Copyright © 2002, American Society for Microbiology. All Rights Reserved.
CCR5 and CXCR4 Usage by Non-Clade B Human Immunodeficiency
Virus Type 1 Primary Isolates
Daniah A. D. Thompson, Emmanuel G. Cormier, and Tatjana Dragic*
Albert Einstein College of Medicine, Department of Microbiology and Immunology, Bronx, New York 10461
Received 29 August 2001/Accepted 30 November 2001
CCR5 and CXCR4 usage has been studied extensively with a variety of clade B human immunodeficiency virus type 1 (HIV-1) isolates. The determinants of CCR5 coreceptor function are remarkably consistent, with a region critical for fusion and entry located in the CCR5 amino-terminal domain (Nt). In particular, negatively charged amino acids and sulfated tyrosines in the Nt are essential for gp120 binding to CCR5. The same types of residues are important for CXCR4-mediated viral fusion and entry, but they are dispersed throughout the extracellular domains of CXCR4, and their usage is isolate dependent. Here, we report on the determinants of CCR5 and CXCR4 coreceptor function for a panel of non-clade B isolates that are responsible for the majority of new HIV-1 infections worldwide. Consistent with clade B isolates, CXCR4 usage remains isolate dependent and is determined by the overall content of negatively charged and tyrosine residues. Residues in the Nt of CCR5 that are important for fusion and entry of clade B isolates are also important for the entry of all non-clade B HIV-1 isolates that we tested. Surprisingly, we found that in contrast to clade B isolates, a cluster of residues in the second extracellular loop of CCR5 significantly affects fusion and entry of all non-clade B isolates tested. This points to a different mechanism of CCR5 usage by these viruses and may have important implications for the development of HIV-1 inhibitors that target CCR5 coreceptor function.
Coreceptors mediate human immunodeficiency virus type 1 (HIV-1) fusion and entry into CD4⫹cells (4, 18). A number of
CC and CXC chemokine receptors, belonging to the seven transmembrane G protein-coupled receptor family, have been shown to act as HIV-1 coreceptors in vitro (39, 41). However, CCR5 and CXCR4 are the major HIV-1 coreceptors in vivo (40, 41). CCR5 is the principal coreceptor for HIV-1 R5 vari-ants that are sexually transmitted and persist within the ma-jority of infected individuals (2, 15, 25, 30, 31; P. Garred, J. Eugen-Olsen, A. K. Iversen, T. L. Benfield, A. Svejgaard, B. Hofmann, et al., Letter, Lancet349:1884, 1997). The appear-ance of R5X4 and X4 variants that use both CCR5 and CXCR4 or just CXCR4, respectively, signals accelerated CD4⫹T-cell loss and disease progression (12, 34). The
phe-notypic switch from R5 to X4 viruses occurs in about 40% of infected individuals and only after several years of infection (16, 29).
Coreceptor usage and switching have been analyzed most extensively for clade B isolates, which predominate in North America and Western Europe (1, 3). The biological and mo-lecular properties of non-clade B viruses, which now cause the vast majority of new HIV-1 infections worldwide, remain largely unknown. Coreceptor specificity of non-clade B HIV-1 isolates is only beginning to be characterized (5, 6, 10, 35–38). Available data suggest that clade C isolates, which predomi-nate in sub-Saharan Africa and Asia, are by and large R5, even when derived from patients with advanced AIDS (5, 6, 10, 35). Not enough isolates from the other HIV-1 clades have been
analyzed to draw firm conclusions about coreceptor usage pat-terns.
The determinants of CCR5 and CXCR4 function have been analyzed extensively for clade B isolates (18). Negatively charged and tyrosine residues dispersed throughout the extra-cellular domains of CXCR4 are important for viral fusion and entry, but each HIV-1 X4 isolate seems to be dependent on a slightly different subset of amino acids (18). In contrast to X4 isolates, all R5 isolates characterized to date interact with the same cluster of negatively charged and sulfotyrosine residues in the CCR5 amino-terminal domain (Nt) (18). We have shown recently that the CCR5 Nt specifically associates with residues in the C4/V3 stem region of gp120 (13, 14). Our data furthermore suggest that the V3 crown binds to another, hith-erto-unidentified region of CCR5. All of these studies were performed with clade B isolates, and very little is known about how the envelope glycoproteins of non-clade B viruses interact with CXCR4 and CCR5.
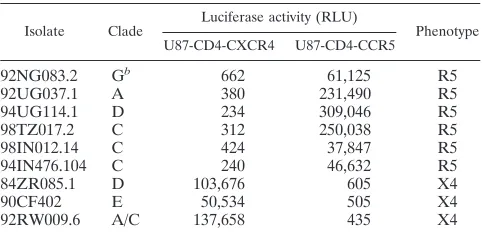
Here, we studied CCR5 and CXCR4 usage by nine non-clade B, primary, full-length molecular clones previously de-scribed by Gao et al. (21–23). We subcloned the envelope glycoprotein genes of these viruses into the SV7D expression vector and used them to pseudotype luciferase-expressing (NLluc⫹env⫺) virions as previously described (19, 24, 32). We were able to generate infectious pseudotyped virions with 9 of the 12 envelope glycoprotein genes. The coreceptor specificity of these envelope glycoproteins was determined by infecting either U87-CD4-CCR5 or U87-CD4-CXCR4 cells with 100 to 200 ng of the p24 of various viral pseudotypes per ml. Lucif-erase activity (in relative light units [RLU]) in cell lysates was determined 48 h postinfection. Six of the nine isolates exclu-sively used CCR5, whereas the other three excluexclu-sively used CXCR4 (Table 1). No dually tropic isolates were observed. More extensive studies of coreceptor usage by non-clade B * Corresponding author. Mailing address: Albert Einstein College
of Medicine, Department of Microbiology and Immunology, 1300 Morris Park Ave., Bronx, NY 10461. Phone: (718) 430-3282. Fax (718) 430-8711. E-mail: tdragic@aecom.yu.edu.
3059
on November 8, 2019 by guest
http://jvi.asm.org/
isolates will have to be performed in order to determine whether the absence of dual tropism is a significant character-istic of these viruses.
Alanine substitutions were introduced into CCR5 and CXCR4 by PCR-based site-directed mutagenesis, as described previously (19, 24, 27, 32). We tested the ability of the core-ceptor mutants to mediate entry of the different non-clade B HIV-1 isolates into U87MG-CD4 cells, a human neuronal cell line that does not express CCR5 or CXCR4 (19, 24, 27, 32). Briefly, cells were transfected by lipofection with wild-type or mutant coreceptor genes and then infected with 100 to 200 ng of the p24 of various pseudotyped NLluc⫹env⫺viruses per ml as described previously (19, 24, 27, 32). Luciferase activity (in RLU) was measured in cell lysates 48 h postinfection. All coreceptor molecules used in this study had a nine-residue hemagglutinin tag as a carboxy-terminal extension to allow detection by dot blotting with an anti-hemagglutinin monoclo-nal antibody (BAbCO, Richmond, Calif.). Expression levels of mutant and wild-type coreceptor proteins were determined in
FIG. 1. Viral entry mediated by CXCR4 mutants. U87MG-CD4 cells transiently expressing wild-type CXCR4 or CXCR4 with alanine substitutions were infected with NLluc⫹env⫺reporter viruses pseudotyped with different non-clade B X4 envelope glycoproteins. The name and clade of each isolate are indicated in the first two columns and the mutated residues are indicated in the top row of each panel. Luciferase activity (RLU) was measured 48 h postinfection and standardized for CXCR4 expression levels (IDV). The coreceptor activity of each mutant is expressed as a percentage of wild-type coreceptor activity, calculated with the formula [(mutant RLU⫼wild-type RLU)⫼(mutant IDV⫼wild-type IDV)]
[image:2.587.43.284.92.209.2]⫻100% Shading indicates values lower than 20%. All values are means of three independent experiments, each performed in quadruplicate. Standard deviations were not more than 20% for each data set and are not shown for the sake of clarity.
TABLE 1. Coreceptor usage by non-clade B HIV-1 molecular isolatesa
Isolate Clade Luciferase activity (RLU) Phenotype U87-CD4-CXCR4 U87-CD4-CCR5
92NG083.2 Gb 662 61,125 R5
92UG037.1 A 380 231,490 R5
94UG114.1 D 234 309,046 R5
98TZ017.2 C 312 250,038 R5
98IN012.14 C 424 37,847 R5
94IN476.104 C 240 46,632 R5
84ZR085.1 D 103,676 605 X4
90CF402 E 50,534 505 X4
92RW009.6 A/C 137,658 435 X4
aU87-CD4-CCR5 and U87-CD4-CXCR4 cells were infected with NLluc⫹env⫺virions pseudotyped with the envelope glycoprotein derived from nine non-clade B HIV-1 molecular clones. Luciferase activity in cell lysates was determined 48 h postinfection; data are from a representative experiment.
b92NG083.2 is a G/A recombinant, but the envelope gene belongs to clade G.
on November 8, 2019 by guest
http://jvi.asm.org/
each experiment as described previously (19, 24, 27, 32). Inte-grated density values (IDV) were used to standardize lucif-erase activities (RLU) with the formula [(mutant RLU⫼ wild-type RLU) ⫼ (mutant IDV ⫼ wild-type IDV)] ⫻ 100%. Expression levels of all mutants were between 10 and 120% of wild-type coreceptor expression as described previously (19, 24, 27, 32).
Alanine mutants of tyrosines and negatively charged resi-dues in the Nt and extracellular loop 2 (ECL2) of CXCR4 were evaluated for their ability to mediate the entry of three non-clade B X4 isolates: 84ZR085.1 (non-clade D), 90CF402 (non-clade E), and 92RW009.6 (clade A/C). No single alanine mutant signif-icantly altered the entry of any of the isolates (Fig. 1). Double and triple mutations decreased entry by 0 to 80%, depending on the combination of mutations and isolate (Fig. 1). Combi-nations of six to eight alanine substitutions of negatively charged and tyrosine residues in the Nt and/or ECL2 of
CXCR4 were required to decrease viral entry by an order of magnitude or more (Fig. 1). Our results suggest that CXCR4 usage by non-clade B X4 isolates depends on the overall con-tent of tyrosines and negatively charged residues in the Nt and ECL2 rather than on a specific amino acid sequence. A similar pattern of CXCR4 usage has been reported for clade B X4 isolates (8, 9, 11, 27).
Alanine substitutions in all four extracellular domains of CCR5 were assessed for their ability to mediate the entry of six non-clade B R5 isolates: 92NG083.2 (clade G), 92UG037.1 (clade A), 92UG114.1 (clade D), 98TZ017.2 (clade C), 98IN012.14 (clade C) and 94IN476.104 (clade C). The entry of all six isolates was strongly dependent on residues Y10, D11, Y14, Y15, and E18 (Fig. 2). In addition, alanine substitutions of Nt residues D2, Y3, Q4, S17, K22, and K26 significantly compromised viral entry. However, considerable variability was observed in the degree of dependence on these residues, FIG. 2. Viral entry mediated by CCR5 mutants. U87MG-CD4 cells transiently expressing wild-type CCR5 or CCR5 with alanine substitutions were infected with NLluc⫹env⫺reporter viruses pseudotyped with different non-clade B R5 envelope glycoproteins. The name and clade of each isolate are indicated in the first two columns and the mutated residues are indicated in the top row of each panel. Luciferase activity (RLU) was measured 48 h postinfection and standardized for CCR5 expression levels (IDV). The coreceptor activity of each mutant is expressed as a percentage of wild-type coreceptor activity, calculated with the formula [(mutant RLU⫼wild-type RLU)⫼(mutant IDV⫼wild-type IDV)]⫻
100%. Shading indicates values lower than 20%. All values are means of three independent experiments, each performed in quadruplicate. Standard deviations were not more than 20% for each data set and are not shown for the sake of clarity.
on November 8, 2019 by guest
http://jvi.asm.org/
since entry was decreased anywhere from 2- to 30-fold, de-pending on the combination of mutation and isolate (Fig. 2). Surprisingly, entry of all six non-clade B isolates was also sig-nificantly reduced by alanine substitutions of residues in the extracellular loops of CCR5. The entry of isolates 92NG083.2 (clade G) and 92UG037.1 (clade A) was reduced approxi-mately 10-fold by alanine substitutions of residues H88 and W94 in ECL1 (Fig. 2). Most striking, however, was the depen-dence of non-clade B HIV-1 entry on a cluster of residues in the CCR5 ECL2. Entry of all six test isolates was suppressed 5-to 100-fold by alanine substitutions of Y176 and K191/N192 (Fig. 2). Furthermore, entry of the test isolates depended sig-nificantly on residues S169, Q170, K171/E172, and T177. How-ever, considerable variability was observed in the degree of dependence on these residues since entry was decreased any-where from 0- to 30-fold, depending on the combination of mutation and isolate (Fig. 2). As expected, alanine substitu-tions of all four cysteine residues in the extracellular domains of CCR5 strongly suppressed the entry of all six test isolates. Here we report on the usage of CCR5 and CXCR4 by nine non-clade B isolates. Even though the number of test isolates per clade was small, we believe that the differences and simi-larities in the pattern of CCR5 usage by clade B and non-clade B HIV-1 isolates are significant. Further testing of larger num-bers of non-clade B isolates will be performed in order to confirm our present findings. A previously described cluster of sulfotyrosines and negatively charged residues in the CCR5 Nt is requisite for the fusion and entry of all test isolates, regard-less of clade. A number of other Nt residues, including D2, Y3, Q4, S17, K22, and K26, variably affect the entry of non-clade B
R5 test isolates. The usage of yet other CCR5 Nt residues, including S6, S7, I9, N13, Q21, and K22, by clade B isolates has been reported (7, 20, 32). Entry of two test isolates, a subtype A and a subtype G, furthermore depends on two residues in ECL1. Most importantly, a cluster of charged and polar resi-dues in ECL2 plays a key role in the entry of all six non-clade B test isolates. Reliance on all ECL2 residues, other than Y176 and K191/N192, is isolate dependent.
[image:4.587.73.522.69.338.2]These observations are in contrast to what was reported previously for clade B isolates. Indeed, in those studies, single alanine substitutions in the extracellular loops of CCR5 did not significantly affect the entry of any clade B R5 or R5X4 isolate (19, 24, 32). This may indicate that the affinity of clade B gp120s is much higher for extracellular loop residues than the affinity of non-clade B gp120s. Thus, the effect of single muta-tions is negligible, and multiple mutamuta-tions might have to be introduced in the extracellular loops in order for a decrease in fusion and entry to be observed. Others have reported, how-ever, that residues in the CCR5 extracellular loops influence fusion and entry of clade B isolates, including Q93 in ECL1 (28), Y184, S185, and R197 in ECL2 (17, 33), D276 and Q280 in ECL3 (17, 20). It should be noted that residues Y184 and S185, like residues R197 and D276, had to be replaced to-gether in order to compromise viral fusion and entry (17, 33). We recently demonstrated that the CCR5 Nt binds to con-served residues in C4 and the V3 stem of a clade B R5 gp120 (13, 14). These regions are strongly conserved between clade B and non-clade B R5 isolates (Fig. 3) and might therefore have similar functions. Our data further suggested that residues in the extracellular loops of CCR5 bind to the V3 crown (13, 14). FIG. 3. Alignment of amino acid sequences in the V3 and C4 regions of gp120. Amino acid sequences of the HIV-1 test isolates were obtained from GenBank and aligned. The first three columns show the name of the molecular clone, clade classification, and coreceptor tropism. The V3 crown is boxed. Residues in bold in the JR-FL sequence were previously shown to be important for gp120/CCR5 interactions. Net charges are on the far right.
on November 8, 2019 by guest
http://jvi.asm.org/
This gp120 region is somewhat less conserved between clade B and non-clade B R5 isolates (Fig. 3). We propose that CCR5 extracellular loop residues that we identified here as being important for viral entry constitute the binding site for the V3 crowns of non-clade B gp120s. These or other extracellular loop residues could also constitute the binding site for the V3 crowns of clade B gp120s. Variability in the V3 loop crown may account for the variability in the usage of these residues (26). X4 isolates exhibit more variability than R5 isolates through-out the V3 loop (Fig. 3) (26), which may explain the entirely isolate-dependent usage of CXCR4 extracellular residues.
The isolate- and clade-associated variability in the usage of CCR5 extracellular loop residues may portend isolate- and clade-dependent differences in the efficiency of CCR5 inhibi-tors. Compounds such as TAK-779 do not inhibit gp120/CCR5 Nt interactions yet inhibit gp120 binding to cell surface CCR5, perhaps by inhibiting gp120 interactions with extracellular loop residues. Significant, isolate-dependent variability is observed for inhibition of HIV-1 entry by TAK-779 (A. Trkola, personal communication). Our work further highlights the need to test the ability of all novel CCR5 inhibitors to block the entry of a large panel of clade B and non-clade B isolates.
We thank Feng Gao and John Moore for their generous gift of the HIV-1 non-clade B molecular clones.
This work was supported by grant AI43847 to T.D.
REFERENCES
1.Bazan, H. A., G. Alkhatib, C. C. Broder, and E. A. Berger.1998. Patterns of CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J. Virol.72:4485–4491. 2.Bennetts, B. H., S. M. Teutsch, M. M. Buhler, R. N. Heard, and G. J.
Stewart.1997. The CCR5 deletion mutation fails to protect against multiple sclerosis. Hum. Immunol.58:52–59.
3.Berger, E.1997. HIV entry and tropism: the chemokine receptor expression. AIDS11(Suppl. A):S3-S16.
4.Berger, E. A., P. M. Murphy, and J. M. Farber.1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol.17:657–700.
5.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo.1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol.71:7478–7487.
6.Bjorndal, A., A. Sonnerborg, C. Tscherning, J. Albert, and E. M. Fenyo. 1999. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res. Hum. Retrovir. 15:647–653.
7.Blanpain, C., B. J. Doranz, J. Vakili, J. Rucker, C. Govaerts, S. S. W. Baik, O. Lorthioir, I. Migeotte, F. Libert, F. Baleux, G. Vassart, R. W. Doms, and M. Parmentier.1999. Multiple charged and aromatic residues in CCR5 amino-terminal domain are involved in high affinity binding of both chemo-kines and HIV-1 env protein. J. Biol. Chem.274:34719–34727.
8.Brelot, A., N. Heveker, K. Adema, M. J. Hosie, B. Willett, and M. Alizon. 1999. Effect of mutations in the second extracellular loop of CXCR4 on its utilization by human and feline immunodeficiency viruses. J. Virol.73:2576– 2586.
9.Brelot, A., N. Heveker, M. Montes, and M. Alizon.2000. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem.275:23736–23744. 10.Cecilia, D., S. S. Kulkarni, S. P. Tripathy, R. R. Gangakhedkar, R. S.
Paranjape, and D. A. Gadkari.2000. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology271:253–258.
11.Chabot, D. J., P. F. Zhang, G. V. Quinnan, and C. C. Broder.1999. Mu-tagenesis of CXCR4 identifies important domains for human immunodefi-ciency virus type 1 X4 isolate envelope-mediated membrane fusion and virus entry and reveals cryptic coreceptor activity for R5 isolates. J. Virol.73:6598– 6609.
12.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau.1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med.185:621–628.
13.Cormier, E. G., M. Persuh, A. D. Thompson, S. W. Lin, T. P. Sakmar, W. C. Olson, and T. Dragic.2000. Specific interaction of CCR5 amino-terminal
domain peptides containing sulfo-tyrosines with HIV-1 envelope glycopro-tein gp120. Proc. Natl. Acad. Sci. USA97:5762–5767.
14.Cormier, E. G., D. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic.2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide inter-action with soluble human immunodeficiency virus type 1 gp120-CD4 com-plexes. J. Virol.75:5541–5549.
15.de Roda Husman, A. M., M. Koot, M. Cornelissen, I. P. Keet, M. Brouwer, S. M. Broersen, M. Bakker, M. T. Roos, M. Prins, F. de Wolf, R. A. Coutinho, F. Miedema, J. Goudsmit, and H. Schuitemaker.1997. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann. Intern. Med.127:882–890.
16.de Roda Husman, A. M., R. P. van Rij, H. Blaak, S. Broersen, and H. Schuitemaker.1999. Adaptation to promiscuous usage of chemokine recep-tors is not a prerequisite for human immunodeficiency virus type 1 disease progression. J. Infect. Dis.180:1106–1115.
17.Doranz, B. J., Z. H. Lu, J. Rucker, T. Y. Zhang, M. Sharron, Y. H. Cen, Z. X. Wang, H. H. Guo, J. G. Du, M. A. Accavitti, R. W. Doms, and S. C. Peiper. 1997. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol.71:6305–6314.
18.Dragic, T.2001. An overview of the determinants of CCR5 and CXCR4 co-receptor function. J. Gen. Virol.82:1807–1814.
19.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon.1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Vi-rol.72:279–285.
20.Farzan, M., H. Choe, L. Vaca, K. Martin, Y. Sun, E. Desjardins, N. Ruffing, L. Wu, R. Wyatt, N. Gerard, C. Gerard, and J. Sodroski.1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunode-ficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol.72:1160–1164.
21.Gao, F., S. Morrison, D. Robertson, C. Thornton, S. Craig, G. Karlson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, et al.1996. Mo-lecular cloning and analysis of functional envelope genes from human im-munodeficiency virus type 1 sequence subtypes A through G. J. Virol.70: 1651–1667.
22.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn.1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol.72:5680–5698.
23.Gao, F., D. L. Robertson, S. G. Morrison, H. Hui, S. Craig, J. Decker, P. N. Fultz, M. Girard, G. M. Shaw, B. H. Hahn, and P. M. Sharp.1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013–7029.
24.Genoud, S., F. Kajumo, Y. Guo, D. Thompson, and T. Dragic.1999. CCR5-mediated human immunodeficiency virus entry depends on an amino-termi-nal gp120-binding site and on the conformatioamino-termi-nal integrity of all four extra-cellular domains. J. Virol.73:1645–1648.
25.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup.1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med.2:1240–1243.
26.Hung, C. S., N. Vander Heyden, and L. Ratner.1999. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J. Virol.73:8216–8226.
27.Kajumo, F., D. A. D. Thompson, Y. Guo, and T. Dragic.2000. Entry of R5X4 and X4 human immunodeficiency virus type 1 strains is mediated by nega-tively charged and tyrosine residues in the amino-terminal domain and the second extracellular loop of CXCR4. Virology271:240–247.
28.Kuhmann, S. E., E. J. Platt, S. L. Kozak, and D. Kabat.1997. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J. Virol.71:8642–8656.
29.Li, S., J. Juarez, M. Alali, D. Dwyer, R. Collman, A. Cunningham, and H. M. Naif.1999. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from ad-vanced stages of disease and comparison to tissue-derived isolates. J. Virol. 73:9741–9755.
30.Meyer, L., M. Magierowska, J. B. Hubert, C. Rouzioux, C. Deveau, F. Sanson, P. Debre, J. F. Delfraissy, I. Theodorou, et al.1997. Early protective effect of CCR-5 delta 32 heterozygosity on HIV-1 disease progression: re-lationship with viral load. AIDS11:F73-F78.
31.Michael, N. L., L. G. Louie, A. L. Rohrbaugh, K. A. Schultz, D. E. Dayhoff, C. E. Wang, and H. W. Sheppard.1997. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat. Med. 3:1160–1162.
32.Rabut, G. E., J. A. Konner, F. Kajumo, J. P. Moore, and T. Dragic.1998. Alanine substitutions of polar and nonpolar residues in the amino-terminal
on November 8, 2019 by guest
http://jvi.asm.org/
domain of CCR5 differently impair entry of macrophage- and dualtropic isolates of human immunodeficiency virus type 1. J. Virol.72:3464–3468. 33.Ross, T. M., P. D. Bieniasz, and B. R. Cullen.1998. Multiple residues
contribute to the inability of murine CCR-5 to function as a coreceptor for macrophage-tropic human immunodeficiency virus type 1 isolates. J. Virol. 72:1918–1924.
34.Simmons, G., D. Wilkinson, J. D. Reeves, M. T. Dittmar, S. Beddows, J. Weber, G. Carnegie, U. Desselberger, P. W. Gray, R. A. Weiss, and P. R. Clapham.1996. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J. Virol.70:8355–8360.
35.Tscherning, C., A. Alaeus, R. Fredriksson, A. Bjorndal, H. Deng, D. R. Littman, E. M. Fenyo, and J. Albert.1998. Differences in chemokine core-ceptor usage between genetic subtypes of HIV-1. Virology241:181–188. 36.Tscherning-Casper, C., D. Vodros, E. Menu, K. Aperia, R. Fredriksson, G.
Dolcini, G. Chaouat, F. Barre-Sinoussi, J. Albert, E. M. Fenyo, et al.2000. Coreceptor usage of HIV-1 isolates representing different genetic subtypes obtained from pregnant Cameroonian women. J. Acquir. Immune Defic. Syndr.24:1–9.
37.Xiao, L., S. M. Owen, I. Goldman, A. A. Lal, J. J. deJong, J. Goudsmit, and
R. B. Lal.1998. CCR5 coreceptor usage of non-syncytium-inducing primary HIV-1 is independent of phylogenetically distinct global HIV-1 isolates: delineation of consensus motif in the V3 domain that predicts CCR-5 usage. Virology240:83–92.
38.Zhang, K., M. Hawken, F. Rana, F. J. Welte, S. Gartner, M. A. Goldsmith, and C. Power.2001. Human immunodeficiency virus type 1 clade A and D neurotropism: molecular evolution, recombination, and coreceptor use. Vi-rology283:19–30.
39.Zhang, L., T. He, Y. Huang, Z. Chen, Y. Guo, S. Wu, K. J. Kunstman, R. C. Brown, J. P. Phair, A. U. Neumann, D. D. Ho, and S. M. Wolinsky.1998. Chemokine coreceptor usage by diverse primary isolates of human immu-nodeficiency virus type 1. J. Virol.72:9307–9312.
40.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore.2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in pri-mary cells. J. Virol.74:6893–6910.
41.Zhang, Y. J., and J. P. Moore.1999. Will multiple coreceptors need to be targeted by inhibitors of human immunodeficiency virus type 1 entry? J. Vi-rol.73:3443–3448.