0022-538X/81/110482-09$02.00/0
Chemical
Methylation of RNA and DNA Viral Genomes
as aProbe
of
In
Situ Structure
MINORUYAMAKAWA, AARON J. SHATKIN,ANDYASUHIRO FURUICHI*
Roche Institute of MolecularBiology, Nutley, New Jersey07110
Received3April 1981/Accepted12June1981
We used[methyl-3H]dimethylsulfatetoprobethegenomestructuresof several
RNAand DNA viruses. Wecompared sites of modification in nucleic acids that
were methylated chemically before and after extraction from purified virions.
With both single-stranded and double-stranded substrates alkylation occurred mainly attheN7 position of guanine. However, adenine NI atomswere differ-entially accessibleinsingle-stranded RNA and DNA. For example, the ratios of 1-methyladenosine to 7-methylguanosine for reovirusmRNAand deproteinized
genome RNA were 0.43 and 0.03, respectively. Members of the Reoviridae
methylated insitu yieldedRNAs with ratios of0.04 to 0.08, indicating that the intraviriongenomes weredouble stranded. Weobtained ratios of 0.26 and0.35for
the RNAs ofdimethyl sulfate-treated brome mosaic and aviansarcomavirions,
respectively,whichwasconsistent with partial protection ofadenine Ni sites by
structural proteins or genome conformation or both. The ratios of
1-methyl-adenosine to7-methylguanosineforvacciniavirus DNAs methylated in situ (0.10)
and after phenol extraction (0.14) wereless than the ratios for 4X174 and M13
DNAs (0.39 to 0.64) but considerably greater than the ratio observed with
adenovirus DNA (0.002 to0.02). The presence ofa single-stranded region(s) in
the vaccinia virusgenomewasconfirmedbySi nuclease digestionof[methyl-3H]
DNA;the released radiolabeledfractionhadaratioof0.41,compared with0.025
for the residualduplexDNA. Inadditiontothestructure-dependent accessibility
ofadenineNi, methylationofadenine N3wasseveralfold lowerin the intravirion
genomes of vaccinia virus, 4X174, and adenovirus than in the corresponding
extracted DNAs. Chemicalmethylation ofvirions and subviralparticles should
be useful for in situanalysesofspecific regionsof RNA and DNAgenomes,such
asthe sites ofproteinbinding duringvirusmaturation.
Animal virus genomes varywidely in compo-sition and structure. Different groups contain
single-stranded DNAs
(parvoviruses),
single-stranded RNAs
(picornaviruses),
double-stranded DNAs (adenoviruses), and
double-strandedRNAs
(reoviruses) (14).
Inmostcasesthe basic structures of these genomes were
es-tablished aftertheywereextracted frompurified
virionsbytreatmentwithphenol,sodium
dode-cyl sulfate, or other protein denaturants. The
importance ofcharacterizing viral genomes in situby directmethods, suchaschemical
modi-fication,has been shownbystudies of the
defec-tive parvovirus adeno-associated virus. The
DNA extracted from purified preparations of this virus is double stranded (25), but intact virions containeitherplusorminusDNAsingle
strands whichreadilyannealtoform
base-paired
duplexes upon deproteinization (18, 24).
Re-cently,weobserved thatpurifiedreovirus cores,
like adeno-associated virus (17), fluoresce red 482
whentheyarestainedwith acridine orange and
examinedunder UV
light.
Underthesamecon-ditionsphenol-extracted reovirus genome RNA
gave agreenishyellow
color,
which istypicalofdouble-stranded nucleic acids (2, 6). Since red
fluorescence is a characteristic that is usually
attributed to single-stranded polynucleotides
(16), thisfindingraised the possibility that the double-stranded genome RNA extracted from reovirus may be derived from polynucleotide
chains that exist in situascomplementary single
strands.
Dimethyl sulfate (DMS),a potent alkylating reagent, reactswithnucleic acids under neutral pH conditions(13, 27).In contrast tomanyother
alkylating agents that react only with
single-stranded nucleic acids, DMS methylates both
single-anddouble-stranded
polynucleotides;
themethylationsitesareconfined tothe base
moi-etyof the targetnucleotides,andtheyvarywith substrate strandedness. For example, DMS
on November 10, 2019 by guest
http://jvi.asm.org/
treatment of double-stranded DNA results in methylation mainly on the N7atomof guanine within the major groove and the N3 atom of adenine within the minor groove of the helix. The adenine Ni position is protected by base pairing. Insingle-stranded DNA the Ni position rather than the N3 position of adenine is readily methylated by DMS, in additiontothe guanine N7 atom (13, 19, 27). By an analogous compari-son of DMS-susceptible sites in phenol-ex-tracted reovirus genome RNA and single-stranded viral mRNA, we found that methyla-tion of the Niposition of adenine is also largely blocked in double-stranded RNA. Byusing the availability of the adenine Ni position for meth-ylation by DMS as an indicator of base pairing, we confirmedthat the reovirus genome is also
double stranded in situ. Studieswith a variety
of otherRNA and DNA viruses indicated that
methylation patterns reflect overall genome
structure,suggesting that this simple method is useful forprobing viruses and subviral particles.
MATERIALS AND METHODS
Viruses. Human reovirus type3Dearing strain was
purifiedfrominfectedmouseLcellsasdescribed pre-viously (26). Reovirus cores were prepared by
digest-ingpurified virions (2 mg/ml) with chymotrypsin (1
mg/ml)at45°Cfor30minin50mM
Tris-hydrochlo-ride buffer (pH 8.0) containing 50 mM KCI. The
resulting viruscoreswerecollected by centrifugation
at10,000xgfor15minat4°C, washed by suspension
in10mMHEPES
(N-2-hydroxyethylpiperazine-N'-2-ethanesulfonicacid) buffer (pH 7.1) containing 50 mM
KCl,centrifuged again,andfinally resuspended in 10
mMHEPES buffer (pH 7.1).
Cytoplasmicpolyhedrosis virus (CPV)wasisolated
from polyhedralinclusion bodies extracted from the midguts of infected silkworms (Bombyx mori). After
purification by successive sedimentations in sucrose
and CsCldensity gradients(3), the finalsuspension of
purified CPV was dialyzed against 10 mM HEPES
buffer(pH7.1).
Aviansarcomavirus(ASV)B77 and brome mosaic
virus (BMV) were kindly supplied by J. M. Bishop
(UniversityofCalifornia-SanFrancisco) and P. Kaes-berg (University of Wisconsin-Madison), respectively.
These viruses were also dialyzed against 10 mM
HEPES buffer (pH 7.1) and stored at
40C.
Adenovirus type 2 and vaccinia virus strain WR
werepurified from infected HeLacellsbypreviously
describedprocedures (7, 11). BacteriophageM13 and
adeno-associatedviruswerekindly provided by Grace
Ju(Roche Institute of Molecular Biology) and Barrie
Carter (National Institutes of Health), respectively.
OX174
virus and DNA were purchased from Miles Biochemicals.Viralnucleic acids. Genome RNAs were obtained
by extracting purified virions with distilled phenol
followed by ether extraction and ethanol precipitation.
To obtain DNA genomes, viruses were dialyzed
against10 mMcacodylate buffer (pH 7.0) and treated
for 30 minat 37°C with a solution containing 0.5%
sodium dodecyl sulfate andproteinase K (1mg/ml)
predigested for 2 hat37°C.Anequalvolume ofphenol
wasthen added, and after three extractionsatroom
temperature, the DNA was ethanol precipitated,
washed twice with 80%ethanol,anddissolved inwater.
Methylation of nucleic acids with
[methyl-3H]DMS.Genome RNAs (30to100,ug)wereincubated
at room temperature in reaction mixtures (0.2 ml)
containing0.1 M phosphate buffer (pH 7.0), 0.2 M
NaCl, and10mM[methyl-3H]DMS (specific activity,
200 tCi/umol;NewEngland Nuclear Corp.) (9). After
3h orlonger, the mixtureswerediluted twofold with
waterandphenolextracted. Theincorporationof
3H-labeled methyl groups into RNA was measured by
precipitating asample ofanextracted mixture in 5%
trichloroacetic acid andcountingonnitrocellulose
fil-ters. Theusual levels of incorporation were -4,000
and -6,000 cpm from 3H-labeled methylgroups per
,ugfor the extracteddouble-stranded reovirus genome
RNAs andsingle-strandedviralmRNA's,respectively.
Incorporationwasabout 20% less for RNAs
methyl-ated in reovirus cores and intact CPV. To eliminate
unreacted[3H]DMSfrom radiolabeledRNAs, reacted
sampleswereethanolprecipitated and passed through
Sephadex G-100 in 20 mM Tris-hydrochloride (pH
7.5). Theradioactive material in the excluded volume
wasethanolprecipitated,and thepurifiedRNAswere
dissolved in5mMsodiumacetatebuffer(pH6.0).
Viral DNAswerealkylatedinsituorafterphenol
extraction from virions in incubation mixtures (0.2ml)
containing 10 mM cacodylate or sodium phosphate
buffer(pH7.0),200
lCi
of[3H]DMS(0.5 mM;specificactivity,2.1 Ci/mmol; NewEnglandNuclearCorp.),
and40 to 50,tg ofextracted DNA oran equivalent
amount asvirions.Incubationwas at0°Cfor17h;in
thecaseof in situmethylation,DNAwasextractedas
described above.
Digestion of RNA and separation of
3H-methyl-labeled ribonucleosides by electropho-resis. 3H-methyl-labeledRNAs (-50,ug)in0.1ml of
5mMacetate buffer(pH6.0) were digestedto
mon-onucleotides by incubating them with 100iLg of P1
nuclease(YamasaShoyu Corp.)at37°Cfor1h.Before
enzyme treatment,duplexRNAswereheatdenatured
(100°C for 1 min and rapid chilling). P1 nuclease
digestswereneutralized byaddingTris-hydrochloride
buffer (pH 8.0) and then incubated with 8.6 U of
bacterialalkaline phosphatase (Worthington
Diagnos-tics) permlfor 90min at 37°C. Theproducts were analyzed byhigh-voltagepaperelectrophoresisin 10%
pyridine-acetate buffer (pH 3.5) containing 1 mM
EDTA(4). Todistinguishbetween7-methylguanosine
(m7G) and 1-methyladenosine
(m'A),
which werepoorly resolvedinthissystem,m7Gwasconvertedto
m7G* [i.e., the ring-opened derivative
2-amino-4-hydroxy-5-N-methyl
carboxamide-6-(N-fi-ribofurano-syl)pyrimidine] by incubation in7NNH40Hfor2h
at room temperature. Under these conditions m7G,
which hadanetchargeof+1,wasconverted
quanti-tatively to thering-opened compound, which had a
netchargeof zero;m'Awaspartially(17%)converted
tom6A. To determine thecontentofmethylated
cy-tosine,whichwas notresolvedfromm'A by
electro-phoresis, radiolabeled RNA samples that were
di-VOL. 40,1981
on November 10, 2019 by guest
http://jvi.asm.org/
gested with P1 nuclease and phosphatase were ana-lyzed by paper chromatography in n-butanol-NH40H
(100:1). Polyadenylic acid, polycytidylic acid, and
polyguanilic acid were also 3H methylated and
ana-lyzed inparallel. TheRfvalues form'A,m6A,
meth-ylated cytidine, and m7G were 0.2, 0.3, 0.44, and 0.04, respectively.
Resolutionof'H-methyl-labeledDNAbases by
columnchromatography.After chemical
modifica-tionby[3H]DMS,DNAwasprecipitated with ethanol,
washed two times with80% ethanol, dried, and
hydro-lyzed in 7 NHCl04 for 1 hat 1000C.Theresulting
digest was analyzed by chromatography on a Dowex
50 x 4column (0.4 by 20 cm); 0.5-ml fractions were
eluted with0.3Mammoniumformate at a flow rate of
4.5ml/h (20).The marker compounds used included
m7G,m'A,and m3A (Vega Fox Chemicals), aswell as
3-methylcytosine and5-methylcytosine (PL
Biochem-icals).
Sinucleasedigestion. Phenol-extractedvaccinia
virus DNA was methylated by exposure to[3H]DMS
and waspurifiedby gel filtration on a Sephadex G-100
column (0.8by25cm) in50mMsodium acetate buffer
(pH6.0)containing 7Murea.The DNAwas
precipi-tated with ethanol, washed two times with 80%
ethanol, dried, and dissolved inwater. For each
en-zymedigestion a reaction mixture (0.5 ml) containing
30mMsodiumacetatebuffer(pH4.5), 0.3 MNaCl, 5
mMZnCl2,[methyl-3H]DNA,and 200 U ofSinuclease
(MilesBiochemicals) wasincubated for2h at370C.
The Si-resistant and -sensitive DNA components
were separatedby gel filtrationon aSephadex G-50
column(0.8by25cm) in10mMammonium
bicarbon-ate(pH 7.6). Fractions(0.5ml) containingthepeaks
ofradioactivity were pooled and lyophilized. Acid
hy-drolysis andanalysisofmethylated bases by Dowex
columnchromatographywereperformedasdescribed
above.
RESULTS
Methylation
of RNAgenomes.Tocharac-terize the sites of DMS
alkylation
indouble-stranded and single-stranded RNAs, genome
RNA thatwas
phenol
extracted frompurified
reovirus and viral mRNAsynthesized invitroby
reovirus cores were incubated with 10 mM
[methyl-3H]DMS
as described above. Underthese conditions
incorporation
of 3H-labeledmethylgroups into virus RNAs was nearly linear
for3to4hand then continuedat aslowerrate.
Thereacted RNAswereisolated from the
incu-bation mixtures
by
Sephadex
G-100gel
filtra-tion, and the
3H-methylated
nucleosides oftheseRNAswere
analyzed by
high-voltage
paperelec-trophoresis after successive
digestions
with P1nuclease and
alkaline
phosphatase (4). Thedi-gests of both the double-stranded genomeRNA
(Fig. 1A) and the single-stranded viral mRNA
(Fig. 1B)
yielded
most of theradioactivity
asmaterial that
migrated
inpositions
correspond-ing
to thepositions
of marker ribonucleosidesmG and
m1A.
Bothm7Gandm1A
contained ax
_
-S
5 ~~~~~~~B
ee
2~ h
E._
5
o
O, ,
5
C C
10FRACTION20
3
FIG. 1. Analysis of methylated residues in RNAs.
(A) Reovirus double-stranded genome RNA was
phenol extracted from virions, purified by Sephadex
G-100column chromatography, and chemically
meth-ylated by exposure to [methyl-3H]DMS, as described
in thetext. (B)Reovirus mRNA synthesized in vitro
by using virus-associated RNA polymerasewas
meth-ylated asdescribed above. The RNA preparations (50
ag in(A)and (B) weredigested withP1nuckeaseand
alkaline phosphatase, and theresulting3H-labeled
nucleosides were analyzed by high-voltage pagper
electrophoresis. (C)and(D)The materialin fractions
2 through 6 of(A) and(B), respectively, was eluted,
treated with 7 NNH4OH,andre-electrophoresed.
positivechargeunder these conditions (pH3.5),
and these two compounds migrated together
toward the cathode. Toidentifythe
3H-methyl-ated residue(s), fractions 2 through 6 (Fig. 1A
and B) were eluted from the paper inwaterand analyzedbyelectrophoresis aftertreatmentwith 7 M
NH4OH
for 2hatroomtemperature. These conditions degraded m7G completely tothe ring-opened, uncharged derivative m7G* and also partially convertedm'A
to m6A (4, 8). As Fig.10
shows, the predominant 3H-methyl-labelednucleoside in DMS-treated reovirus
double-stranded RNA was m7G (97%), whichwas con-verted by alkali treatment to the ring-opened derivative
m7G*.
A small portion of theon November 10, 2019 by guest
http://jvi.asm.org/
[image:3.497.291.442.63.365.2]METHYLATION OF VIRAL GENOMES IN SITU 485
activity (3%) migrated in the original position (Fig. 1C, fractions 3 and 4). This minor 3H-la-beled compound presumably was m1A since it comigrated with m6A after alonger treatment
with 7 MNH4OH(datanotshown).
Incontrast tothedouble-stranded RNA, the
3H-methyl-labeled materialderived from
reovi-rus single-stranded mRNA was resolved into three prominent, distinct peaks by electropho-resis aftertreatmentwith P1nuclease,
phospha-tase,and alkali(Fig. 1D). Theradioactivitynear
the origin (fractions 20 through 24) was m7G*
(61% of the total), and the small peak of radio-activity (7%) in fractions 15 and 16 corresponded
to m6A. The radioactive material (31%) that comigratedwithauthenticm'Atoward the cath-ode consisted of m1A andmethylated cytidine. The m'A was converted to m6A by prolonged alkline treatment (87% conversion after 18 h [datanotshown]). Under thesameconditions,
morethan75% ofthemethylated cytidine
deriv-ative, whichwasthe mainproduct inchemically
methylated polycytidylic acid, remained
un-changed. The methylated cytidine derivative did
notcomigrate with marker3-methylcytidineor
5-methylcytidine during paper electrophoresis atpH 3.5orduringpaperchromatographyin
n-butanol-NH4OH (100:1). Althoughthe methyl-ated cytidine derivative was not identified, its contributiontothemethylated nucleoside com-position of the RNAswasassessedby chroma-tographyasdescribed above(Table 1). Like the otherminorradioactivecomponents, the
3H-la-beledmaterial in Fig. 1D, fractions9 and10, in thepositionof5-methylcytidinewasnot identi-fied.
These data demonstratedaclear difference in the chemical methylation patterns of double-stranded and single-stranded RNAs. Although
the plus strands of reovirus double-stranded
RNAareapparently identical insequencetothe corresponding single-stranded viral mRNA's (12), thepresenceofthesestrands inbase-paired duplexes limits methylation almost exclusively
to the guanosine N7 position, compared with additional accessible sites on adenosine and cytidine residues in the mRNA. The ratio of m1Atom7G fordouble-strandedRNAwas0.03
(i.e., 14-fold less than the ratio obtained for mRNA). Sincetheextentofguanosine methyl-ationwasnearly identical in bothtypesofRNA
(5% ofthe totalguanosineresidues), the 1.7-fold increase in methylation of mRNA compared
withgenomeRNAresulted fromthe differential incorporation ofmethyl groups intoadenosine residues and,toasmaller extent, cytosine
resi-duesinthesingle-stranded molecules. The
[image:4.497.249.446.83.211.2]lim-itedextentofmethylation presumablywasdue
TABLE 1. Methylation of virus RNAs in situ'
Total % Ofradioactivity in amtof thefollowing
prod-radio- ucts: Ratio
Sample activity ofm'A
ana- mA + 7 to
m7G
lyzed mC mC m7G
(cpm) m
Reovirus double- 3,345 3.0 <0.1 97.0 0.03 strandedRNA
Reovirus mRNA 5,827 38.6 12.0 61.4 0.43
Reovirus cores 3,808 7.0 93.0 0.08
CPVvirions 11,298 3.4 96.6 0.04
BMVvirions 21,467 25.1 5.4 74.9 0.26 ASVvirions 1,593 29.3 4.8 70.7 0.35 aPhenol-extracted reovirusdouble-strandedgenome RNA, reovirus mRNA, reovirus cores, and purified virions were labeledbyincubatingmaterialequivalent to 30 to 100 ug of RNA with[methyl-3H]DMSasdescribed inthe text. After purification,the extents oflabeling were asfollows:reovirus double-stranded,3,100cpm/g,reovirus mRNAsynthesized invitro, 6,950 cpm/llg; reovirus cores, 4,300cpm/pg;CPV, 4,825 cpm/pg, BMV, 4,474cpm/g,and ASV, 880cpm/yg. Sampleswereanalyzedformethylated nucleosidesby paper electrophoresis after P1 nuclease andphosphatasedigestion andquantitativeconversion ofm7Gtom7G in 7 NNH40H. mAincludedm'A and itsalkalinedegradationproductm"A, which were separated by paperelectrophoresis.The methyl-ated cytidine derivative (mC) whichcomigrated with m'A during electrophoresiswasquantitatedby paper chromatog-raphy of samples digested with P1 nuclease and phosphatase asdescibed in the text.
to a
decreasing concentration
ofDMS,
whichwascaused
by evaporation
anddegradation
dur-ing the modification reaction.
Methylation
by DMSwas nextusedwithge-nome RNAs in virus
particles.
Reovirus coreswereprepared from
purified
virionsby
chymo-trypsin
digestion (1).
Another member of theReoviridae,
insectCPV,
wasusedasintact,
pur-ified virions. BMV and ASV werealso treated.
After the
particles
wereincubatedwith[methyl-3H]DMS
for24hat roomtemperaturetoobtainmaximum
modification,
viralRNAs
werephenol
extracted and freed ofresidual
alkylating
agentby
Sephadex
G-100gel
filtration and ethanolprecipitation.
Thepurified
RNAswereanalyzed
fortheir
methylated
nucleosidecompositions bypaper
chromatography
andelectrophoresis
afterdigestion
with P1 nuclease and alkalinephos-phatase.Ineachcasethe
electrophoretic
patternwassimilartothe
pattern
shown inFig. 1A;
i.e.essentially
all of the3H-labeled ribonucleosidesmigrated in fractions2
through
6, thepositions
ofm7G and
m'A
markers. The material fromeach
digest
waselutedfrom the paper, treatedwith alkali to convertm7G to the
ring-opened
structure, and
reanalyzed by
electrophoresis.
The resultswithreoviruscores
(Fig. 2A)
couldbe
compared
directly
withthe resultsobtainedwithdeproteinized reovirus genome RNA
(Fig.
1C) since
equivalent
amounts of nucleic acidVOL. 40,1981
on November 10, 2019 by guest
http://jvi.asm.org/
(W)
2
x
0 2
'~8
cn 6
4
2
1.0 D
0.5-0 to 20
[image:5.497.77.243.62.482.2](0 FRACTION
FIG. 2. Methylation ofgenomeRNAs in situ.
Pur-ified particles were methylated by using
[methyl-3HJDMS,asdescribedin the text.RNAswerephenol
extracted andpurified by gel filtration, andsamples
weredigestedtothe nucleosidelevel withPlnuclease
andphosphatase. m7Gwasconverted tom7G*, and
m'Awaspartiallyconvertedtom6Abyexposureto7
NNH40H beforepaperelectrophoresis. (A)Reovirus
cores. (B)CPV.(C)BMV. (D)ASV.
were reacted with DMS. The methylated
nu-cleosidepatternswerealmostidentical forRNAs
modified in situ and after extraction. In each
case more than 90% of thealkylation occurred
at the N7 position of guanine. Similar results
wereobtained withintact CPV(Fig.2B). Thus, genome RNAs packaged within reovirus cores
or intact CPV yielded patterns of chemical
methylation similartothose obtained with
de-proteinized double-stranded RNAs.
Further-more,theextentsofguanosine methylation were
similar (-5% oftheguanosine residues) for
reo-virusand CPV genomeRNAsalkylatedin situ
andafter extraction. Incontrast to the two
rep-resentative members of the Reoviridae,
meth-ylation of BMV and ASV resulted in the
for-mation of significantly higher levelsof
methyl-ated adenosine relative to methylated
guano-sine, whichwas consistent with thepresence of
single-stranded genomes in these two viruses
(Fig. 2C and D). It should be notedthatthere
was no apparent disruption of virus particles
under the conditions whichwe used forchemical
methylation and that the mRNA
guanylyltrans-ferase and nucleotide phosphohydrolase
activi-ties associated withpurified reoviruscoreswere
notaffected by the DMStreatment (30).
The methylated nucleoside compositions of
the RNAsamples are summarized in Table 1.
Thediagnostic ratios of
m'A
tom7Gwere0.03,0.08,and0.04 forphenol-extracted reovirus
ge-nome RNA, intact reoviruscores, and CPV
vi-rions,respectively. These valueswerewellbelow
thevalue of0.43 obtained with reovirus
single-stranded mRNA. The genome RNAs of BMV
and ASV yielded ratios of0.26 and 0.35 (i.e.,
values which were intermediate between the
value for double-stranded reovirus RNA [0.03]
and the value for
single-stranded
viral mRNA[0.43]). The viral genomes of BMV and ASV
apparently
contained some adenosine residuesthat were inaccessible to the
methylating
re-agent, possibly dueto shielding by
intramolec-ular base
pairing
or intermolecular structures(23) in the case of ASV
diploid
RNA. Alsoassociation of thegenomeRNAs with structural
proteins in virions
probably
accounted forsomeprotectedsites (10, 21).
Analyses
ofchemically methylated
viralDNAs. To assessthe generality of using DMS
methylation
patterns todetermine viralgenomestrandedness, weexposed a
variety
of DNAvi-ruses tothe radiolabeled
alkylating
agent. Nu-cleic acids wereextracted and acidhydrolyzed,and the
resulting
baseswereanalyzed by
Dowexcolumn
chromatography.
Wecompared
viral ge-nomes methylated in situ and viral genomesafterextraction from virions. AsFig.3Ashows,
adenovirusDNAwas
methylated mainly
ongua-nineresiduesatthe N7position
and,
to alesserextentontheadenine N3
position.
Theseresultsand the
striking
absence of1-methyladenine
(m'A)
incontrast totheprominent
m1Apeak
inDMS-reacted
4X174
DNA (Fig.3B andD) areconsistent with a
completely
double-strandedstructure for DNA extracted from adenovirus.
Interestingly,
DMS treatmentof intact adeno-J.on November 10, 2019 by guest
http://jvi.asm.org/
virions yieldedDNA that containedasignificant
m'A fraction (Fig.30). Thissuggestedthat the genome in situ contained destabilized regions whichwereabsent from thedeproteinized DNA.
The methylation patterns of phenol-extracted and intravirion
OX174
DNAswerealsonoticea-bly different. Therewas anincreased
accessibil-ity of the adenine N3 sites after deproteinization (Fig. 3B), suggesting that there was some pro-tective effect of the virion proteins, either
di-rectly by shieldingorindirectly byalterationof thegenome confonnation. Similarresultswere obtained with M13, another single-stranded DNA bacteriophage (Table 2). As this table shows, in each of the genomes examined the accessibility of the N3 atom of adenine was enhancedinpurifiedDNA.
In theparvovirus,adeno-associatedvirus,the relativeamounts of m1A andm3A confirmedthe predominantly single-strandednatureof the
ge-E
C~~~~~~~~~~~~~~~~~~~~~~~~~~C
50,2-
B
D 50300
120
4
-~~~~~~~~~~~~~~~~~~~~~~~~~-2~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~1
20 40 60 100 20 40 60 100
[image:6.497.70.432.193.437.2]FRACTION
FIG. 3. Separation of31H-methyl-labeled bases byDowex column chromatography. ViralgenomeDNAs
weremethylated,acidhydrolyzed, andanalyzedasdescribed in thetext.AdenovirusDNAwasmethylated
after extraction (A) and in situ(B). X174 virus DNAwasmethylated afterextraction(C)and in situ(D).A26o, Absorbanceat260nm.
TABLE 2. Methylation ofvirusDNAsa
Totalamtof radioac- % Ofradioactivityin the follow- Rtio
Sample tivity analyzed per 40 ing products:
Agof DNA(cpm)
m1A
m3A m7Gm1A/m7G
m3A/m7GAdenovirus DNA 66,396 0.2 7.6 92.0 0.002 0.082
Adenovirus 62,142 2.2 0.8 97.0 0.022 0.008
4X174virusDNA 143,969 26.4 6.1 67.5 0.391 0.090
4X174 113,327 35.7 1.7 62.6 0.570 0.027
M13 virusDNA 219,686 29.7 6.0 64.3 0.461 0.092
M13 virus 154,271 38.1 4.5 58.9 0.646 0.076
Adeno-associatedvirus DNA 45,976 7.9 7.4 84.7 0.093 0.087
Adeno-associatedvirus 40,168 36.8 1.1 62.1 0.593 0.017
Vaccinia virus DNA 23,558 11.7 6.7 81.6 0.143 0.082
Vaccinia virus 22,106 8.9 1.5 89.6 0.100 0.017
aChemicalmethylationofviriongenomesin situorafter phenol extractionwas
performed
as described in thetext.3H-methyl-labeled baseswereanalyzed by Dowex chromatography. 40,1981
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.497.59.451.506.635.2]AND FURUICHI
nome in virions and the duplex structure of extracted DNA. A high proportion of m1A was
obtained inDNAfrom DMS-treatedintact
vir-ions; isolated adeno-associated virus DNA
yieldedanincreased amount ofm3Aand a low
percentage of
m1A
(Table 2).Vaccinia virus DNA exposed to DMS either
in the virionor after phenol extraction yielded
consistently high levels ofm1A compared with
the amounts obtained with adenovirus
double-stranded DNA (Table 2). The higher ratio of
m'A
tom7Gwasapparentlynotdue tocontam-inating RNA or single-stranded DNA since the
same value (0.14) was obtained after RNase
treatmentand repurificationof
high-molecular-weight vaccinia virus DNA. This was
accom-plished bytreatmentwith pancreaticRNase (10
,ug/ml,
370C,
15min) followed by sedimentation(SW27rotor, 52,000 x g, 3 h,
2000)
through a 5to20%sucrosegradient(0.05 MTris buffer, pH
8, 1M NaCl, 0.001 MEDTA, 0.15% Sarkosyl)
onto a 50% glycerol cushion. (Under the same
conditions
4X174
DNA remained nearthe topof the gradient.) These results suggested that
vaccinia virusgenomesin situ containunpaired
adenine residues that retain DMS-accessible
Ni
sites after extraction, possibly dueto their
pres-ence in
single-stranded
or readily denaturablestretches
(for
example,
in duplex regionsdis-torted by superhelical twisting or in the loop
structures attheDNA termini [5]). To test for the presence ofsingle-stranded regions,
depro-teinized vaccinia virus DNA was radiolabeled
with [methyl-3H]DMS, treated with
single-strand-specific nucleaseSi, and filtered through
Sephadex G-100. Most of the radiolabeled DNA
wasresistanttoSidigestion andwasrecovered
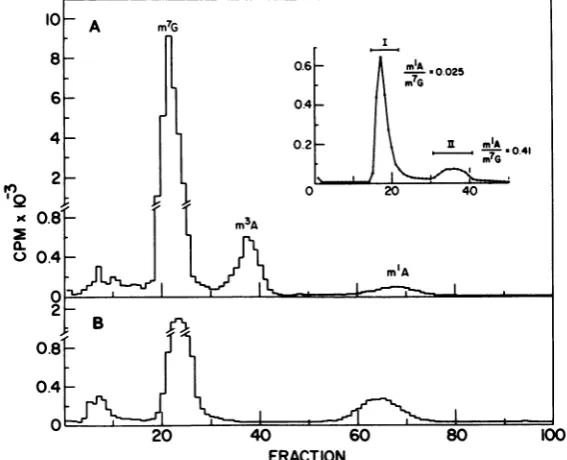
in the excluded volume (Fig. 4, inset, peak I).
However, approximately 20% of the
radioactiv-itywasreleasedaslow-molecular-weight
mate-rial thatwasretardedduring gel filtration (peak
II).Peaks Iand IIwerepooledseparately, con-verted totheirconstituent bases by acid
hydrol-ysis, andanalyzed by Dowex column
chromatog-raphy. Peak I contained radiolabeled m3A and
alow level of
m'A,
in additiontothe mainpeakof7-methylguanine (Fig. 4A). PeakIIyieldeda
higher proportionof
m'A
andnodetectable m3A(Fig. 4B).The ratios of
m'A
to7-methylguanineforpeak I (0.025) and peak II (0.41) were
con-sistent with the values expected for
double-stranded and single-stranded DNAs,
respec-tively.
DISCUSSION
Alkylation of nucleic acids by exposure to
DMS at a neutral pH is an extremely useful
A m7G
-L m3A
0.6 0.4
0.2
I
.0.025
m1A
0---
4m7G.0.41
-%I
0 20 40
mIA
B J-~
20 40 60 80 100
FRACTION
FIG. 4. Chromatography ofanSI nucleasedigest of 3H-methylatedvaccinia virusDNA. Vaccinia virus
DNA (45pg) wasmethylated and separated fromunreacted[3H]DMS by Sephadex G-100filtration. The
DNAwasdigestedwith SInuclease andfilteredthrough SephadexG-50asdescribed in the text(inset).The
Si nuclease-resistantDNAfraction (peak I)and the included materialfromthedigestedDNA(peak II)were
pooled separatelyand lyophilized.Methylatedbases obtainedbyacidhydrolysiswereseparated byDowex
columnchromatography. (A)PeakI. (B) peakII.
I0
8
6
41
0.8
0.4
)Q
x
a.
C-0.8
0.4
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.497.111.395.376.606.2]METHYLATION OF VIRAL GENOMES IN SITU 489
procedure for molecular structure studies and
has provided the basis for rapid chemical se-quencing of DNA (15) and,morerecently, RNA (22).We used this method forananalysis of the
structures of viral genomes. Both RNA and
DNAweremodified chemically in situ by
expos-ing virus particles to [methyl-3H]DMS. Al-though onlya small fraction of thetotalbases were modified under the experimental
condi-tions whichweused, the availability ofareagent
withahigh specific radioactivity facilitated the
studies.
Methylation occurred mainlyonpurine
resi-dues. In eachcasem7Gwasthemajor 3H-labeled
product, together with m1A, minoramountsof methylated cytosine, and (in DNA) m3A. As previously observed for DNA (13), single-stranded and double-single-stranded RNAs could be distinguished onthe basis oftheaccessibility of
the adenine Ni positions. In double-stranded molecules, hydrogen bonding of adenine-uracil base pairs shielded the adenine Ni sites. Con-sequently, the yield of m'A from phenol-ex-tracted reovirusgenomeRNAwasconsiderably
lower than the yield obtained with single-stranded RNA. Our results confirmed that the RNA genomes of human reovirus type 3 and insect CPV are double stranded in situ. The genomeRNAsof intact BMV and ASVyielded relatively high amounts ofradioactivity in the
m1A fraction, as expected for single-stranded RNAs, but the valueswereless than thevalue
observed for reovirus mRNA. Since reovirus mRNA contains extensive amountsof intramo-lecular structure (28), which tendsto decrease the availability of adenine Ni sites, it seems
reasonabletosuggestthat the loweraccessibility of adenine Ni atoms in the single-stranded RNAs ofBMVand ASVwasduetoprotection by structural proteins. Clearly, it should be pos-sible by comparative sequence analyses of ge-nomes modified in situ and after deproteiniza-tion(10, 21, 23)toestablish whetherdiminished adenine Ni accessibility is primarily due to
shielding byassociated proteinsordue to
con-fornational effects.
The potential of this approach for probing specific regionsofaltered structure inviral
ge-nomes is suggested by our preliminary results
with vaccinia virus DNA. Obviously, resolution
canbeimproved markedly by using cloned DNA
fragments (29), as has been done with simian
virus40origins of DNA replication andT anti-gen (R. Tjian, Curr. Top. Microbiol. Immunol., in press). Extension of the DMS methylation methodtostudies of RNA viruses and subviral nucleoprotein complexes should allow identifi-cation ofbinding sites of structuralproteins and virion-associatedenzymes.
ACKNOWLEDGMENT
We thank Alba LaFiandra for purified reovirus.
LITERATURE CITED
1. Banerjee,A.K., and A. J.Shatkin.1970.Transcription
in vitroby reovirus-associated ribonucleic acid-depend-entpolymerase. J. Virol. 6:1-11.
2.Darzynkiewicz, Z.,F.Traganos,T.Sharpless,and M. R.Melamed. 1975.Conformation of RNA in situ as studiedbyacridine orange staining and automated cyto-fluorometry. Exp. Cell Res. 95:143-153.
3. Furuichi,Y.,and K.-I. Miura. 1973. Identity of the 3'-terminal sequences in ten genome segments of silkworm cytoplasmic polyhedrosisvirus.Virology55:418-425. 4.Furuichi, Y., M. Morgan, S. Muthukrishnan, and
A. J.Shatkin. 1975. Reovirus messenger RNA con-tains a methylated, blocked 5'-terminal structure:
7mG5ppp5'GmpCp....Proc.Natl.Acad. Sci.U.S.A. 72:
362-366.
5. Geshelin, P., andK. L Berns. 1974. Characterization andlocalizationof thenaturally occurring cross-linksin vaccinia virus DNA. J.Mol. Biol. 88:785-796. 6. Gomatos, P. J., I.Tamm,S.Dales, andR. M.
Frank-lin.1962.Reovirus type3:physicalcharacteristics and interaction with Lcells.Virology17:441-454. 7. Green, M.,and M.Pina. 1963.Biochemicalstudies on
adenovirus multiplication. IV. Isolation, purification, and chemical analysis of adenovirus. Virology 20:199-207.
8. Hail, R. H. 1971. Specific procedures for isolation of
modifiednucleosides,p. 232-253. In The modified nu-cleosides in nucleic acids. ColumbiaUniversity Press, New York.
9. Jeng,Y.-H.,andR.H. Doi. 1975. Newtransfer ribonu-cleic acidspeciesduringsporulationofBacillussubtilis. J. Bacteriol.121:950-958.
10.Johnson, A., B. J. Meyer, and M. Ptashne. 1978. Mechanismof action of thecroprotein ofbacteriophage X. Proc.Natl. Acad.Sci.U.S.A.75:1783-1787.
11. Joklilk,W. K.1962.Thepreparationand characteristics
of highly purified radioactively labelled poxvirus. Biochim.Biophys.Acta 61:290-301.
12. Joklk,W. K. 1974.Reproduction of Reoviridae, p.
231-334. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 2. Plenum Press, New York.
13. Lawley, P. D., and P.Brookes.1963. Further studies
onthealkylationof nucleic acids and their constituent nucleotides. Biochem. J.89:127-138.
14. Luria,S.E.,J. E.Darnell, Jr.,D.Baltimore,and A.
Campbell.1978.Generalvirology,3rd ed. JohnWiley &Sons, Inc.,New York.
15. Maxam,A.M.,and W.Gilbert1977.Anewmethod for
sequencingDNA. Proc.Natl. Acad. Sci. U.S.A. 74:560-564.
16. Mayor,H.D.,and A. R. Diwan.1961.Studiesonthe acridineorangestainingoftwopurifiedRNA viruses:
poliovirusand tobacco mosaic virus.Virology14:74-82.
17. Mayor,H.D.,and J. LMelnick.1966.Small deoxyri-bonucleicacid-containing virumses(picodnavirusgroup). Nature(London)210:331-332.
18. Mayor,H.D.,K.Torikai,J.LMelnick,and M.
Man-del.1969.Plus and minussingle-strandedDNA sepa-rately encapaidatedinadeno-associated satellite viri-ons. Science166:1280-1281.
19. Mirzabekov,A.D.,and A. M.Kolchinsky. 1974. Lo-calizationofsomemolecules withinDNAgroovesby methylation of theircomplezeswithdimethylsulphate. Mol. Biol.Rep. 1:379-384.
20. Mirzabekov,A.D.,D. F.San'Ko,A.M.Kolchinsky, and A. F.Melnikova.1977. Proteinarrangement in the DNAgroovesinchromatin and nucleoprotamine in vitro and in vivo revealed by methylation. Eur. J.
VOL. 40,1981
on November 10, 2019 by guest
http://jvi.asm.org/
Biochem.75:379-389.
21. Ogata, R.T.,andW. Gilbert.1978. Anamino-terminal fragment of lacrepressorbindsspecificallytolac
op-erator.Proc.Natl. Acad. Sci. U.S.A.75:5851-5854. 22. Peattie, D. A.1979.Directchemical method for
sequenc-ing RNA. Proc.Natl. Acad. Sci. U.S.A. 76:1760-1764. 23. Peattie, D. A., and W. Gilbert. 1980.Chemicalprobes
forhigher-order structurein RNA. Proc. Natl. Acad. Sci. U.S.A. 77:4679-4682.
24. Rose,J. A., K. L. Berns,M. D. Hoggan, andF. J.
Koczot. 1969. Evidence forasingle-stranded
adenovi-rus-associated virusgenome:formationofaDNA
den-sity hybridonreleaseof viral DNA. Proc. Natl.Acad. Sci. U.S.A. 64:863-869.
25. Rose,J.A., M. D. Hoggan, and A. J. Shatkin.1966.
Nucleic acid fromanadeno-associated virus: chemical andphysical studies. Proc.Natl.Acad. Sci. U.S.A. 56:
86-92.
26. Shatkin, A. J., and A. J. LaFiandra.1972. Transcrip-tion by infectious subviral particles of reovirus. J. Virol. 10:698-706.
27. Singer, B. 1975. The chemical effects of nucleic acid alkylation and their relationtomutagenesisand
carci-nogenesis. Prog.Nucleic Acid Res.Mol. Biol.
15:219-284.
28. Warrington, R.C., C. Hayward, and A. M. Kapuler. 1973. Conformational studies of reovirus single-stranded RNAs synthesized in vitro. Biochim. Biophys. Acta 331:231-242.
29. Wittek, R., andB.Moss.1980.Tandemrepeatswithin the inverted terminalrepetition of vaccinia virus DNA. Cell21:277-284.
30. Yamakawa, M., Y. Furuichi, K. Nakashima, A. J. LaFiandra, and A. J. Shatkin.1981.Excess synthesis of viral mRNA 5'-terminaloligonucleotides by reovirus transcriptase. J. Biol. Chem. 256:6507-6514.