNAfrom controlASV-infected cells;laneB, polysomalRNA
from3-deazaadenosine-treatedASV-infectedcells.
RNAs from the gradient fractions allowed an independent quantitative comparisonof the
rel-ative amounts of genomic and subgenomic RNAs. Hybridization values in therangeof0to
40%canbe assumedtoberoughly proportional
toCrtand, therefore, foraconstanthybridization time proportional to viral RNA concentration (9). These dataaresummarized in Table3.We
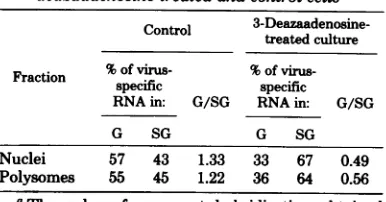
observedthat therewasadecrease in the ratio
ofgenomic RNA tosubgenomic RNA in both thenuclear RNA andthepolysomal RNA from 3-deazaadenosine-treatedcells.Therefore, these
datasupportthe data inFig.5andsuggestthat the relative concentration of the 35S-38S RNA
FIG. 6. Glycerol gradient sedimentation of
virus-specific nuclear RNA (A) andpolysomalRNA (B)
from 3-deazaadenosine-treated and control cells. Four100-mmplatesofconfluent cellsinfected with B77ASVwere treated with either MEMorMEM
containing100
AM
3-deazaadenosine for19h. The cells were fractionated into nuclei and cytoplasm,and thepolysomes were isolated on discontinuous sucrosegradients. The RNAsfrom the nuclear and
polysomalfractionswerepurifiedasdescribed
pre-viously(8). Thepoly(A)J RNAwasisolatedby
oligo-deoxythymidylic acid-cellulose chromatography. Ap-proximately equalamountsof RNA were applied to 5 to30%glycerol gradients. Thegradientswere sed-imentedbycentrifugationat24,000 rpmfor16h ina BeckmanSW41 rotor. Thegradientswere
fraction-ated, and the RNA in eachfractionwasprecipitated with2volumesof95% ethanol. Theprecipitateswere collected and dissolved in sterile distilledwater(100 and50AIfor the polysomal RNAs from thecontrol and3-deazaadenosine-treatedcells,respectively; 50 and25Alfor the nuclear RNAsfromthe controland 3-deazaadenosine-treated cells, respectively). Sam-ples (10
#1)
fromeachfractionwerehybridized for42 hat68oCto32P-labeledB77ASVcDNA. Agradient containing 38S viral genome RNA and 28S and 18S rRNA'swas runinparallel,and thepositions of the peaksaremarkedonthefigure. Symbols: O,3-dea-zaadenosine-treatedcells;0,control cells.
wasindeedreducedin3-deazaadenosine-treated cells.
DISCUSSION
Weshow in thispaperthat,in the presence of 100 uM 3-deazaadenosine, there is a selective
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.494.101.199.85.390.2]TABLE 3. Distributionofvirus-specificRNA in genomic 4nd subgenomic size classes from
3-deazaadenosine-treated and controlcellsa
Control
3-Deazaadenosine-treated culture
Fraction %of virus- %of
virus-specific specific
RNAin: G/SG RNA in: G/SG
G SG G SG
Nuclei 57 43 1.33 33 67 0.49
Polysomes 55 45 1.22 36 64 0.56
aThe values for percent hybridization obtained
from theresults in Fig. 6 were assumed to be propor-tional to concentration. This is only an approximate relationship and is valid between values of 0 and50%
hybridization (9). Genomic (G) and subgenomic (SG) RNAs were defined as the material sedimenting be-tweenapproximately 32Sand 408 and between 18S and 30S,respectively.
inhibition of the synthesis of the ASV proteins pr769'a andpr1809`9
1".
Thesynthesis ofa poly-peptide presumed to be pr92env appears to be affected less. In general, host cell proteinsyn-thesis is also less sensitive to inhibition. It ap-pears that the inhibition of viral gag protein synthesis, coupled withadecrease of30 to50% in the rate of overall protein synthesis, could explain the inhibitionof more than 90% inthe production of virions previously observed by Baderetal. (2) and confirmedbyushere.
We found that atleast part of the selective effect of 3-deazaadenosine treatment on viral protein synthesis is dueto aselective decrease in therelative amounts of viral mRNA's. The
Crtl/2
values for therateofhybridizationof viral cDNA toinfected cell RNA indicated that the concentration of viral RNA in treated cellswasto approximately 40% of the concentration in
control cells. If the inhibition of the rate of synthesis ofradioactivelylabeled cellular RNA (=40 to 50%) is also reflected in a decreased steady-state level of cellular RNA in 3-deazaa-denosine-treated cells,the absoluteamountsof viral RNAmaybereducedtolevels whichare aslittle as20% of theamounts incontrol cells.
Itshould be stated that the cDNA used in the hybridization kinetics experiments (Fig. 4) was
not a representative copy of the viral genome, and therefore we cannot be certain from this experiment whether the concentrations of all
viralsequences are reducedequally.From anal-yses of the viral RNA species onagarose gels containing methylmercury hydroxide and on
glycerolgradients,itappearedthat in
3-deazaa-denosine-treated cells, there were relatively larger decreases in the concentrations of the genome 38S and 35S mRNA's than in the
con-centrations of the 28S env and 21S src mRNA's. These relative decreases in the levels of the putative mRNA's for pr76Wg and prl80gYg P' mayexplainthe greatereffectof the
3-deazaa-denosine treatment on the synthesis of these
polypeptides compared with the presumptive
pr92env polypeptide (Fig.2).
The mechanism by which the steady-state
levels of viral RNAarereduced in
3-deazaaden-osine-treated cells remains obscure. Sucha de-creasecould be causedeither by apreferential decrease in therate ofvirus RNA synthesisor
byapreferential increaseintheturnoverof viral RNA. Baderet al. have proposed that since
3-deazaadenosineis an inhibitor of S-adenosylho-mocysteine hydrolase and since the intracellular concentration of S-adenosylhomocysteine
in-creases dramatically in treated cells,
methyla-tion reacmethyla-tions are inhibited. In particular, the failure to methylate viral RNA may render it
nonfunctional (2). We have shown previously
that another in vivo inhibitor of methylation,
cycloleucine, inhibits the internal N6-methyla-denosine and 2'-O-methyl ribose cap methyla-tions of both viral genome RNA and cellular mRNA'sby80 to90%.However, the production ofASV under these conditions is inhibited only slightly. This ledus to proposethat the
meth-ylations inhibited by the cycloleucinetreatment are notessential for virusreplication (7). There-fore, if the effectsonviral RNA levelsaredueto
inhibition of RNA methylations, we would ex-pect that those methylations which are not
blocked in thepresenceofcycloleucine(i.e., the
5'-terminal N7-methylguanosine methylations) would be affected. We have tested for suchan
effectonthe bulk cellular poly(A)+ RNA from 3-deazaadenosine-treated ASV-infected chicken embryo fibroblasts and have found little or no
inhibition ofN7-methylguanosine methylations (Stoltzfus, unpublished data). Wehavenotyet
tested fora possible specific effect on the N7-methylguanosine methylation of the various
viralmRNA's. However, it would be surprising
ifsuchwerethe case, sincecellularmethylating
enzymes arepresumablyusedtomethylateviral RNA.
As analternativehypothesis,wepropose that
3-deazaadenosine may selectively inhibit the
synthesis of viral RNA. It has been shown that theproductionof avian retrovirions isextremely
sensitive to the effects of thetranscription inhib-itor actinomycin D. Even at concentrations of
actinomycin D as low as 0.1 ,ug/ml, virus pro-duction is inhibitedmorethan 90%(22). Under theseconditions,thesynthesisof hostpoly(A)+
RNA is inhibited only about 30% (Stoltzfus, unpublished data). Whether this sensitivity to
lowconcentrationsofactinomycinD isdue
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.494.49.242.106.207.2]182 STOLTZFUS AND MONTGOMERY
tirelytoeffectsonviral RNAsynthesis hasnot
been established yet. We have also obtained preliminary evidence that treatment of ASV-infectedcells with a-amanitin (a specific
inhibi-torof RNApolymerase II)at aconcentrationof
10,ug/ml inhibits cellularpoly(A)+RNA synthe-sis by approximately 30%, whereas virion
pro-duction is inhibited by about 70% (Stoltzfus, unpublished data).Forbothinhibitors,theeffect
onvirus production and presumablythe effect
on viral RNA transcription are considerably
greater than the effect onthe synthesis of the bulk cellular mRNA. We have shown that
3-deazaadenosine inhibits bulk cellular poly(A)+ RNAsynthesis by40 to50%.Therefore, it would
not be surprising if the 3-deazaadenosine inhi-bitionof viral RNAlevelsweredueto aselective inhibition of viral RNA synthesis. Additional studiesonthekinetics of viral RNAsynthesis in 3-deazaadenosine-treated cells will be required
to testthishypothesis.
The reasonfor the selective decrease in the
amountof35S-38S RNA relativetotheamounts
of 28S and 21S mRNA's in 3-deazaadenosine-treatedcells is alsonotclear. Itisthought that the38S RNA isaprecursorofthe 28S and 21S mRNA's and that thesesubgenomic RNAsare
derivedbyasplicingprocess(6, 13, 20).Ifthis is indeed thecaseand ifa constant amountof viral 38S RNA issplicedperunittime,adecrease in the rate of transcription of 38S RNA would result inalarger proportion ofsplicedRNA and
anincrease in the ratio ofsubgenomic RNAto
genomic RNA.Therefore, the alteration inthe steady-state levels of the various viral RNA species observed in 3-deazaadenosine-treated cellsmayreflect the effect ofthe analogon the rate ofviralRNA transcription.
Finally,wepointoutthatthepartial reversion
ofcellsinfected with the Bryan strain ofRous sarcomavirus fromatransforned phenotypeto amorenormalphenotypeupon treatmentwith 3-deazaadenosine, as reported by Bader et al. (2), could also be explained by inhibition ofviral
RNA transcription. This would result in the
production of less src mRNA and, therefore, presumably would lead to a reduction in the
intracellular levels of the protein which is
re-quired for transformation, pp60' (5). We have
not yet determined the rate of synthesis of
pp6OBrc
in3-deazaadenosine-treatedcells.Basedonacomparisonof the relative amounts of the
putative 21S src mRNA in
3-deazaadenosine-treated and control cells (Fig. 5), we anticipate
that thereduction in the amount ofpp60O' is not asprofoundasthereduction in the amount
of
pr76g'.
To our knowledge, 3-deazaadenosine is the only reported example ofa substance that
ex-hibits selectivity in the inhibition of retroviral RNA andproteinsyntheses. Forthisreason,it
maybeausefultool foralteringthe intracellular levels of retroviral mRNA's and proteins and determining theconsequencesof these changes
oncellularand viralfunctions.
ACKNOWLEDGMENTS
Wethank PaulaSnyderandElizabeth Lanshe for technical assistance.
This research was supported by Public Health Service grant CA-28051 fromthe National Cancer Institute. C.M.S. was supported by Public Health Service Research Career Development Award CA-00659 from the National Cancer Institute.
LITERATURE CITED
1.Alwine, J. C., D. J.Kemp, and G. R. Stark. 1977. Methodfor detection ofspecific RNAsinagarosegels bytransfertodiazobenzyloxymethyl-paperand hybrid-ization with DNAprobes. Proc. Natl.Acad.Sci.U.S.A. 74:5350-5354.
2. Bader, J.P.,N. R.Brown,P.K.Chiang,andG.L. Cantoni.1978.3-Deazaadenosine,aninhibitorof ade-nosylhomocysteinehydrolase,inhibitsreproductionof Roussarcoma virus and transformation of chickembryo cells.Virology89:494-505.
3. Bailey,J.M.,and N. Davidson. 1976.Methylmercury
as areversible denaturingagentfor agarosegel electro-phoresis.Anal. Biochem. 70:75-85.
4. Bonner,W.M.,and R. A.Laskey.1974.Afilmdetection method for tritium-labelledproteinsandnucleic acids inpolyacrylamide gels.Eur. J. Biochem.46:83-88. 5. Brugge, J.S.,andR.L. Erikson. 1977. Identification of
atransformation-specific antigeninducedbyanavian sarcoma virus. Nature(London)269:346-348. 6. Cordell, B., S.R.Weiss, H. E.Varmus, and J. M.
Bishop.1978. Atleast 105 nucleotidesaretransported from the5'-terminusof the aviansarcoma virus genome tothe 5'-termini of the smaller mRNAs.Cell 16:79-91. 7. Dimock,K., and C. M. Stoltzfus. 1978. Cycloleucine blocks5'-terminal and internalmethylationsof avian sarcomavirus genome RNA. Biochemistry 17:3627-3632.
8. Dimock, K.,andC.M.Stoltzfus.1979.Processingand function ofundermethylatedchickenembryofibroblast mRNA. J.Biol. Chem.264:5591-5594.
9.Fan,H.,and D. Baltimore.1973.RNAmetabolism of murine leukemia virus: detection ofvirus-specific RNA sequences in infected and uninfectedcelisand identifi-cationofvirus-specificmessengerRNA. J. Mol. Biol. 80:93-117.
10. Hayward, W. S. 1977. Size and genetic content of viral RNAs in avianoncovirus-infected cells. J. Virol. 24:47-63.
11. Kessler,S.W. 1975.Rapidisolation ofantigensfrom cells with astaphylococcal protein A-antibody adsorbent: parameters of the interaction ofantibody-antigen com-plexes with protein A. J. Immunol. 115:1617-1624. 12. Laemmli, U. K. 1970. Cleavage of structural proteins
duringtheassemblyof thehead of bacteriophage T4. Nature(London)227:680-685.
13.Mellon, P., and P. H. Duesberg. 1977. Subgenomic cellularRoussarcoma virusRNAs contain
oligonucle-otides from the3'-halfandthe5'-terminusof thevirion RNA. Nature(London)270:631-634.
14.Moelling, K., and M. Hayami.1977.Analysisof precur-J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
sorstotheenvelope glycoproteins of avian RNAtumor viruses in chicken and quail cells. J. Virol 22:598-607. 15. Montgomery, J.A.,A.T.Shortnacy, andS.D.
Clay-ton.1977. Acomparisonof two methodsfor the
prep-arationof 3-deaza purine ribonucleosides. J. Heterocycl. Chem. 14:195-197.
16. Oppermann,HE, J. M. Bishop,H.E. Varmus, and L
Levintow.1977. Ajoint productof thegenesgagand pol of avian sarcoma virus: a possible precursor of reversetranscriptase. Cell 12:993-1005.
17. Pawson, T., R. Harvey, andA.E.Smith. 1977.The
izeof Roussarcoma virusmRNA's activeincell-free
translation. Nature (London) 268:416-419.
18. Purchio, A. F., E. Erikson, J.S.Brugge, and R. L
Erikson.1978. Identification ofapolypeptideencoded
by the aviansarcomavirussrcgene.Proc.Natl Acad. Sci. U.S.A. 75:1567-1571.
19. Stacey,D. W., V. G.A1lfrey,andH.Hanafdbsa 1977.
Microinjection analysis of envelope activities of avian
leukosis viral RNAs. Proc.Natl. Acad. Sci. U.S.A. 72: 489564899.
20.Stoltzfs,C. ML, and L K. Kuhnert.1979.Evidence for
the identify of shared 5'-terminalsequencesbetween genomeRNA andsubgenomic mRNA's of B77 avian
sarcomavirus. J. Virol. 32:536-45.
21.Stoltzfus, C. IL,andP. N.Snyder.1975.Structure of
B77 sarcoma virus RNA. stabilization of RNA after
packaging. J. Virol. 16:1161-1170.
22.Temin,H.1963.Theeffects ofactinomycin Dongrowth of Roussarcomavirusin vitro.Virology 20:577-582. 23.Vogt, V. ML,R.Elsenman,andH.Diggelmann. 1975.
Generationof avianmyeloblastosisvirus structural
pro-teinsby proteolytic cleavageofaprecursorpolypeptide. J.Mol. Biol. 96:471493.
24.Weiss,S.R.,H.E. Varmus, and J. ML Bishop. 1977.
Thesize and geneticcomposition ofvirus-specificRNAs
inthecytoplasm of cells producingavian
sarcoma-leu-kosis viume. Cell 12:983-992.
on November 10, 2019 by guest
http://jvi.asm.org/