JOURNAL OFVIROLOGY,JUIY1991, p.3932-3937 0022-538X/91/073932-06$02.00/0
Copyright C)1991, American Society forMicrobiology
Analysis of the Transcription
Pattern and
Mapping
of
the
Putative
rev
and
env
Splice
Junctions of
Bovine
Immunodeficiency-Like
Virus
M. STEVEN OBERSTE, JOHN D. GREENWOOD, AND MATTHEWA. GONDA*
Laboratory ofCell and MolecularStructure, Program Resources, Inc., NCI-Frederick Cancer Research andDevelopment Center, Frederick, Maryland 21702-1201
Received 15 June 1990/Accepted23March 1991
The bovine immunodeficiency-like virus (BIV) genome contains the obligatory structural genes of all retrovirusesand,inaddition,thecomplexcentralregionoflentiviruses;thisnovelregionmaycode for atleast fivenonstructural/regulatorygenesinBIV(K. J. Garvey,M.S.Oberste, J.E.Elser,M.J.Braun,and M.A. Gonda, Virology 175:391-409, 1990). As a prelude todeterminingthe function of these novel open reading frames,thetranscriptional patternof BIVwasstudiedbyNorthernanalysisofRNAfrom BIV-infectedcells. Five sizeclasses ofBIV-specificRNAs of 8.5,4.1,3.8,1.7,and 1.4 kbweredetected. The 8.5-kbRNAcontains
sequencesfromall regionsofthegenome;it isthe virion RNA andprobablyserves asthegag-pol transcriptas
well. Byusinggene-specific probes, subgenomicviral RNAs of3.8, 1.7,and 1.4 kbweretentatively identified
astheenv,tat,andrevsplicedmessages,respectively. The 4.1-kb RNAcould not beunambiguouslyidentified butmayencode
vif.
Thehybridization patterns of theputativetatandrev mRNAssuggestthatthey aretheproducts ofmultiple splicing events. Discrete transcripts for the BIV W and Y central regionopen reading frameswerenotdefined.The characterization ofpartialcDNA clones haspermittedthemappingof theenvand
putativerevsplice junctions.
Members of the lentivirus subfamily of retroviruses are
responsible foranumberof chronic, multisystemic diseases
ofhumans and othermammals(21, 23). This family includes the agents ofAIDS, human immunodeficiency virus type 1 (HIV-1) and HIV-2 (2, 5, 19), as well as the simian
immu-nodeficiency viruses (10, 26), visna virus (41), ovine
progres-sive pneumonia virus (9, 27), caprine arthritis encephalitis virus (7), feline immunodeficiency virus (33), equine infec-tious anemia virus (30), and bovine immunodeficiency-like virus (BIV) (3, 22, 42).
BIV resembles otherlentiviruses in certain aspects of its pathogenesis, ultrastructure, genome organization, and
in-fectiouscycle in culture (3,20, 22). The complete nucleotide
sequence of BIV has been determined; the BIV genome
containstheusualretrovirus structuralgenes, gag,pol,and
env, as well as open reading frames (ORFs) similar to the nonstructural/regulatory genesvif, tat, andrevfound in the
lentivirus central region (20). BIV lacks the nefgeneseenin
theprimatelentiviruses (25) but containstwouniqueORFs, Wand Y,inagenomelocationanalogoustothat ofthe R, U, and XORFsofHIVs and SIVs(20).Theorganization of the BIVgenomesuggeststhat multiple transcriptsareexpressed
during the replicative cycleofthe virus, as seenwith HIV
and other lentiviruses (12, 25, 31, 32, 34, 35, 38, 44). To determine whether BIV has a complex transcriptional
pat-tern, we have analyzed viral mRNAs related to BIV infec-tion inbovinecells, and wealsoreportmappingdataonthe
splicejunctions of putative revandenv transcripts.
The molecular cloning of biologically active proviruses (BIV 106 and BIV 127) from the genomic DNA of BIV-infected bovine cells has been reported (3). BIV 106, BIV 127, and parental BIV stocks were propagated in bovine
leukocyte adherent cells (BLAC-20cells) as described
pre-*Corresponding author.
viously(20). BIV-infected culturesweremonitored for
max-imum syncytium induction before being split in a 1:2 ratio
(infected:uninfected) with uninfected BLAC-20 cells. Total cellular RNA was extracted from BIV-infected and
unin-fectedBLAC-20 cells 24to48 h afterpassagebythemethod of Chomczynski and Sacchi (4). RNA (5 ,ugper lane) was
fractionated on 1% agarose-formaldehyde gels (13) and
visualized by ethidium bromide staining priortotransferto GeneScreen Plus(Du Pont NEN).
A near-genome-length DNA probe representing nucleo-tides (nt)695to8716of the BIV 106proviruswasgenerated by digesting a cloned 9.6-kb SmaI fragment derived from
BIV 106 (3) with SmaI and BglI and isolatingthe 4.6- and 3.4-kb fragments on a gel(Fig. 1A). Equimolar amounts of these fragments were 32p labeled by nick translation. A
gag-specific1.0kbHindIII-BamHI restriction fragment and
a BIV long terminal repeat (LTR)-specific probe (AvaIl-Narl) were gel purified and32p labeled by random priming
using the Klenow fragment of Escherichia coli DNA
poly-merase I (Pharmacia). Probes specific for other regions of
thegenome weresynthesized bypolymerase chain reaction
(PCR) using Amplitaq (Perkin-Elmer Cetus) and primers specific forthetargetgene(Table 1;Fig. 1A).Thesynthesis
reaction for PCR probes contained 10 mM Tris-HCl (pH 8.3), 3.0 mM MgCl2, 50 mM KCl, 0.01% gelatin, 200 ,uM eachdeoxynucleoside triphosphate, 0.3 p.Meach oligonucle-otideprimer, 20ngof BIV106 SmaIfragment, and 0.5 U of Amplitaq in atotal volume of100 ,ul. The reaction mixture
wasoverlaid withmineraloil, and synthesiswascarriedout
inaDNAThermalCycler(Perkin-Elmer Cetus) by using 30
cyclesof94°Cfor 1min, 55°C for 2.0 min, and 72°C for 2.0 min. The PCR products were extracted with
chloroform-isoamyl alcohol (24:1) and purified by low-melting-point
agarosegel electrophoresis. Probefragments were excised
from the gel and labeled with 32P by the random priming method. Probeswerehybridizedtotheimmobilized RNAat
3932
Vol. 65,No.7
on November 10, 2019 by guest
http://jvi.asm.org/
NOTES 3933
A
iVI
L
U3 RU5pol
v IVV VT v
2 | env U
F-il
lat U3 RUS5[]70 4[
6EDOa
B
RNA _
cag
pol-erv_
v;'.W
_-vIa'rev_
:a, ev 'ev
8 5vb
8.5 38
4.1
1.7 1.4
la 1b 1c ld 2 3 4 5 6
V;47
H.8.5- q* _ r
...mOW
4.1_ I
3.8
1.7 1
1.4
0.-0
7 8 9
__
___s___L
-_F | | __
-wF-w-^
X .j w1..:w .w
_W .zi 'S ..
* '" '}X wF
w ',,b'. I ^
W.sti'.- s
.
t:.
..'. '' -_
_ffi.N
nW:.z ....
W:r
§-w-.>. .s
': ..
-8.5
_4.1
-3.8
-1.7
-1.4
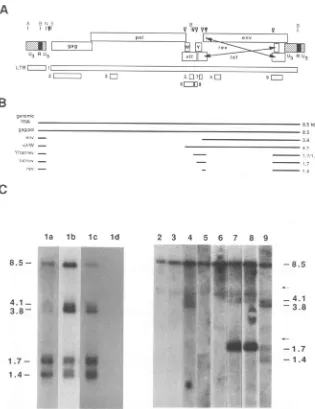
FIG. 1. Northern analysis ofBIV-specific transcripts in infectedcells. (A)Geneticmapof theBIVgenome,withmajor ORFsindicated.
Potentialsplice donors (filled triangles) andacceptors(opentriangles)aremarked abovetheORFmap.Restriction sites: A,Avall;B,BglI;
N,Narl;S,SmaI. Thefollowing probes (shownasstippled rectangles)wereusedforNorthernblothybridization:LTR,AvaIl-Narl;1,full length(SmaI-Bg1IplusBgII-BgII); 2,gag;3,pol;4,env;5,vif; 6,viflW;7,viflY;8, Y/tatexon1;9,envlrevexon2/tatexon2. (B)Proposed
BIVtranscriptionpattern.The splicingpatternwasdeduced fromNorthern analysis (Fig.1C; Table 2) and is compared withthetranscription patternsofHIV-1, visna virus, and equine infectious anemia virus (20, 25, 31, 32, 35, 38, 44). The sizes (inkilobases) of the predicted spliced mRNAsareshownontheright. (C) Northern blot hybridization using full-length and subgenomic probes.Lanes lathroughd;BLAC-20cells infectedwith BIV106, BIV 127,orparental virusand uninfected control, respectively, with probe 1; lanes 2 through 9; BIV 106with the indicatedprobe. Arrows indicate the positions of 28S (upper arrow) and 18S (lower arrow) rRNAs,asvisualized byethidium bromide staining
priortoblotting.
42°C in 50% formamide-5x SSC (lx SSC is 0.15 MNaCl plus 0.015 M sodiumcitrate [pH7.01)-ix Denhardt's solu-tion (0.02% Ficoll, 0.02% polyvinylpyrrolidone, 0.02% acetylated bovineserumalbumin)-0.02Msodiumphosphate (pH 6.8)-0.2% sodium dodecyl sulfate (SDS)-0.005 M EDTA-10 pugofyeasttRNAperml-10,gof sheared salmon
sperm DNA (5'->3') per ml. Following hybridization, the filter was washed at 55°C in O.1x SSC-0.5% SDS and exposedtoX-rayfilm. Forhybridizationwithgene-specific probes (probes 2 to 9 in Fig. 1A), 50 ,ug of total RNA extracted from BIV106-infected BLAC-20 cells wasloaded into a 7-cm-wide well in a 1% agarose-formaldehyde gel.
Following electrophoresisandtransferof the RNAto Gene-Screen Plus, the membrane was cut into 10 strips ofequal width. Eachstripwashybridizedwithoneof thesubgenomic
probes. The results were virtually identical when BIV 127 RNAwas used (datanot shown).
Tomap splicejunctions, cDNA clones were synthesized
according to procedures described elsewhere (32a, 39). Briefly, BIV cDNAs were synthesized from total cellular RNAextracted from BIV 106-infected BLAC-20cells. Neg-ative-strand cDNA synthesis was initiated with the phos-phorylated primer GATAGGCGTCGTTCTTGCTT (nt7818 to 7799) or ACTGCCAACTATCCTTGTCT (nt 5712 to
C
VOL.65, 1991
_IS _j-d" s
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.156.471.75.484.2]3934 NOTES
TABLE 1. Synthetic oligonucleotidesusedas PCRprimers forprobesynthesisa
ORF(strand) Sequence(5'to3') Position in BIV Length (nt)
127RNA(nt)
po/(+) GATGGAGGATGCTAATGG 2248-2265 351
pol(-) CCATCCTTGTGGTAGAAC 2598-2581
env(+) AACTCTCACTATGGCAGGAA 5747-5766 138b
env(-) CACCAATCTATGGTATCTCT 5971-5952
vif(+) CCACTGAAGAAATAGGTCACTTAG 4887-4911 174
vif(-) AGCGTTGGAGGGAGTGTCTAGGGG 5060-5037
Vij7W(+) AGTTCGTGAGGCAGACAGAA 4734-4753 327
viftW(-) AGCGTTGGAGGGAGTGTCTAGGGG 5060-5037
Yltatl (+) GATAATGTTGCCACAGCCCA 5251-5270
Yltatl (-) CATATGGGACAACAACAATGTGGACAG 5364-5338 114
viflY (+) GTCTCTGTTGGTACCCGTTA 5091-5110 134
vifY(-) GCCACAATCTATGAATGTTACGACGAG 5224-5198
envlrev2ltat2 (+) AGTGGACTGGCTCAATAGCT 7481-7500
env/rev2ltat2 (-) GATAGGCGTCGTTCTTGCTT 7818-7799 338
aBIV106 DNA was used asthePCRtemplate.
bThe envprobespans the87-bpdeletion in BIV 106relativetoBIV127. WithaBIV 127template DNA,thePCRproductwould be 225bplong.
5693), which correspondstotheenvlrevor envregionof the BIV genome, respectively (20). BIV-specific cDNAs were
PCR amplified with the phosphorylated primers GATAG
GCGTCGTTCTTGCTT(nt 7818 to7799)orACTGCCAAC TATCCTTGTCT (nt 5712 to 5693) and TAAGAGAGAC TCGGCTCGAG(nt231 to250; common 5'-leader sequence [20]) and were cloned into the phage vector
M13mplO
for furtheranalysis. DNAsequencingandanalysiswerecarriedoutasdescribed previously (20, 22).
BIV-specific RNAs were mapped by hybridization with the whole-genome probe and a series ofgene-specific sub-genomic probes. The whole-genome probe detected five
BIV-specificRNAs of 8.5, 4.1, 3.8, 1.7, and 1.4 kb(Fig. 1C; Table 2). The BIVLTR-specificprobe, whichwas expected to hybridize with all viral mRNAs, recognized the same
transcriptsasthefull-lengthprobe,demonstrating that each mRNA contained a common
5'-leader
sequence (data notshown).
Allprobeshybridizedwith the 8.5-kb RNA,indicatingthat it encompasses all regions of the genome and is most likely thefull-lengthviral RNA. In retroviruses, the genome-length RNAacts asthe mRNAforbothgag andpol (43).
All
probes alsodetectedanRNAof about 5.0 kb whichmigratesattheleading edge of the 28S rRNA band. We believe this to be an
artifactdue to thelargeamountof 28S rRNA. The BIV gag TABLE 2. Summary of probehybridization in
Northern blotanalysisa
Hybridizationwithprobe: RNA (kb)
1 2 3 4 5 6 7 8 9
8.5 + + + + + + + + +
4.1 + + +b +b + + +
3.8 + + +
1.7 + + + +
1.4 + +
aResultsfromNorthern blots(Fig.1) using a single lot of RNA,exceptas
noted.
bResults fromseparateexperimentswith other lots of RNA.
and pol probes (probes 2 and 3, respectively) hybridized
exclusively with the 8.5-kb RNA, confirming its identity as theonly putative gag-pol message (Fig. 1C; Table 2). In the human lentiviruses (25, 31, 34), visna virus (12, 38, 44), and
equine infectious anemia virus (32, 35), the mature env transcript is derived from the genome-length RNA by a
single splice which generates a 4- to 5-kb subgenomic
message. These env mRNAs are processed from theprimary
transcript by using a splice donor between the 5' LTR and the gagORF and a splice acceptorimmediately5' of the env ORF. Probe 4 (env) hybridizedtothe8.5-, 4.1-, and 3.8-kb RNAs (Fig. 1C; Table 2), suggesting that the BIV env message is a 3.8- or 4.1-kb spliced mRNA containing the untranslated 5' leader fused to the 3' portion of the genome (Fig. 2A). Probe 9 (envlrev2/tat2) also hybridized to the 3.8-kbRNA;weexpected this result, since the envtranscript contains untranslated sequences for the rev and tat ORFs (Fig. 1C; Table 2).
We previously predicted the location ofapotentialsplice acceptor site,immediately 5' of theenvORFat nt5396,that would be used byboth env and revtranscripts (20). cDNA cloning of BIV transcripts was undertaken to derive exper-imental data to support this assignment. We selected two oligonucleotide primers(one from sequences of the common 5' leader and the other from the middle of theoverlapping revexon2 andenvORFs) to synthesize and amplify cDNAs encompassing this region. Ourpreliminary characterization ofputative env- and rev-specific partial cDNAsindicates that nt5396 is anauthentic splice acceptorfor thesetranscripts (Fig. 2A andB,respectively).The useof the splice acceptor at nt 5396(Fig. 2A),immediately upstreamof the env ORF, would result in a mature transcript of 3376 nt (without
polyadenylation);the firstAUG 3'ofthisspliceacceptor site is the firstAUG of the envORF. Sinceenvand thefirst rev
codingexoncoincide in thesamereadingframe,it is possible that Rev translation begins at the first AUG while Env translation initiates at a downstream AUG. Baculovirus
expressionofanenvlrevconstructwhose first AUG is that of revfails to produce detectable Env protein (34a). A synthetic env construct which begins with an AUG created at pre-J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.56.558.90.293.2] [image:3.612.56.297.610.716.2]A
1 101
* . . g~~~~~~~1.9.
TAAGA,GAGACTCGGCTCGAGTA^'AGAAGJLCCCAGCTCGAACGAGAAGACTCCGGACAGAAAAATCTAGGAATCAACTATGGATCAGGACCTAGACCGCG start env - M D Q D L D R A CGGAACGC G CAAGAACTGCTTCAGGAGGAGATCAACGAAGGGAGGCTGACAGCCAGAGAAGCTTTACAAACATGGATCAA
E R ZE R aG G S E E L L Q E E I N E G R L T A R E A L Q T W I N 201 TAACGOTGAGATCCACCCTTGGGTCCTGGCAGGAATGCTGTCCATGGGAGTAGGAATGCTACTAGGAGTATATTGTCAGTTACCAGACACACTGATTTGG
N GiZIr P W v L A G M L S M G V G M L L G V Y C Q L P D T L I W
301 ATACTAATGTTTCATTATGCCTTTATTGGGGTTTGGGTGACATCTAGAGAATTAGACAAGGATAGTTGGCAGT 376 I L N F Q L C L Y W G L G E T S R E L D K D S W Q
100
200
300
B
1 101 201
ttcagctcgtgtagctcattagctccgagctccccaacctacagcctgagaggcactggctcggttgggtagccagcctttcgggtaataaaggcttgtt
ggcattcggcatctacccgtgcctcctgt;ttgtcttactcgagcgaac;cacaactccgtcctgctgagstcacagctcgcggggcggtgaagaacacc,
caacagttggcgeccaacgtggggctcgagtaAGAGAGACTCGGCTCGAGTAAAAGAAGACCCAGCTCGAACGAGAAGACTCCGGACAGAAAAATCTAGG
g
3 01 AATCAACTATGGATCAGGACCTAGACCGCGCGGAACGCGGGGAAAGGGGAGGAGGATCCGAAGAACTGCTTCAGGAGGAGATCAACGAAGGGAGGCTGAC start rev - K D Q D L D R A E R G E R G G G S E E L L Q E E I N E G R L T
* . .cS
401 AGCC AC A A G C A TTCCTAGGTATGTTAAGAAGCTGCGCCAAGGTCAGCCAGAATTACCAACATCTCCCGGCGGA
A R E A L Q T W I N N D S P R Y V K K L R Q G Q P E L P T S P G G
M L R S C A K V S Q N Y Q N L P A E 501 GGAGGAGGAC C CAGAAAGCTCCCCGGCGAGAGGAGACCCGGCTTCTGGAAGTCTCTACGAGAATTGGTTGAACAAAATAGGAGAAAGC
G G G R G E R A R K L P G E R R P G F W K S L R E L V E Q N R R K Q
EE D G D T E P E S S P A R G D P A S G S L Y E N W L N K I G E S
,601 AAGAACGAlu:gctatcgggtctggacaga'agaatacaacagcttgaggatcttgttcgccacatgtcgctgggatctcctgacccctcaactccttcagc
E R R L S G L D R R I Q Q L E D L V RH M S L G S P D P S T P S A K N D A Y R V W T E E Y N S L R I L F A T C R W D L L T P Q L L Q L 701 ttccgttctttctgttaaccctcctgctcaaactcctttgggacatcttccgccacgctcctattttaaacttaaaagggtggactgtggggcagggtgg
S V L S V N P P A Q T P L G H L P P R S Y F K L K R V D C G A G W P F F L L T L L L K L L W D I F R H A P I L N L K G W T V G Q G G
801
DL R XT TA A P G L P I C E L D W I Q G T K *
T S G Q Q Q P P D F P Y V N W T G S R E Q N N P E G G L D S G A W 901
tatgaaggc;tgagaggtt;tcagtagattgtaagtcttcggcgagactg'catgtctgcacgtagacaggaaatgtttatcttctcagctgattgtggtt
Y EN L R 8 Q a
1001
aggcccgatacctggaaactgacaacctgttctttagtg'gttaagattatgcataagtgctcgcaatgatgtagctgcttacgcttg0ttactccgccc
1101 tgaaacgcctaccttaacacgcaacacgcccacctgtaagaatatataaaccatatcttcactctgtacttcagctcgtgtagctcattagctccgagct 1201 ccccaacctacagcctgagaggcactggctcggttgggtagccagcctttcgggtaataaaggcttgttggcattcggca 1280100 200
300
400
500
600
700
800
900
1000 1100 1200
FIG. 2. Sequenceanalysis ofBIV cDNAs. (A) Partial sequence ofaBIV cDNAcontainingtheenv codingexon. Theenv cDNAwas
amplified with primersTAAGAGAGACTCGGCTCGAG (nt231to250)andACTGCCAACTATCCTTGTCT(nt5712to5693) (see text).(B)
Sequence ofaBIV cDNAcontainingputativerev exons.TherevcDNAwasamplifiedwithprimersTAAGAGAGACTCGGCTCGAG(nt231 to 250) andGATAGGCGTCGTTCTTGCTT (nt 7818to7799) (see text). The region sequencedis shownin uppercase letters;the region
inferredfromthe BIVnucleotidesequenceisshown in lowercase letters(20).The aminoacidtranslation of themajorORFs is shown below thesequence.Verticalarrowsindicatesplicejunctions.Where the cDNA sequence inpanelBdiffersfromthepreviouslypublishedBIV 127 sequence(20), the nucleotidefrom thepreviouslypublished sequenceisshown above the cDNAsequence.
dictedenvamino acid 50(CTG-*ATG; Leu--Met) produces
immunologically detectable BIV Env. This artificial start codon isjust upstream of the second env AUG, near the amino terminus of theputative Env signal sequence (20).
The 4.1-kb RNA hybridized with probes 1, 4, and 7
through 9 but not with probe 2 or 3 (Fig. 1C; Table 2), suggesting that it arises froma splicingevent which deletes the gag-pol region and joins the central region to the 5' untranslated leader. In someexperiments, with other
prep-arationsofRNA, probes5 and 6hybridized weaklywith the
4.1-kb RNA(Table2 and datanot shown). Because of this ambiguity, the identity of the 4.1-kb RNA cannot be
un-equivocally determined. Ifhybridization with probe 5 (viJ)
and probe 6 (vifiW) does occur, the 4.1-kb RNA may representthevif mRNA,but it could also directsynthesisof a protein from BIV ORF W(Fig. 1A and B; Table 2). The
4.1-kbband isprobablynotthe mRNA foranyother central
region ORF, because the large number ofpotential AUG
start codons upstream of the authentic translation start site would result in very inefficienttranslation ofthese
proteins.
Although the 4.1-kb RNA could contain the entire env
codingsequence,webelieve that it doesnotcode for theenv
ORF products, since it has nine
potential
start codons upstream of the envORF(20).
With the
exception
ofvif,
vpr, and vpu(1,
40)
(and
possiblyWand Y),the
nonstructural/regulatory
genesof the lentivirus central region are encodedby
multiply
spliced
mRNAs,usuallyof less than2kb
(12,
25, 31,
32,
34, 35,
38,
44). The 1.7-kb RNA isrecognized by
probes
7(Yltatl),
8(viflY),
and 9(envlrev2ltat2)
(Fig. 1C;
Table2).
The broad-nessof this band suggests that it may encompass morethan oneRNAspecies
ranging
from 1.7to 1.8kb.ThesemRNAsprobably represent the tat or Yltat
transcript(s)
becauseprobes from the tat and YORFs
hybridized
to the 1.7-kb RNA but not to the 1.4-kb RNA(Fig.
1C;
Table2).
Theproposed Y
splice
acceptor is54ntupstream ofthepredicted
gaectcaggacaacagcagcccccggacttcccatatgtgaattggactggatccagggaacaaaataacccagaagggggattagactctggggcttggon November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.88.525.71.455.2]3936 NOTES
tat
splice
acceptor,suggesting
that a 1.8-kb RNAmight
encode
Y,
while the 1.7-kb RNA is believed to be the tat message(20).
The tatmessage ofprimate
lentiviruses often contains ashortnoncoding
exonderived from the 3' end ofthe
pol
region
(25),
butnoneof theprobes
used in thisstudy
would havedetected such an exon.
The 1.4-kb RNA
hybridized
only
withprobes
1 and 9,indicating
thatits 5'-mostsplice
acceptor
is 3'to nt5364(the
3' end ofprobe
7).
Inlight
of thisfinding
and the datadiscussed
below,
we believe that the 1.4-kb RNA is therev message.Coding
exon 1ofBIVrev(Fig.
1)
wasidentifiedinthe BIV sequence
by
its location(overlapping
and in thesame
reading
frame asenv)
andby
thesimilarity
(in
charac-ter, ifnot
identity)
ofits deduced amino acid sequence tothose ofotherlentivirus Rev
proteins
(20).
The locationof BIVrevcoding
exon 1isnotwithoutparallel;
thefirstcoding
exonof visna virus rev
occupies
asimilarposition
atthe 5' end of and in the samereading
frameas env(11, 29).
The secondcoding
exon ofvisna virus rev is near the 3' endof the genome,overlapping
env, but in a differentreading
frame. In the cDNA
sequencing
studiesreported here,
wehaveidentifieda
doubly spliced
BIVtranscript (Fig. 2B)
with thesamestructureasthevisna virusrevmRNA(11, 29).
Theboundaries ofthefirstintronofBIVrevareidenticaltothose
ofthe
single
intron intheenvtranscript,
sincethey
share thesame
splice
donor andacceptor for thisregion (Fig. 2).
Therev
transcript
isspliced
asecondtime(post-rev
coding
exon1),
deleting
sequences(rev
intron2)
foundin the 3' endofthe genome. Wehadpreviously
predicted
thesplice
donorandacceptor
for the second intron ofrevtoresideat nt5395andnt
7698,
respectively
(20).
HIowever,
ourpresent dataindi-cate that the
splice
donor and acceptor for the second revintron areat nt5540 and
7636,
respectively (Fig.
2B).
Figure
1B
shows theputative
BIVtranscription
pattern,deduced from Northern
(RNA) analyses (Fig. 1C;
Table2),
preliminary
cDNAsequencing (Fig. 2),
and acomparison
between BIV and other lentiviruses. The pattern of BIV
transcription
shows acomplexity
characteristic ofthelenti-virus
subfamily
ofretroviruses. Alltranscripts presumably
initiateatthesamesiteatthe 5'end ofthe R
region
of the leftLTR andterminate atthe 3' end of theR
region
of theright
LTR(20).
Thegenome-length
RNAencodesthevirionRNA and thegag-pol
polyprotein,
which in lentiviruses is believedtobe translated
through
a-1-ntframeshift,
since thesetwoORFs are in different but
overlapping reading
frames(20,
43).
Theremaining
proteins
appear to betranslated
fromspliced
messages.BIV has
recently
been shown to transactivate its LTR(32c),
presumably
through
the action ofthetatgeneproduct.
The
protein
potentially
encodedby
sequences in the BIV1.7-kb mRNA contains a
Cys-rich
region
that ishighly
conserved among lentivirus Tatproteins
(20). This con-served motif hasbeen showntoberequired
fortheactivity
ofHIV-1 Tat
(16, 36, 37).
Thisregion
has also been impli-cated in theformationofmetal-linkedTathomodimers (17,18).
TheHIV-1 Revprotein
hasbeen shown toregulatethetranslation ofthegag-polandenv
transcripts
by specificallycontrolling
thetransport ofthese mRNAsfromthe nucleus to thecytoplasm
(14, 15, 24,
28). The putative rev geneproduct
of
BIVdoesnotalign
well withotherRevproteins, but itsposition
in the genome andsplicing
pattern (1.4-kbdoubly
spliced
message)
areanalogous
to those of visna virus rev(11, 20, 29).
Thepredicted
BIV Vifprotein also bears littlesimilarity
toitscounterparts in otherlentiviruses,
except
in itsgenomelocation(20)
and thepresenceofashortamino acid motif which is highly conserved among all lentiviruses (32b).
Thetranscriptionalmapof theputativeBIVnonstructural/
regulatory genes (vif, tat, and rev) derived here
closely
corresponds to those of the lentiviruses HIV-1 and visna virus, supporting the tentative identification of each of these ORFs(8, 11,38, 44). Transcripts specifictothe BIV W and Y ORFs could not be identified in the presentstudybecause of thesignificantoverlap in centralregionORFs. However,the location of the BIV W and Y ORFs in the central
region
of the genome is similartothe location of theR, U, and X genes ofprimate lentiviruses (20). In HIV-1, the R and U genes are singly spliced messages of about 4kb,
slightly
smaller than thevif mRNA(1, 40).The BIV W and Y ORFs may be somewhatanalogous to the HIV-1 R and U
ORFs,
respectively. Thishypothesisis basedonseveral features of these putative ORFs. First, predicted splice acceptor sites arelocatedimmediatelyupstreamof both W and Yat nt4714 and 5057, respectively (20). Second, a singly spliced W message,using asplicedonorat nt289 andaspliceacceptor at nt 4714, would be slightly smaller than a singlyspliced
BIVviftranscript.Two splicing events,removing intronson eitherside of the BIV YORF, would result ina YmRNA of approximately 1.4 to 1.7kb;thelengthof this message would depend upon the exact location of the borders of the second intron.Finally, the predicted BIV W and Yproductshavea numberof basic amino acids, suggesting a nucleic acid- ornucleotide-binding domain and thus a possible interaction with nucleic acids. Interestingly, the HIV-1 R gene product
has recently been shown to be a virion-associated protein
and to interact in trans with HIV-1 promoter sequences in the LTR to enhanceexpressionof viral proteins (6). Direct identification of BIV regulatory proteins must await the
developmentofspecific immunologic reagents, as well as the characterization of complete viral cDNAs and their func-tionalstudy.
We thank L. Pallansch and G. Tobin for helpful discussions during the course of this work, K. Nagashima for photographic assistance, and G. Serig for assistance in the preparation of the manuscript.
This project was funded in part with federal funds from the Department of Health andHumanServicesunder contractnumber NO1-CO-74102 with Program Resources,Inc.
REFERENCES
1. Arrigo,S. J., S. Weitsman, J. A. Zack, andI.S. Y. Chen.1990. Characterization and expression of novel singly spliced RNA species of human immunodeficiency virus type 1. J. Virol. 64:4585-4588.
2. Barre-Sinoussi,F., J. C. Chermann, F. Rey, M. T. Nugeyre,S. Chamaret, J. Gruest, C. Dauguet, C. Axler-Blin, F. Vezinet-Brun,C. Rouzioux, W.Rozenbaum, and L. Montagnier. 1983. Isolation ofaT-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868-871.
3. Braun,M.J., S.Lahn, A. L. Boyd, T. A. Kost, K. Nagashima, and M. A.Gonda.1988.Molecular cloningof biologicallyactive proviruses of bovine immunodeficiency-like virus. Virology 167:515-523.
4. Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolationby guanidinium thiocyanate-phenol-chloroform extraction.Anal. Biochem. 162:156-159.
5. Clavel, F., D. Guetard, F. Brun-Vezinet, S. Chamaret, M.-A. Rey, M. 0. Santos-Ferreira, A. G. Laurent, and C. Dauguet. 1986. Isolation ofa new humanretrovirus from West African patientswithAIDS. Science233:343-346.
6. Cohen,E. A., G.Dehni, J. G. Sodroski, and W. A. Haseltine. 1990. Humanimmunodeficiency virus vpr product is a virion-J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
associated regulatory protein. J. Virol. 64:3097-3099.
7. Crawford, T. B., D. S. Adams, W. P. Cheevers, and L. C. Cork. 1980. Chronic arthritis in goats caused by a retrovirus. Science 207:997-999.
8. Culien, B. R., and W. C. Greene. 1990. Functions of the auxiliary gene products of the human immunodeficiency virus type 1. Virology 178:1-5.
9. Cutip, R. C., and G. A. Laird. 1976. Isolation and characteri-zation of a virus associated with progressive pneumonia (maedi) of sheep. Am. J. Vet. Res. 37:1377-1382.
10. Daniel, M. D., N. L. Letvin, N. W. King, M. Kannagi, P. K. Sehgal, R. D. Hunt, P. J. Kanki, and R. C. Desrosiers. 1985. Isolation of T-cell lymphotropicHTLV-III-like retrovirus from macaques. Science 228:1201-1204.
11. Davis, J. L., and J. E. Clements. 1989. Characterization of a cDNA clone encoding the visna virus transactivating protein. Proc. Natl. Acad. Sci. USA 86:414-418.
12. Davis, J. L., S. Molineaux, and J. E. Clements. 1987. Visna virus exhibits a complex transcriptional pattern: one aspect of gene expression shared with the acquired immunodeficiency syn-drome retrovirus. J. Virol. 61:1325-1331.
13. Davis, L. G., M. D. Dibner, and J. F. Battey (ed.). 1986. Basic methods in molecular biology, p. 143-146. Elsevier Science Publishing, Inc., New York.
14. Emerman, M., R. Vazeux, and K. Peden. 1989. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell 57:1155-1165.
15. Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Cope-land, and G. N. Pavlakis. 1989. rev protein of human immuno-deficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. 16. Frankel, A. D., S. Biancalana, and D. Hudson. 1989. Activity of
synthetic peptides from the tat protein of human immunodefi-ciency virus type 1. Proc. Natl. Acad. Sci. USA 86:7397-7401. 17. Frankel, A. D., D. S. Bredt, and C. 0. Pabo. 1988. Tat protein from human immunodeficiency virus forms a metal-linked di-mer. Science 240:70-73.
18. Frankel, A. D., L. Chen, R. J. Cotter, and C. 0. Pabo. 1988. Dimerization of the tat protein from human immunodeficiency virus: a cysteine-rich peptide mimics the normal metal-linked dimer interface. Proc. Natl. Acad. Sci. USA 85:6297-6300. 19. Gallo, R. C., S. Z. Salahuddin, M. Popovic, G. M. Shearer, M.
Kaplan, B. F. Haynes, T. J. Palker, and P. D. Markham. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500-503.
20. Garvey, K. J., M. S. Oberste, J. E. Elser, M. J. Braun, and M. A. Gonda. 1990. Nucleotide sequence and genome organi-zation of biologically active proviruses of the bovine immuno-deficiency-like virus. Virology 175:391-409.
21. Gonda, M. A., A. L. Boyd, K. Nagashima, and R. V. Gilden. 1989. Pathobiology, molecular organization, and ultrastructure of HIV. Arch. AIDS Res. 3:1-42.
22. Gonda, M. A., M. J. Braun, S. G. Carter, T. A. Kost, J. W. Bess, Jr., L. 0.Arthur, and M. J. Van Der Maaten. 1987. Character-ization and molecular cloning of a bovine lentivirus related to human immunodeficiency virus. Nature (London) 330:388-391. 23. Haase, A. T. 1986. Pathogenesis of lentivirus infections. Nature
(London)322:130-136.
24. Hammarskjold, M.-L., J. Heimer, B. Hammarskjold, I. Sang-wan, L. Albert, and D. Rekosh. 1989. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 63:1959-1966.
25. Haseltine, W. A. 1988. Replication and pathogenesis of the AIDS virus. J. Acquired Immune Defic. Syndr. 1:217-240. 26. Kanki, P. J., M. F. McLane, N. W. King, Jr., N. L. Letvin,
R. D. Hunt, P. Sehgal, M. D. Daniel, and M. Essex. 1985. Serologic identification and characterization of a macaque T-lymphotropic retrovirus closely related toHTLV-III. Science
228:1199-1201.
27. Kennedy, R. C., C. M. Eklund, C. Lopez, and W. J. Hadlow. 1968. Isolation of a virus from the lungs of Montana sheep affectedwith progressivepneumonia.Virology 35:483-484. 28. Malim, M. H., J. Hauber, S.-Y. Le, J. V. Maizel, and B. R.
Cullen. 1989. The HIV-1 rev trans-activator acts through a
structured target sequence to activate nuclear export of
un-spliced viral mRNA. Nature(London) 338:254-257.
29. Mazarin, V., I.Gourdou, G. Querat, N. Sauze, and R. Vigne. 1988. Geneticstructure and function ofan early transcript of visnavirus.J. Virol.62:4813-4818.
30. McGuire, T. C., K. O'Rourke, and W. P. Cheevers. 1987. A review ofantigenic variation by the equineinfectious anemia virus.Contrib. Microbiol. Immunol. 8:77-89.
31. Muesing, M. A., D. H.Smith, C. D. Cabradilla, C. V.Benton, L. A.Lasky, and D. J. Capon. 1985.Nucleic acidstructureand expression of the human AIDS/lymphadenopathy retrovirus. Nature(London)313:450-458.
32. Noiman,S.,A.Yaniv, L.Sherman, S. R.Tronick,and A.Gazit. 1990.Pattern oftranscriptionofthe genomeofequineinfectious anemia virus. J. Virol. 64:1839-1843.
32a.Oberste, M. S., et al. Unpublished data.
32b.Oberste,M. S., and M. A. Gonda. Unpublisheddata. 32c.Pallansch, L., et al. Unpublished data.
33. Pedersen, N. C., E. N.Ho, M. L. Brown, andJ. K. Yamamoto. 1987. Isolation of a T-lymphotropic virus from domestic cats
withanimmunodeficiency-like syndrome. Science235:790-793. 34. Rabson, A. B., D. F. Daugherty, S.Venkatesan,K.E.Boulukos, S. I. Benn, T. M. Folks, P. Feorino, and M. A. Martin. 1985. Transcription of novel open reading frames of AIDS retrovirus duringinfection oflymphocytes. Science 229:1388-1390. 34a.Rasmussen, L., J. D. Greenwood, and M. A. Gonda.
Unpub-lisheddata.
35. Rasty, S., B. R.Dhruva, R. L.Schiltz,D.S.Shih,C.J.Issel,and R.C.Montelaro. 1990. ProviralDNAintegration and transcrip-tionalpatterns of equine infectious anemia virusduring
persist-ent andcytopathic infections. J. Virol. 64:86-95.
36. Rice, A. P., and F. Carlotti. 1990. Mutational analysis ofthe conservedcysteine-rich region of thehumanimmunodeficiency
virus type 1 Tatprotein. J. Virol. 64:1864-1868.
37. Ruben, S., A. Perkins, R. Purcell, K.Joung,R.Sia,R.Burghoff, W. A. Haseltine, and C. A. Rosen. 1989. Structural and func-tional characterization of human immunodeficiency virus tat
protein. J. Virol. 63:1-8.
38. Sargan, D. R., and I. D. Bennet. 1989. Atranscriptional map of visna virus: definition ofthesecond intron structure suggests a
rev-like gene product. J. Gen. Virol. 70:1995-2006.
39. Schwartz, S., B. K. Felber, D. M.Benko,E.-M.Fenyo,andG.N.
Paviakis. 1990. Cloning and functional analysis of multiply
spliced mRNA species ofhuman immunodeficiency virus type 1. J. Virol. 64:2519-2529.
40. Schwartz, S., B. K. Felber, E.-M. Fenyo, and G. N. Pavlakis. 1990. Env andVpuproteins ofhumanimmunodeficiency virus type 1 areproducedfrommultiple bicistronic mRNAs. J. Virol. 64:5448-5456.
41. Sigurdsson,B., and P. A. Palsson. 1958. Visnaofsheep.Aslow, demyelinating infection. Br. J. Exp. Pathol. 39:519-528. 42. Van DerMaaten, M. J., A. D. Boothe, and C. L. Seger. 1972.
Isolation of a virus from cattlewithpersistentlymphocytosis.J. Natl. Cancer Inst. 49:1649-1657.
43. Varmus, H., and R. Swanstrom. 1984. Replication of retrovi-ruses, p. 369-512. In R. Weiss, N. Teich, H. Varmus, and J. Coffin (ed.), RNA tumorviruses, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
44. Vigne, R., V. Barban, G.Querat, V. Mazarin,I. Gourdou, and N. Sauze. 1987. Transcription of visna virus during its lytic cycle: evidence fora sequentialearlyand lategene expression. Virology 161:218-227.