Copyright© 1975 AmericanSociety forMicrobiology Printed inU.SA.
Relationships
Among the
Polypeptides
of Newcastle Disease
Virus
LAWRENCE E. HIGHTOWER,1 TRUDY G. MORRISON, AND MICHAEL A. BRATT* Department ofMicrobiology, University of Massachusetts Medical School, Worcester, Massachusetts 01605
Received for publication 18 July 1975
We have studied the relationships among the polypeptides of Newcastle
disease virus by using both kineticand tryptic peptide analyses. The results of
our tryptic peptide analyses suggest that there are at least six unique viral
polypeptides-L,HN,
Fo
(F),NP, M,anda47,000-daltonpolypeptide.The smallvirion glycopolypeptide F is related to
Fo,
a glycopolypeptide found only ininfected cells. In addition, several smaller polypeptides, including a
53,000-dalton polypeptide found both in purified virions and in infected cells, are
related to the nucleocaspid protein. Kinetic analysisofeach viral polypeptide
reveals thatall of the major viralpolypeptides, with thepossibleexception of L,
arestable after an aminoacid chase. Aprecursor-product relationshipbetween
Fo
and F was notdemonstrable bypulse-chase experiments. Also, almost thesame relative amount of F, the putative product, was present in infected
cultures after either 5 or30minofradioisotopic labeling.Theseresults suggest
that
Fo
isprocessedrapidly.Newcastle disease virus (NDV) is a
represent-ativeof the paramyxovirus group ofenveloped,
negative-stranded RNA viruses. Its genome
consists of 5.2 x 106 to 5.6 x 106 daltons of
continuous single-stranded RNA (13). A
ge-nome of this size has a maximum coding
capac-ity of about 5.4 x 105 daltons of protein.
Six to sevenpolypeptides have been detected
in purified virions by sodium dodecyl sulfate
(SDS)-polyacrylamide gel analysis (2, 7, 9, 17,
19). By the suggested nomenclature for
para-myxoviruses (24), these polypeptides include
two glycosylated membrane proteins (HN,F),
one non-glycosylated internal membrane
pro-tein(M), anucleocapsid protein (NP), and two
tothreeminorpolypeptides.The smaller virion
glycoprotein has not beendirectly implicatedin
cellular fusion induced by NDV; however, we
havetentatively adoptedthedesignationFfor
thisglycoproteinbasedonthe close similarities
between Sendai virusandNDVinprotein
com-position and biological activities. Infected
chicken embryo cell cultures contain an
addi-tionalglycopolypeptide,
Fo
(23; J. Kaplan andM. A.Bratt,Abstr.Annu. Meet. Am. Soc.
Mi-crobiol. 1973, V291, p. 243), whichis not found
invirusparticles (1, 10, 16). The total molecular
weight of these proteinsis approximately 6 x
106,
which slightly exceeds the theoreticalcod-ing capacity of the genome. Thus, if all of these
proteins are virus coded, they may not all be
unique geneproducts.
I
Presnt
address: Microbiology Section, Univeresity of Connecticut,Storrs,Conn. 06268.Several recent studies suggest that
relation-shipsdoexist among theproteinsof
paramyxo-viruses. Studies on the effect of proteolytic
cleavage on the size and biologicalfunction of
the virion proteins of Sendaivirussuggest that
the smallestvirion glycopolypeptide isderived
from a larger precursor (12, 24). The putative
precursormoleculehas recentlybeen detected
inSendai virus-infected cells (26; R. A. Lamb,
Abstr. Annu. Meet. Am. Soc. Microbiol. 1975,
S145, p. 237). Kinetic evidence has been
pub-lished that suggests that the NDV
nonstruc-tural glycopolypeptide
Fo
isunstable and maybe converted to one of the virion structural
polypeptides (22).
Thestrongpossibilitythat,like Sendaivirus,
at least one of the NDV virion
glycopolypep-tidesmightbe acleavage productpromptedus
to studythe relationships among the
polypep-tides of NDVbyboth trypticpeptide andkinetic
analysis. We present evidence for six unique
viral polypeptides. Our data suggest that the
nonstructural glycopolypeptide
Fo
isrelated to the small virion glycopolypeptide F. Inaddi-tion, thenucleocapsid polypeptideisrelatedto
several smaller polypeptides, including a
53,-000-dalton polypeptide found in both purified
virions and infected cells.
MATERIALS AND METHODS
Viruspreparation and cell culture. Preparation and cultivation of primary and secondary chicken
embryocell cultureshave beendescribedpreviously
(3). Secondary cultures grownineither60- or
100-mmtissueculture plates for 48 h at 40 C in 5% CO2 1599
on November 10, 2019 by guest
http://jvi.asm.org/
were used in all experiments. Growth in embryo-natedeggs and purification of the AV (Australia-Victoria, 1932) strain of NDV have also been de-scribed (5, 10).
Chemicals and isotopes. Mixtures of 15 14C0 labeled amino acids (-0.1 Ci/mmol) and [35S]methi-onine (211 Ci/mmol) were purchased from New England Nuclear Corp. Trypsin (containing TPCK) was purchased from Worthington Biochemicals Corp. Acrylamide and N,N'-methylene-bis-acryla-mide were purchased from Gallard-Schlessinger Corp. Other sources of chemicals used in the prepa-ration ofpolyacrylamide gels weredescribed previ-ously (10).
General protocolfor pulse-chase experiments. Cell cultures were infected at an inputmultiplicity of 5PFU/cell. After anabsorption period of 45 min,
cultures were incubated at 40 C in 5%CO2for 6 h.
Allexperiments to be described werepetformedat 6 hafter infection. We have shown previously thatthe rate ofviral proteinaccumulationismaximaland constantbythis time (10, 11). At6hpostinfection,
theculturemedium was removed and the cells were
rinsed with Hanks balanced salt solution, pH 7.2.
Radioisotopic labelmedium (amino acid-freeEagle
minimal essential medium [MEM] supplemented
with0.01mCiof14C-labeledamino acid mixture per
ml and 2% dialyzed calf serum adjusted to pH 7.4 with NaHCO3 and prewarmed to 40 C) was then
added. The cultures were incubated at 40 C for 5
min. Next, the labeling medium was removed and
chasemedium(MEM containing five times [1 mM]
thenormalconcentrations of amino acidsplus1 mM
inalanine,asparticacid,glycine, proline, and
ser-ine, supplemented with 2% calf serum,adjusted to pH 7.4 withNaHCO3andprewarmedto40C)was
added. Under these chaseconditions, total
radioiso-topic incorporation into acid-precipitable material stops almost immediately, and incorporation into viralpolypeptides stops afterapproximately 5 min (11). Aftervarying periods ofchase, cultureswere
washed with cold Hanks salts and the cells were
solubilized directly in polyacrylamide gel sample
buffer (0.05 M Tris [pH 6.71, 1%mercaptoethanol, 1% SDS). Sampleswere stored at -20 C and were
notdialyzedbeforeelectrophoresis (10).
General protocol for accumulation experi-ments.Theproceduresused forinfectingand
label-ing cultures are the same as those described for
pulse-chase experiments, except that the
radioac-tivelabel medium contained5%ofthe normal con-centration ofMEM aminoacids. After either5or30
min of labeling, the radioactive medium was
re-moved and thecells were washed with cold Hanks salts. The cellsweresolubilized directly in polyacryl-amide gel sample buffer and stored at -20 C.
Polyacrylamidegelelectrophoresis.
SDS-discon-tinuouspolyacrylamide gelelectrophoresis was car-ried out by the method of Laemmli (14) aspreviously described(10). Gel concentrations of 8.5% were rou-tinely used, except where 6% gels wereemployedto
clearly separate the large (220,000-dalton) viral
polypeptide from thegelorigin.
Although the gel procedureused inthe present
studyissimilar tothat usedinourprevious studies
(10, 11), we haveonlyrecentlybeen abletoroutinely separate the F and NPpolypeptides. Carefulcontrol
J. VIROL.
ofseparatinggelpolymerization times (between 20
and 30min)has been a significant factor in improv-ingboth gel resolution and reproducibility.
Sample preparationfor electrophoresis has been
described previously(10). Samples were boiled for2
minbefore theywere layered on gels. After
electro-phoresis, the gels were fixed in 10% acetic acid,
sliced longitudinally,and dried, and the
radioactiv-ity was analyzed by autoradiography with Kodak
Royal X-omat film (12-h to 1-week exposure). The
resulting autoradiograms were scanned with an
Or-tecmicrodensitometer.
Tryptic peptide analysis. (i) Sample
prepara-tion. Cellcultures wereinfected and incubated as
described above. At 6 h postinfection, infected
cul-tures werelabeled with 0.5 mCi of[35S]methionine perml in methionine-free Eagle MEMfor 30 min.
Cultures were solubilized directly in gel sample
buffer. Extracts derived from 6.7 x 105 cells were
subjectedtoelectrophoresison asinglegel.
Radioactive virus particles were prepared by in-fecting approximately 108 cells as described above. At 2 to 3 h postinfection, the cells were washed with Hanks salts, and Eagle MEM containing 1% the normal concentration of methionine and supple-mented with 0.125 mCi of[35S]methionine per ml and 2% dialyzed calf serum was added. After an incubationperiod of approximately 15 h at 40 C in 5%CO2,theculture medium was collected and the viruswaspurified as follows. After centrifugation at 27,000 x g for 10 min to remove cell debris, the supernatant was collected andsedimentedat82,500 xg (25,000 rpm) for 1 h in a BeckmanSW27 rotor at 4Cthrough a 20% sucrose layer onto a 65% sucrose-deuterium oxide pad. The virus band at the inter-face of the sucroselayers was thencollected,diluted threefold with cold standard buffer (10), and layered on a 20 to 65%sucrose-deuterium oxide linear gra-dient. Centrifugation was carried out in a Beckman SW27 rotor at 82,500 x g for 16 h. The peak of
radioactivitywaspooled and stored at -20 C.
Ap-proximately one-halfof the entire preparation was
subjectedtoelectrophoresison asinglegel.
(ii)Analysis.Radioactiveviralpolypeptidesfrom virions and infected cultures were separated on
SDS-polyacrylamide gels. Thegelswerethen
proc-essed forautoradiographyasdescribed above. After the viral bands were located by autoradiography, discretebandswereexcisedfromthegelsand incu-bated overnight at 37 C in 1 ml of 1% ammonium bicarbonate containing 100,ugof trypsin. The tryp-sin-bicarbonate solution was removed and the gel slice was reincubated for4 h at 37 Cin fresh 1% ammonium bicarbonate-trypsin solution. The two solutions werecombined andlyophilized.The result-ing peptides were washed twice in water and re-solved by paper ionophoresis at pH 3.5 as described previously (8, 15). After electrophoresis, the paper was either cut into 1 cm strips and analyzed by liquid scintillationcountingorexposed to X-ray film for 2 weeks. In the lattercase, theresulting
autora-diogramswere then scanned withan Ortec
micro-densitometer.
RESULTS
Stability of viral polypeptides.
Precursor-productrelationshipsaresometimes
on November 10, 2019 by guest
http://jvi.asm.org/
NDV PROTEIN SYNTHESIS 1601
ble by kineticanalysis of proteinaccumulation. Wehaveusedtwodifferentexperimental
proto-cols to study the kinetics of protein
accumula-tion. In each experiment, the proteins from
infected cultures were separated on
polyacryl-amide gels to monitorthe incorporationof
radio-active aminoacids into viralpolypeptides. We
have shownpreviously that these gels also
con-tainradioactive cellular proteins,thesynthesis
of whichisonly partiallyinhibitedduring
infec-tion (10, 11). Therefore, electropherograms of
the proteinsofuninfected cultures were
super-imposed on the remainingcellularbackground
in the gel patterns of proteins from infected
cultures to estimate thehost-cellcontribution.
The fraction of the total radioactivity in agelin
each viralpolypeptide wasestimatedby
meas-uring the peak area that remained after
sub-tracting out the host-cell contribution. This
method of quantification, which utilized
densi-tometer tracingsof autoradiograms,isless
accu-rate than the double-label difference analysis
(10) ofpolyacrylamide gels; however, it is
ade-quatewhen theproportionofhost-cell proteins
issmall.
In the firstprotocol, infected and uninfected
culturesweregivena5-minpulseof radioactive
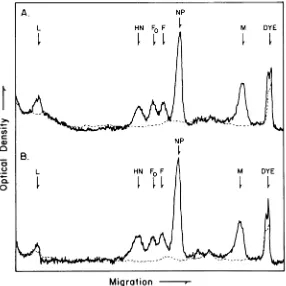
aminoacidsfollowed by eithera2-min(Fig. 1A)
or a 60-min (Fig. 1B) chase with excess
unla-beled amino acids.
Fo,
the putative precursorofF, the small virion glycopolypeptide, did not
decrease significantly during the chaseperiod.
Infact, all of the majorviralpolypeptides(HN,
Fo,
F,NP,M),withthe possible exceptionoftheL(large) polypeptide, were stable. The L region
has been more clearlyresolved on 6%gels(not
shown). Under these conditions, wehave con-sistently found an approximately 50% decrease
in the relative amount of L after a 30-min
chase, with little further reduction after a
60-minchase. Therefore, it is possible that at least
part oftheLregion is unstable.
Using thesecond protocol, we determined the
effect of varying the length of the radioisotopic labeling period on the accumulation of viral
polypeptides. Infected and uninfected cultures
were labeled with radioactive amino acids for
either 5 min (Fig. 2A) or 30 min (Fig. 2B). After
thelonger labeling period, the relative amount
of
Fo
decreasedby approximately 30%, whereasI.
CL0)
[image:3.502.107.393.328.614.2]Migration
FIG. 1. Gel electropherograms of polypeptides from infected and uninfected cultures after amino acid chases. Infected (solid line) and uninfected (broken line) cultures were labeled for 5 min with radioactive amino acids andthen chased for either 2 (A) or 0 (B)min with unlabeled amino acids. Samples of the solubilized culturesweresubjectedtoelectrophoresison8.5%SDS-disc gels. The densitometer tracings of the autoradiograms of gels run inparallelhave been superimposed and the cellular protein backgrounds have been normalized.
16, 1975
on November 10, 2019 by guest
http://jvi.asm.org/
B.
~~~~~~~~~NP
o7 L HN FQF M DYE
[image:4.502.118.403.71.440.2]Migration
FIG. 2. Gelelectropherogramsofpolypeptides from infectedanduninfected cultures after different labeling
periods.Infected (solidline)anduninfected (broken line)cultureswerelabeledforeither 5 (A) or 30 (B)min
with radioactive aminoacids. Samples ofthe solubilized cultures were subjected to electrophoresis and
processedasdescribedinthelegendtoFig.1.
that of F increased by the same amount. The
relative proportions of HN, NP, and M
re-mained constant during this period, while L
increased two- to threefold inrelative amount.
Because of its size (approximately four times
larger than the other viral polypeptides), L
shouldachieve maximum specific activity more
slowly after addition of radioisotopes. This lag
probably accounts for its apparent increase in
relative amount.
Tryptic peptide analysis. No evidence for a
precursor-product relationship between
Fo
andF wasfoundin pulse-chase experiments;
how-ever, studies in which thelabeling period was
varied revealeda weak reciprocal kinetic
rela-tionship between these glycopolypeptides. It
was particularly important to determine the
relationship of L to other viral polypeptides
since a 220,000-dalton polypeptide could be a
possibleprecursorfor,or aggregateof, several
of the smaller polypeptides. To obtain more
conclusive evidence for unique and related
pro-teins, we carriedouttryptic peptide analyses.
Radioactively labeled polypeptides from both
purified virusparticles (Fig. 3A) and infected
cultures(Fig. 3B)wereseparatedon
SDS-poly-acrylamide gels. Discrete bands were excised
from the dried gels anddigested withtrypsin.
The resulting tryptic peptides were separated
by paperionophoresis atpH 3.5.
Evidence for six unique gene products.
The tryptic peptidepatternsofpolypeptides
iso-lated from virions and infected cultures are
comparedin Fig. 4 and 5. Figure4 shows the
on November 10, 2019 by guest
http://jvi.asm.org/
NDV PROTEIN SYNTHESIS 1603
trypticpeptide patternsofpolypeptidespresent
in large enough amounts to be conveniently
analyzed by autoradiography. The tryptic pep-tide patternsof the 47,000-dalton polypeptides
that were present in small amounts and
re-quired analysis byliquid scintillation counting
are shown in Fig. 5. Six distinctly different
tryptic fingerprints were obtained from the
viral proteins L, HN, F, NP, M, and a
47,000-daltonpolypeptide. The trypticpeptidepattern
of this 47,000-dalton polypeptide is different
from thepatternobtained for the46,000-dalton
actin-like polypeptide, which is a major
poly-peptideinuninfected cells (notshown).
There-fore, this polypeptide does not correspond to
anydetectablepolypeptideofsimilarsize inthe
uninfected cell andisprobablyof viral origin.
Pairs ofpolypeptides of similar
electropho-reticmobilityisolatedfrom virions andinfected
cellshavealso beencomparedin Fig. 4 and5.
Polypeptides of similar size fromeither source
havesimilar tryptic fingerprints, thus
confirm-ingtheir identity.
Related viral polypeptides. Several
rela-tionships among the viral polypeptides have
been revealedby tryptic peptide analysis.
Fo,
thenonstructural viral glycopolypeptide, is
re-lated to F, the smaller virionglycopolypeptide
(Fig. 6A). The six majorpeptides of F all have
counterparts amongthe peptides of
Fo.
Inaddi-tion,
Fo
contains two to three methionylpep-tides notpresent in F. There is some ambiguity
in the correspondence between minor tryptic
peptidesofhigher mobilityinthe pattern ofF
and those derived from
Fo.
These species mayarise by contamination of the F preparation
with NP, which migrates very close to F on
gels. Alternatively, there may be changes in the mobility of some of the F peptides as a
result ofprocessing by either glycosylation or
cleavage.
The 53,000-dalton polypeptide (designated
NP1 in Fig. 3) isolated from both purified
vi-rions and infected cultures is related to the
nucleocapsidprotein NP (Fig. 6B). Inaddition,
several minorpolypeptidesofmolecularweight
45,000(NP2) and 43,000(NP3) isolated from
in-fected cultures have tryptic peptide patterns
that are very similar to NP (Fig. 7).
DISCUSSION
Trypticpeptideanalysis suggests that
Fo
andFare related; however, kinetic studies did not
(n,
c]0
B.BFo
NP 47KNP3
O|L HN F [ NP NP2 M DYE
0.
[image:5.502.106.389.358.623.2]Migration
FIG. 3. Gelelectropherograms ofpolypeptides from purifiedvirionsand infectedculturesusedfor tryptic peptide analysis. Polypeptides from purified virions (A) and infected (B) cultures labeled with [3S]methionine
weresubjectedtoelectrophoresis on8.5%SDS-discgels. Densitometer tracings weremade from
autoradi-ograms in which the major viralpolypeptides were overexposed to detectpolypeptides present in small
amounts. Thisaccountsfor the distorted proportions of themajorviral peaks.
A.
L HN F NP NP 47K M DYE
AIAAA
I
I
I
l
nA~~~~~A
A
VOL. 16, 1975
on November 10, 2019 by guest
http://jvi.asm.org/
HNC
HNV v
Fc
F
NPC
NPv
47KV
Mc it
mtit5
J _ _ w0
[image:6.502.72.458.62.497.2]Migration
FIG. 4. Trypticpeptidepatterns of the major unique viral polypeptides. The majorpolypeptides were
excisedfromthe driedgelsshown inFig.3 anddigested with trypsin, and the resultingmethionyltryptic
peptides wereresolved bypaperionophoresis. The densitometer tracings of the autoradiograms oftryptic
peptides derivedfrompolypeptides ofsimilar sizeisolated frompurifiedvirions (subscript v) and infected
cultures(subscript c) are compared. The polypeptide47K,was notpresent in large enough amounts to allow
analysisofitstrypticpeptidepatterns byautoradiography.Theanalysis of this minor polypeptide by liquid
scintillationcounting is shown in Fig. 5. All of the peptidepatterns shown in each figure were analyzed in parallel.
reveal a clear product-precursor relationship.
The fact that almostthesamerelativeamount
ofFispresent after either5or30min of
label-ing may indicate that the processing occurs
quickly.Iftheprocessingisfast, it couldescape
detection in pulse-chase experiments since
ra-dioactive viralproteinscontinuetoaccumulate
for at least 5 min after addition of chase
me-dium (11). Pulse-chase experiments do show
that arelatively stable population ofFoexists
that is either not processed or processed very
slowly. Cultures maycontain twopopulations
ofcells, one capable of processing Fo and the
othernot. Alternatively, onlyaportion of the
Fo glycopolypeptides may be processed in the
infected cell. W
G)
.2_
m
on November 10, 2019 by guest
http://jvi.asm.org/
Our findingofarelatively stablepopulation
of
Fo
isdifferent fromaprevious
report
(22)
thatshowed adramatic chase of
Fo
intoeither NP orF (these polypeptides were not resolved). The
difference is apparently not due to virus strains
since we obtained similar results with either
strain AV or strain L-Kansas, the virus used in the previous study. This discrepancy may be
due tophysiological differences in the cell
cul-tures used in the two studies, particularly if
processing ismediated by cellular enzymes.
20
0
xX
E
15-,, 10 C
0
c0
I 0 20 40 60
[image:7.502.51.243.202.313.2]Fraction
FIG. 5. Trypticpeptide patterns of minor unique viralpolypeptides. The47,000-daltonregion was
ex-cised from thegels shown in Fig. 3 and processed for
trypticpeptide analysis as described in the legend to Fig. 4, except thatthe amount of radioactivity was
determinedby liquid scintillation counting instead of
by autoradiography. Thetrypticpeptide patterns of
the47fKK)-daltonpolypeptide isolated from both
vi-rions (0) and infected cultures (0) are compared.
A, Fo
B, NPc
'C'
NP,c0 NP,v
O
The molecular mechanism fortheprocessing
of
Fo
and F in NDV infection is not known.There is no directevidence for proteolytic
cleav-age, such as exists for Sendaivirus. It islikely
that processing occurs by analogous
mecha-nisms of proteolytic cleavage in both viruses;
however, there are several otherpossible
mech-anismsfor generating relatedpolypeptidesthat
migratedifferentlyonpolyacrylamidegels.For
example, thesamepolypeptide mightbe glyco-sylated to varying extents in the cell.
Differ-encesin glycosylation could alterthemobility
of apolypeptideinagel. Alternatively,asingle
mRNA could have twoinitiation or termination
sites for protein synthesis. The translation of
such an mRNA wouldyieldtwopolypeptides. It
is also possiblethat viral transcriptionresults
intwo mRNAswith overlapping sequences.
We foundatleastthree smallerpolypeptides
invirions andinfected cultures that arerelated
tothenucleocapsid protein. It is known that the
nucleocapsid proteins of paramyxoviruses are
susceptibletospecific cleavages atlimited sites
(18, 20). The size of the cleavage product
de-pends upon the proteolytic enzyme used, and
the cleavage can apparently be accomplished
by cellular proteases in the absence of
exoge-nousenzymes. It islikely that at least some of
the nucleocapsid fragments NP1_3 are
gener-ated bycellular proteases withdifferent
specific-ities, althoughother mechanisms such as
pre-mature termination during transcription or
Migration
FIG. 6. Trypticpeptidepatternsof themajorrelatedviral polypeptides.Polypeptides wereprocessed for
trypticpeptide analysis and analyzed by autoradiographyasdescribedinthe legendtoFig.4. (A)Foand F from infected cultures arecompared; (B) NP isolatedfrom infectedcultures is compared with the53/X
dalton region (NP1)isolated from both virions and infected cultures.
1'.
II - '
l _
19
i
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.502.56.450.409.620.2]c c
.r_
z
0 20 40 60
Fraction
FIG. 7. Trypticpeptidepatternsof minor related viralpolypeptides. Polypeptides wereprocessed for
trypticpeptide analysis and analyzed by liquid scin-tillation countingasdescribed in thelegendtoFig.5. The NP polypeptide is compared with the 45,000-dalton (NP2) and43,000-dalton (NP3) polypeptides isolatedfrom infected cultures.
translation havenotbeen ruledout.
The tryptic peptide analysis of the L
polypep-tide suggests that this polypeptide is unique
andnotanaggregateof smaller viral
polypep-tides. Infected cells contain a plus-stranded
viral RNAlarge enough (35S) tocode forL (4;
B. Spanier-Collins and M. A. Bratt,
manu-scriptinpreparation). RecentlyLhas been
syn-thesized in a cell-free systemprogramed with
the 35S RNA from infected cells (B.
Spanier-Collins, C. W. Clinkscales,M. A. Bratt, andT.
G.Morrison,manuscriptinpreparation).These
data support the hypothesis that L isa
virus-codedpolypeptide.
The possibility that at least part of the L
regionmay beunstable is interesting because
atleastoneviral activity, replication (butnot
transcription), inparamyxovirus-infected cells
requires continuous protein synthesis (21, 25).
A polypeptide ofapproximately the same size
as L is also found associated with
nucleocap-sids isolated from NDV virions (R. J. Colonno
and H. 0.Stone,Abstr. Annu. Meet. Am. Soc.
Microbiol. 1975,S223,p. 250).Recently,alarge
polypeptide has also been detected in
ribonu-TABLE 1. Unique virala polypeptides
Polypeptide Mol wt
L 220,000b
HN 67,000'
Fo 60,000c
NP 56,000
47K 47,000
M 41,000
Total 491,000d
aPolypeptides found in virions and infected cells
but not inuninfected cells are considered virus spe-cific. The possibility that some of these proteins are induced cellular polypeptides has not been rigor-ously eliminated.
bComigrates with rabbit myosin on6% polyacryl-amide gels.
cEstimates for non-glycosylated form based on
size of in vitro translation product on gels (T.G. Morrison, S. R. Weiss, B. Spanier, L. E. Hightower,
and M. A. Bratt, unpublished data).
dApproximate coding capacity is 540,000+54,000 daltons.
cleoprotein complexes isolatedfrom Sendai
vi-rus-infectedcells (26).
We have presented evidence for six unique
viral polypeptides. These polypeptides have a
total molecular weight of 491,000 according to
our best size estimates (Table 1). This total
approaches the maximum coding capacity of
the genome. The number of unique
polypep-tides found here isingood agreementwith the
number predicted from both studies on
tempera-ture-sensitivemutants (five to six
complementa-tion groups; J. Ebel-Tsipis and M. A. Bratt,
manuscriptinpreparation) and the analysis of
viral mRNA on gels (five to six different size classes; 6).
ACKNOWLEDGMENTS
We gratefully acknowledge the technicalassistanceof Eivor Houriand thehelp ofGayle Hightowerinthe prepara-tionof themanuscript.
This work was supported by research grant BMS 75-05024 to M. A. BrattfromtheNational ScienceFoundation, Public Health ServicegrantAl12467-01 to M. A. Brattfrom the National InstituteofAllergy andInfectious Disease, and grant VC-167toTrudyG. Morrisonfrom the American CancerSociety.
LITERATURE CITED
1. Alexander, D.J., andP. Reeve. 1972.Theproteinsof Newcastle disease virus.2. Virus-inducedproteins. Microbios 5:247-257.
2. Bikel, I., and P.H.Duesberg. 1969. Proteins of
Newcas-tle disease virus and of the viral nucleocapsid. J. Virol. 4:388-393.
3. Bratt, M.A., and W. R. Gallaher. 1969. Preliminary analysisof therequirementsfor fusionfrom within and fusionfrom withoutby Newcastlediseasevirus.
Proc.Natl.Acad.Sci. U.S.A. 64:536-543.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.502.65.254.73.331.2] [image:8.502.264.458.82.191.2]NDV PROTEIN SYNTHESIS 1607
4. Bratt, M. A., andW. S. Robinson. 1967. Ribonucleic acid synthesisincells infected with Newcastle dis-easevirus. J.Mol. Biol.23:1-23.
5. Clavell,L.A., and M. A. Bratt. 1972.Hemolytic interac-tionofNewcastle disease virus and chicken erythro-cytes. II. Determining factors. Appl. Microbiol. 23:461-470.
6. Collins, B.S., andM. A. Bratt. 1973.Separationof the messengerRNAs ofNewcastle diseasevirusbygel electrophoresis. Proc. Natl. Acad. Sci. U.S.A. 70:2544-2548.
7. Evans, M. J., and D. W.Kingsbury.1969.Separationof Newcastledisease virus proteins ofpolyacrylamide gelelectrophoresis. Virology37:597-604.
8. Goldman, E., and H. F. Lodish. 1971. Inhibition of replication ofribonucleic acid bacteriophage f2 by superinfection with bacteriophage T4. J. Virol. 8:417-429.
9. Haslam,E.A., I. M.Cheyne, andD.0. White. 1969. Thestructural proteinsof Newcastle disease virus. Virology39:118-129.
10. Hightower,L. E., and M. A. Bratt. 1974.Protein synthe-sis in Newcastle diseasevirus-infected chicken
em-bryocells. J.Virol. 13:788-800.
11. Hightower,L.E.,andM. A.Bratt.1975.Protein metab-olism during thesteadystate of Newcastledisease virusinfection.I.Kinetics ofaminoacid and protein accumulation.J. Virol. 15:696-706.
12. Homma,M., and S.Tamagawa.1973.Restoration ofthe fusion activity ofLcell-borneSendaivirusbytrypsin. J.Gen. Virol. 19:423-426.
13. Kolakofsky,D., E. Boy de la Tour, and H. Delius. 1974. Molecularweight determinationofSendaiand New-castle diseasevirus RNA. J.Virol. 13:261-268. 14. Laemmli,U. K. 1970. Cleavage of structural proteins
during theassembly of the head ofbacteriophageT4. Nature(London)227:680-685.
15. Lodish,H. 1968.Bacteriophage f2 RNA: control of
trans-lation andgeneorder. Nature(London) 220:345-349. 16. Lomniczi, B., A.Meager,and D. C.Burke.1971. Virus
RNA andproteinsynthesisincellsinfectedwith dif-ferent strains ofNewcastle disease virus. J. Gen. Virol. 13:111-120.
17. Meager, A., and D. C. Burke. 1973. Studiesonthe structural basis of the RNApolymerase activityof Newcastle disease virus particles. J. Gen. Virol. 18:305-317.
18. Mountcastle,W.E.,R. W. Compans, L.A.Caliguiri, and P. W.Choppin. 1970.Nucleocapsidprotein sub-units of simian virus5,Newcastle disease virus,and Sendai virus. J. Virol.6:677-684.
19. Mountcastle,W.E., R.W.Compans, andP. W. Chop-pin.1971.Proteins andglycoproteinsof paramyxovi-ruses: a comparison ofsimian virus 5, Newcastle diseasevirus,andSendai virus. J. Virol. 7:47-52. 20. Mountcastle,W.E.,R. W.Compans,H.Lackland,and
P.W.Choppin.1974.Proteolyticcleavageofsubunits ofthenucleocapsidoftheparamyxovirus simian
vi-rus5.J.Virol.14:1253-1261.
21. Robinson,W.S.1971.SendaivirusRNAsynthesisand nucleocapsid formation in the presence of cyclohexi-mide.Virology44:494-502.
22. Samson,A.C.R.,and C. F. Fox. 1973. Precursor pro-teinforNewcastle diseasevirus. J. Virol.12:579-587. 23. Samson,A. C.R., andC. F. Fox. 1974. Selective inhibi-tion ofNewcastledisease virus-inducedglycoprotein synthesisby D-glucosamine hydrochloride. J. Virol. 13:775-779.
24. Scheid, A.,and P. W.Choppin. 1974. Identification of biological activities ofparamyxovirusglycoproteins. Activation of cell fusion, hemolysis, and infectivity byproteolytic cleavage of an inactive precursor pro-teinofSendaivirus.Virology 57:475-490.
25. Weiss, S. R., and M. A. Bratt. 1975. Effect of cordycepin (3-deoxyadenosine) on virus-specific RNA species synthesized in Newcastle diseasevirus-infectedcells. J. Virol. 16:1575-1583.
26. Zaides,V.M.,L.M.Selimova, 0. P. Zhirnov, and A.G. Bukrinskaya. 1975. Proteinsynthesisin Sendai vi-rus-infectedcells. J. Gen. Virol. 27:319-327. VOL. 16,
on November 10, 2019 by guest
http://jvi.asm.org/