0022-538X/91/116137-07$02.00/0
Copyright © 1991, American Society for Microbiology
Identification
of Cell
Membrane Proteins
That
Bind Visna Virus
SHARON E. CRANE,1 JEANINEBUZY,2 ANDJANICE E.
CLEMENTS'*
Division of Comparative Medicine, Department of Molecular Biology and Genetics1 and NeuroscienceDepartment,2
TheJohns Hopkins University School of Medicine, Baltimore, Maryland 21205
Received 16 April1990/Accepted 31 July 1991
Visna virus infects cells of ovine origin by attaching to a cell surface receptor via its envelopeglycoprotein. Theidentity of the visna virus receptor is not known. To identify the molecule responsible for binding the virus to targetcells, virus overlay protein blot assays were used to examine the molecular weights of cell surface molecules which bind purified virus. Molecules on the surface of goat synovial membrane (GSM) cells and sheep choroid plexus (SCP) cells ofapproximately 15, 30, and 50 kDa bound to visna virus. Thebinding ofvisna virus to these proteins was reduced by preincubating virus with neutralizing antibodies. '251-labeled cell membranepreparations of GSM and SCP cells were used to affinity purify these virus-binding proteins. These proteins were analyzed by sodium dodecylsulfate-polyacrylamide gel electrophoresis and had molecular masses of15, 30, and 50 kDa. Antibodies to the 50-kDa protein bound to the surface ofbothliveSCP and GSMcells in immunofluorescence assays. In addition, antibodies to the 50-kDa protein blocked the binding of
[35S]methionine-labeled visna virus to SCP cells in culture. Antibodies raised against the 15- and 30-kDa
proteins did not block virus binding to cells. The blocking activity of antibody to the 50-kDa protein provided data that this protein is the molecule which visna virus recognizesand binds to on the surface of targetcells.
Visna virus is a sheep lentivirus which causes a
progres-sivepneumoencephalitis andarthritis in sheep (26). In vivo,
the virus infects cells of the monocyte-macrophage lineage (19). In primary cells from sheep and goats, visna virus binds toits receptor and probably enters the cell through fusion of the viral membrane with the cell membrane, as does a related lentivirus, human immunodeficiency virus (15). The
identity ofthe receptor for visna virus has not yet been
determined. Neutralizingantibodiestothevisna virusinhibit
thebinding of virus toits receptor on sheep cells in culture
(12). In contrast, neutralizing antibodies facilitate binding,
penetration, and uncoatinginmacrophages in culture,owing
tobinding ofthevirus-antibody complextoFc receptors (10,
11). The spread of virus from macrophage to macrophage
maybeamechanism forviral persistence in the host (5, 12).
Enveloped viruses such as visna virus attach to cells
through the binding of their envelope glycoprotein to a
specific cell surface molecule. Cells must have the virus
receptor on their surface to be susceptible to infection. However, the presence of the receptor on the cell surface
doesnotnecessarilyrender thecell susceptibletoinfection;
it has been shown in other viral systems that there are
additional species-specific and cell-specific factors which
allow the virus to continue its lifecycle and replicate in a
permissive cell (14). The receptor can be a ubiquitous
molecule such as a sialic acid-containing glycoprotein for
influenza virus (3) or a specific molecule such as the C3d
receptorfor Epstein-Barrvirus (8), the intercellular adhesion molecule ICAM-1for rhinovirus (9,29),ortheCD4 molecule
for human
immunodeficiency
virustype 1(14,25).
Anumberof the specific molecules found to be viral receptors are membersof theimmunoglobulinsuperfamily, asdetermined by theirconserved amino acids and domain structures (18).
These receptors may beimportant in the therapeutic treat-ment ofviraldiseases (13, 27).
To characterize the cell surface molecules which bind
visnavirus,weutilized theaffinity ofthevirusfor molecules
* Correspondingauthor.
onthe surfaceof sheepchoroidplexus(SCP) cells and goat
synovial membrane(GSM)cells. Virus overlay protein blot
assays(VOPBA)wereused to identify moleculeswhichthe virusrecognized (2). Three sizes of proteins wereidentified,
15,30, and 50kDa,towhich visna virus bound.Neutralizing
antibodiestovisnavirus reducedbindingof the virustothe
cellsurface molecules in the VOPBA. Affinitypurificationof theseproteinswasused to isolate them and raiseantibodies
against them. Antibody to the 50-kDaprotein reacted with
the surfaces ofboth live SCP and GSM cells in
immunoflu-orescence assays and blocked the binding of radiolabeled
virus to the surfaces ofintact cells. Therefore, the 50-kDa
protein isimportant in thebinding of visnavirus to
suscep-tible cells and may in fact be the virus receptor or a component ofthe receptor molecule.
MATERIALS ANDMETHODS
Cells and virus. SCP and GSM cells were obtained as
previouslydescribed(21,22, 30). L cellswereobtainedfrom
theAmericanType CultureCollectionand grown inminimal
essentialmedium (MEM) (GIBCO)
supplemented
with 10%fetal bovine serum. Purified visna virus strain 1514 and
caprine
arthritis-encephalitis
virus strain COwereprepared
aspreviouslydescribed (4, 23,24). Visna viruswas
radiola-beled with
[35S]methionine
aspreviously
described(12).
Viruswaspurifiedfrom the mediumbyclarifyingat10,000 x
gfor 15 min and centrifugingat 100,000 x gfor 2h. VOPBA. Virus binding assays were done as
previously
described (2) with the
following
modifications. A1-mg
sample of cell
lysate
orcell membranewasrun on a5to20%gradient sodium dodecyl sulfate
(SDS)-polyacrylamide
gel
and transferredtonitrocellulose membranes. Nitrocellulose
was stained with ponceau S (Sigma), cut in
strips,
and blocked in VOPBA buffer (50 mM Tris HCl[pH
7.4],
150mM NaCl, 1 mM EDTA, 0.05% Tween 20, 0.1% bovine
serum albumin [BSA])
containing
5% BSA and 5%chickenserumfor 1h with
rocking.
Strips
werewashed three times for 5 min each in VOPBAbuffer. A2-mIsample
of10 ,ugofvirus per mldiluted inMEM-10mM HEPES
(N-2-hydroxy-6137on November 10, 2019 by guest
http://jvi.asm.org/
ethylpiperazine-N'-2-ethanesulfonic acid)-0.5% fetalbovine
serum was incubated with each strip for 1 h. For some assays,5 x 104 cpmof35S-labeled viruswereused, and the
strips were washed and autoradiographed. The strips were
washed three times, incubated with antivirus antibody for 1 h, and washed three times. The stripswereincubatedfor 1 h
in secondary antibody (peroxidase conjugated) which was
diluted 1:200 in VOPBAbuffer. Thestripswerethenwashed
andstained in 0.06%4-chloro-1-naphthol-0.014% H202-20% methanol in phosphate-buffered saline (PBS).
Cell lysates and cell membrane preparations. Twenty-five 150-cm2 flasks of SCP,GSM, or mouseLcellswerewashed
inPBS, and thecellswere scraped and pelleted at1,000 x g
for 10min.Thepelletwaswashed in PBS andpelleted again. The cells were lysed in lx lysate buffer (1% Zwittergent,
0.5% sodium deoxycholate, 0.15 M NaCl, 1 mM
phenyl-methanesulfonyl fluoride, 0.1% SDS, 1 mM EDTA, 25 mM Tris [pH 7.6], 0.02% sodium azide) and sonicated on ice.
Protein concentration was determined by modified Lowry assays (16) orbicinchoninic acid assays (Pierce).
Cell membranes were prepared by differential
centrifuga-tion ofSCP and GSM cellsgrownin25850-cm2 roller bottles
orLcellsin 25150-cm2flasks(7). Cellswerewashed inPBS,
scraped, andpelletedat 1,000 x gfor 10min. Thepelletwas
washed in PBS and repelleted. The pellet was then
sus-pended in 4 ml ofhomogenizing buffer (0.25 M sucrose, 5 mM TrisHCl [pH 7.4], 1 mMMgCI2, 0.1mgof
phenylmeth-ylsulfonyl fluoride per ml) (STM) per g of cell pellet and
homogenized in a Dounce homogenizer. The lysed cells werecentrifugedat 1,000 x gtopellet nuclei, and the pellet
was washed in STM and repelleted. The supernatant was
diluted to 40 ml in STM and centrifuged at 10,000 x g to
remove lysosomes and mitochondria, and the pellet was
washed and repelleted. This final supernatant was then
centrifugedat100,000 x gtopelletmembranes. Allfractions
were saved, and 5' nucleotidase (Sigma) assays were
per-formed to localize plasma membranes. Bicinchoninic acid
assays were doneto determine protein concentration. lodination ofcell membranes. Cell surface moleculeswere
iodinated as previously described (1). Intact SCPcells (107) wereresuspended in 10ml ofPBS (pH 7.4) which contained
10mCi of
Na1251,
20 p.goflactoperoxidase perml,and100,ulof 0.03%H202.Thereactionmixturewasincubatedatroom
temperaturefor15min, and 100,ul offresh 0.03% H202was
added at 1, 5, and 10 min. At 15 min, the reaction mixture
wasdiluted in 15 ml of cold PBS containing 10 mMNal and the cellswere centrifugedat 1,000 x gfor 10min.
The 125I-labeled SCP cells(107cellsin10ml ofPBS)were
incubated with 500 ,ug ofpurified unlabeled visna virus for 1 h at 37°C. The virus was cross-linked to any bound
pro-teins (see below), and then one-half was incubated with
preimmune serum or serum against purified gpl35. The
immune complexes were isolated with protein A-Sepharose
(Pharmacia) and analyzed by SDS-polyacrylamide gel elec-trophoresis (PAGE). The labeled proteinsweredetected by
autoradiography.
Affinity purification. Cell surface virus-binding molecules
wereaffinity purifiedaspreviously described(17).Cells from 25 flasks of SCP cells were suspended in 12.5 ml ofMEM containing 50 mM BES (N,N-bis[2-hydroxyethyl]-2-amino-ethanesulfonic acid) (pH 6.5) and 1 mg of BSAper ml and
incubated with1.25mgofpurified visna virus for1hat37°C.
Cells were pelleted, washed, and resuspended in 25 ml of
PBS (pH 8.3) containing 50 p.g of dithiobis(succinimidyl)
propionateperml, 1 mM MgCl2,and0.02%NaN3for 1 hat room temperature. This cross-linked the virus to the cell
proteins it bound. Cells were again pelleted, washed, and
thenlysed in 25 ml of 0.02 M Tris (pH 8.0)-0.2 M NaCl-5 ,ug
of aprotinin per ml-0.2 mM EGTA-0.2 mM NaF-0.2%
sodium deoxycholate-0.5% Nonidet P-40. Nuclei were
re-moved by centrifugation at 1,000 x g for 10 min. Serum
raisedagainstvisna virusgp135, wasadded to the superna-tant at adilution of 1:400 for 1 hat 37°C. Protein A-Sepha-rosebeads(125 mg)wereaddedovernightat4°C. The beads
were centrifuged, washed three times in lysate buffer, and
applied to a 5 to 20% gradient SDS gel. Labeled proteins
werevisualizedbyautoradiography, and unlabeledproteins
were visualizedby staininginCoomassie blue.
Immune serum. Neutralizing antibodies to visna virus were obtained from a hyperimmune sheep as previously
described (20).
Preparationand purificationof antibodiestovirus-binding
protein. Antibodies to purified virus-binding proteins from
cell membranes were made by immunizing rabbits with
proteins affinity purified by cross-linkingtovirus (described
above) andseparated by SDS-PAGE (6). A 1-mg sample of
purified proteinfrom cellmembraneswas run on anSDSgel
and stained with Coomassie blue. Bands at the molecular
weightindicated inFig.3wereexcised andequilibratedwith
150 mMNaCI-10mMNa2PO4 (pH 7.4) andhomogenizedin 5 mlofsaline inaDouncehomogenizer.Thecrushedgelwas
lyophilized and resuspended in 1 to 5 ml of saline and
emulsified in Freund's complete adjuvant, and 2 ml was
injected intradermally into rabbits. Rabbits were boosted
every 4 weeks with an intramuscular injection of 2 ml of
crushed gel emulsified in Freund's incomplete adjuvant.
Immunoglobulins from the sera of the rabbits (20 mg of
proteinperml)were purified by precipitationwith saturated
ammoniumsulfate and resuspended at aconcentration of5
mg ofprotein per ml. Sera were tested by
enzyme-linked
immunosorbent assay(ELISA)(28)againstSCP membranes (200 ng perwell) purifiedas describedabove.
Immunofluorescence. GSM and SCP cells were plated in
35-mm2tissue culturedishesandallowedto growto
conflu-ency. Live-cell immunofluorescence was done, using no fixation or permeabilization of the cells. Primary antibody
from rabbit 2-21 pre- andpostimmunization (15-kDa protein)
andrabbit 2-23(50-kDaprotein)wasdilutedto1:20in MEM
supplementedwith2%lambserumand addedtoslidesfor20
min at 25°C. The cells were washed three times in PBS.
Fluorescein-conjugated swine anti-rabbit antibody was
added in MEM-1% lamb serumfor 30 min at a dilution of 1:20 inPBS, and the cells were washed threetimes in PBS andexaminedby fluorescence microscopy.
Virus binding assays.
[35S]methionine-labeled
virus was bound to SCP cells in 96-well plates. Cells werecooled to4°C
and washed in cold Hanks' balanced salt solution.Labeled viruswasdilutedto50ng/,ulin cold MEM
contain-ing0.5%fetal bovineserumand 25 mM HEPES andserially
diluted, and 100 ,ul was added per well. The plates were
incubatedat4°Covernight.Supernatants were removed, and
the cellswere washed twice with PBS (pH 7.4). Atotal of 100,ul of1%SDSwas added toeachwell, lysed cells were removed, and the plates were washed twice with PBS. The
35S-labeled
virus in the supernatants and cells was countedseparately in Liquiscint (National Diagnostics) on a
scintil-lation counter. Unlabeled virus competitions were done in the same manner, butplates were preincubated with unla-beled visna virusfor2hat4°C. Antibody competitionswere
done by preincubating plates for 2 h at 4°C with 5 mg of
purified immunoglobulinperml from rabbits.
Western (immunoblot) analysis. Cell membrane (1 mg
on November 10, 2019 by guest
http://jvi.asm.org/
a b c d ef
92"-I
691
i-46
I
9 h i
Lcells SCP GSM GSM
FIG. 1. VOPBA with 2 mg of protein from L cells (lanes a to d), SCP cells (lanes e to h), and GSM cells (lanes i to r). Visna virus was bound tolanes c, d, g, h, k,1, and o to r and was detected with either preimmune serum (negative control) (lanes b, d, f, h, j,1,n, p, and r) orpostimmune serum (lanes a, c, e, g, i, k, m, o, and q). Lanes a, b, e, f, i, and j were incubated in the absence of virus. Lanes q and r were preincubated with hyperimmune serum at a dilution of 1:100 before visna virus was added. Numbers on the left show size standards (in kilodaltons).
prepared as described above) from SCP, GSM, and L cells wasdiluted in 2x protein buffer (20% glycerol, 50 mM Tris
[pH 7.4], 2%SDS, 0.01%bromophenol blue), run on a 5 to
20% gradient SDS-polyacrylamide gel, and transferred to
nitrocellulose. The nitrocellulose was stained with ponceau
S, and strips were cut and incubated in blocking buffer containing5%BSA, 10 mM Tris (pH 7.4), and 0.15 M NaCl.
The strips were incubated with purified immunoglobulin
from pre-and postimmune rabbits for 2 h at concentrations
of0.25, 0.5,and1.0mg/mlinbinding buffer containing0.25%
gelatin, 20% chick serum,0.05% NonidetP-40,50 mMTris
(pH 7.4),5 mM EDTA, and0.15%NaCl. The nitrocellulose
strips werewashed three times in buffercontaining 0.15 M
NaCl,10 mMTris(pH 7.4),0.005%Tween 20,and1% BSA.
Thestrips were thenincubated with peroxidase-conjugated
swine anti-rabbit antibody (Dako)at adilution of1:200for 2
h and washed three times. Substrate containing
4-chloro-1-naphtholandhydrogenperoxide in PBS was added, and the
reaction was stopped by rinsinginwater. RESULTS
Identification of visna virus-binding proteins on cells. To
identify the molecules on susceptible cells which bind to
visna virus, the virus was bound to cell lysates and
mem-braneswhichhadbeenrunonSDS-PAGE and transferredto
nitrocellulose. Virus wasbound directly to the transferred
proteins, antivirus antibody was added, and a
peroxidase-conjugated secondary antibodywasaddedtovisualize bands
on thenitrocellulose which bound virus. Visna virus bound
to molecules of
approximately
15and 30kDain celllysates
from both SCP and GSM cells
(Fig.
1). To control fornonspecific binding of
antibodies,
we ran a control in theabsence of virus. Preimmune serum was also used as a
control forserumfrom virus-immunizedanimals. No bands were seeninthe absenceofvirusorin the presenceofvirus
plus preimmune serum. In
addition,
noprotein
bandswerevisualized in celllysates frommouse L
cells,
whichare notsusceptibletoinfection
by
visnavirus. Proteinbandswhichbound to visna virus were diminished in
intensity
whenstrips of nitrocellulosewere
preincubated
withhyperimmune
serumfromasheepatadilutionof 1:100 for1h
(Fig. 1,
lanesqandr). Proteins
corresponding
tothebands boundby
virusdonotappeartobedisulfide-linked
subunits,
asnonreducing
gel conditions giverisetoprotein bands of the same
molec-ularweight(data not shown).
To demonstrate direct binding of virus to the protein
bandson nitrocellulose,we used an assay with
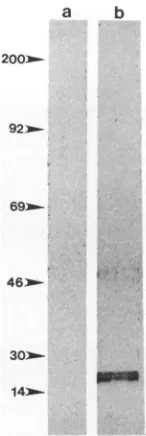
[35S]methio-nine-labeled visna virus. Labeled virus bound to molecules
of 15 and 50 kDa in SCP cell lysates (Fig. 2, lane b). No directbindingof labeled virustoLcells was detected(Fig.2, lane a). The detectionof the 50-kDa protein may bedue to the increased sensitivity of the labeled virus procedure,
whichincludes fewerstepsand therefore appears to be more
sensitive.
Affinitypurification and antibody response to
purffied
mol-ecules.Thecell membranes of SCP cellswereiodinatedwithlactoperoxidaseto labelsurface molecules. Visna viruswas
allowed to bind to the iodinated cells, and the virus was
cross-linkedtotheboundproteinsasdescribed inMaterials
a b
200-921m
6g10
FIG. 2. VOPBA with [35S]methionine-labeled visna virus (105 cpm)on 2mgofproteinfromLcells(lanea)and SCP cells(lane b).
Numbersonthe left show size standards(inkilodaltons).
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.112.502.79.223.2] [image:3.612.401.474.480.698.2]a
b
(<50
>
>
<~--30
^ <15
-FIG. 3. SDS-PAGE of '25I-labeled surface proteins recognized by visna virus. Iodinated cell membranes from SCP cells were incubated with visna virus and preimmune serum (lane a) or antiserumto visna virusgp135(lane b). The arrowheads indicate the 15-,30-,and50-kDa bands. The bands above and below the 50-kDa band arerecognizedby preimmune serum.
and Methods. The labeled cells and virus were pelleted at 1,000 x g,and unbound virus was removed by washing and
repelleting the cells. The cells were lysed (described in
Materials and Methods, "Affinity purification"), and
anti-bodiesto thevisna virusgp135wereused to
immunoprecip-itate the virus-protein complexes. The complexes were analyzed on SDS-polyacrylamidegels, and125I-labeled pro-teins were detected by autoradiography. Three species of
125I-labeled
proteins with molecular masses of approxi-mately 15, 30, and 50 kDa were identified. These proteins were not observed when preimmune serum was used (Fig. 3). Preimmune serum did recognize bands above and below the 50-kDa protein. Noniodinated cells were then used topurify larger quantities of the three protein species (see
Materials and Methods for details) for injection into rabbits. The rabbit immunized with the 15-kDa band (rabbit 2-21) developeda high titer of antibody after the first inoculation when its serum was tested against SCP membranes in an
ELISA.This titer diminished after the next boosterinjection
and did not increase after subsequent inoculations. Rabbit 2-22did not developappreciableamountsofantibodytothe 30-kDa protein, and rabbit 2-23 responded weakly to the initial inoculation of the 50-kDa protein. The ELISA is
againstall the components ofthe SCP membranes and thus
the relative amount of any protein in the membrane will
affect the assay.Although rabbit 2-23respondedonlyweakly
in the ELISA, these antibodies were found to recognize a component of the SCP cell surface in an
immunofluores-cence assay (seebelow).
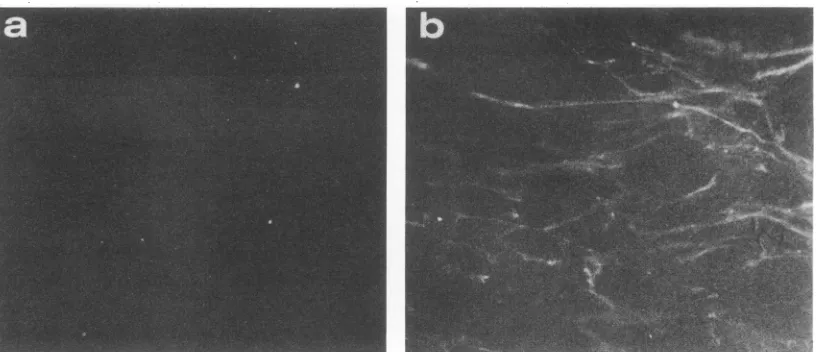
Immunofluorescence. To determine whether the immune rabbit serarecognized molecules on the surface oftheSCP
cells,wedidlive-cellimmunofluorescenceonSCP and GSM
cells. Antibodies to the 50-kDa protein
recognized
surfacemoleculesonboth GSM and SCP cells (datashownforSCP
cells,Fig. 4b),and preimmuneserum wasnegative (Fig.4a).
The antibodies to the 15-kDa protein werealso
positive
inthelive-cell immunofluorescence assay (datanot shown).
Binding ofvisna virus to SCP and GSM cells. 35S-labeled visna virus binds to the surface of both SCP and GSM cells (Fig. 5aandb).Competitionexperiments in whichunlabeled
visna virus was preincubated with SCP cells before the
addition oflabeled visna virus showed thata150-foldexcess
ofunlabeled visna virus competes for35%of thebindingof labeled virus (Fig. Sc). To examine which of the three
affinity-purified moleculesweremostimportantinbindingto
visnavirus, wepreincubated SCP cells withpurified
immu-noglobulinfrom rabbits 2-21 (15-kDaprotein), 2-22 (30-kDa
protein), and 2-23(50-kDa protein)at5mg/ml before adding
labeled virus. Only immunoglobulins fromrabbit 2-23 inhib-ited the binding of visna virus. Although the antibody
responseof this rabbit to SCP membranes was low,purified antibody blocked 34.7%of labeled virus binding or 99% of the inhibition by cold virus (Fig. Sd and e). An equal concentration ofimmunoglobulins directed against the 30-kDa banddid not block binding. However, since there was such a low antibody response against this protein, no con-clusionscan be made about theroleof this protein in virus
binding. Further, antibodies directed against the 15-kDa
band did not block binding despite the good response of
serum from the animal to SCP membranes in the ELISA.
FIG. 4. Immunofluorescenceassay ofSCP cells using preimmune serum from rabbit 2-23 (50-kDa protein) (a) or postimmune serum bleed 5(b), eachat adilution of1:20.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.115.523.528.704.2]a
0.20 ,
0.15-C
0
a
C
a
30 0.10
IIL
a.W I , . . ..
0
C 10007
x
0
E
a 0
er
CS
la
.5
.5
is 900
-800
-
700-0
c
0
0
E
30
0
0-I ,
200 400
BvirmcpmonSCP
600
100 200
unWlbd vIu viru (ug)
1:80 1:40 1:20 1:10
SwumDiktlon
ba
0
E a
U.
800
d
X
c
0
E
IL 0 S C
i
.5
0
a
'0
Therefore, the 50-kDa protein appears to be an important
molecule inthe binding of visna virus tocells.
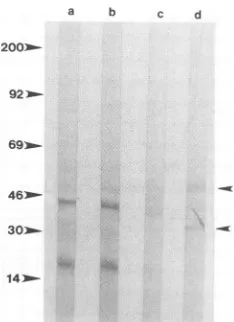
Western analysis of antibodies to the 50- and 15-kDa membrane proteins. Purified SCP and L-cell membranes wereprepared to determine whether the antibodies to the 50-and15-kDaproteinsbind to theappropriate proteins. West-ern blots containing purified cell membranes were reacted
with purified immunoglobulin from the rabbits. L cells
showednospecific reactivitytopostimmune rabbit
antibod-ies from rabbits immunized with the 15- or50-kDaprotein. Immuneserumfrom rabbit2-21, immunized with the 15-kDa
band, recognizedalargegroup of moleculesat a molecular
massrange of200 kDa(datanotshown). This couldbe due to the fact that the 15-kDa molecule is actually a cleavage
product of a larger molecule important in virus binding.
Antibodies tothe 50-kDaprotein (postimmune serumfrom
0.05'
0.07
0.08
0.05
0.04I
0
~~~~10000
20000avim
cpm
anGSM
400
300
200
100
-1:80 1:40 1:20
SwumDikhtlon
1:10
FIG. 5. Binding of35S-labeledvisna virus(specific activity,7 x
104cpm/,ug)tothesurface of cells. (a) Scatchard plot of visna virus boundtoSCP cells.(b)Scatchard plot of visna virus boundtoGSM cells. (c) Competition of unlabeled visna virus preincubated with SCP cells for2 hbeforetheaddition of5 x 104 cpmof35S-labeled visna virus. (d)Inhibition of1 x 104cpmof35S-labeledvisna virus bindingtoSCP cellsbyserumfrompreimmune rabbit2-23(50-kDa protein).
E,
preimmune; *, postimmune. (e) Inhibition of labeled visnavirus binding toSCPcellsby postimmunerabbit2-23bleed4 (50-kDa protein) serum divided by inhibition of labeled virus by unlabeled virus x 100. B/F, bound/free.rabbit 2-23) specifically recognized molecules at 50 and 30 kDa in the SCP membranes (Fig. 6, lane d). This demon-stratesthat theantibodiestothe50-kDaproteinwereagainst
cell membrane proteins and also that the antibodies to the 50-kDaproteinrecognized aprotein oftheappropriate size.
DISCUSSION
Virus overlay assays have been used successfully to
identifythe receptormolecule whichbindsto murine
hepa-titisvirus(2). Thismoleculewasfoundtobe presentonlyon
cellswhich weresusceptibletoviral infection. The VOPBA
was utilized in this study to identify molecules which bind
visnavirus.UsingtheVOPBA,itwasshown that visna virus
binds to molecules of approximately 15 and 30 kDa on
susceptible SCP and GSM cells. Reduced binding ofvisna
virus was demonstrated by preincubating the virus with
hyperimmune serumbeforebinding.
Bindingof visna virustoLcellswas not
observed,
exceptwhen radioactively labeled virus was used in the assay,
which appears to increase the sensitivity of the assay.
Although visna virus does appear to bind L cells at a low
0
-.- I .
-lq
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.612.63.293.71.558.2]a b c d
200:-
9210-
46_-303t 14
FIG. 6. Western blot with 1 mg of protein from cell membrane preparation ofL cells (lanes a and b) and SCP cells (lanes c and d). Purified immunoglobulin was diluted to 0.5 mg/ml. Lanes a and c contain immunoglobulin from preimmune rabbit 2-23 (50-kDa protein), and lanes b and d contain immunoglobulin from postim-munerabbit 2-23 (bleed 4). Numbers on left show molecular mass markers (in kilodaltons).Arrowheads on right show 50- and 30-kDa proteins.
antibody were made to the 15-kDa band; these antibodies reacted in immunofluorescence assays with surface mole-cules on SCP and GSM cells; however, they were not active ininhibiting virus binding to cells. Therefore, this molecule or portion of a molecule does not seem to be required for virus binding. However, antibodies to the 50-kDa band, which never reached high titers against SCP cell membranes,
blocked bindingof visna virus to the surface of SCP cells and
did so to the same extent as unlabeled virus. In addition, these antibodies recognized cell surface molecules in an
immunofluorescence assay. This result demonstrates that
thebinding of the antibody to the cells is highly specific and blocks the virus binding to the cell. Thus, the 50-kDa protein appears tobe the molecule or an essential component of the molecule which serves as the cellular receptor for visna virus.
ACKNOWLEDGMENTS
Wethank MaryannBrooks for invaluable assistance in prepara-tion of the manuscript.
This work was supported by Public Health Service grants NS16145, NS23039, and A128748 from the National Institutes of Health.
level, L cells are not susceptibleto infection. This may be
duetothefact that L cells have only low levels of expression ofthereceptormolecule for visna virus.Morelikely, there is
an intracellular species-specific block which prevents the completion of the life cycle of visna virus in a murine cell
(14).
The sizes of the virus-binding proteins were verified by affinity purification of purified cell membranes.Inadditionto the 15- and 30-kDa proteins identified in the VOPBA, a
50-kDabandwasobserved. Itwas thoughtthat the 15-kDa
molecule mightbe a subunit ofthe 30-kDa molecule; how-ever, no disulfide linkages could be demonstrated, since nonreducing gels showed identical 15- and 30-kDa bands in theVOPBA. It ispossiblethat the 15- and30-kDamolecules
are cleavage products of the larger 50-kDa protein. This is supported by the fact that antibody to the 50-kDa band recognizes not only the 50-kDa band but also the 30-kDa bandonnitrocellulosetowhich cell membraneproteinswere
bound.However,thisantibodydidnotrecognizethe 15-kDa band. Thiscould be due tomany reasons. Two possibilities
are(i)that theantibodydirectedtothe 50-kDa band bindsto
anepitopeorepitopespresenton the 30-kDaportionof the larger50-kDa molecule andnot tothe 15-kDaportionand(ii) that the 15-kDa molecule is a portion of another larger molecule which is associated with the virus-binding mole-cule but isnotactuallythe high-affinity virus-binding mole-cule or receptor. The notion that the 15-kDa band is a
cleavage productofalargermoleculewassupported bythe fact that antibody to the 15-kDaprotein recognized larger molecules onthe Western blot.
Protein moleculespurified by gel electrophoresisand used to immunize rabbits may not be optimal for inducing anti-bodies to the biologically active molecule. These proteins have been denatured and therefore maynotinduce antibod-ies against epitopes whichdepend on secondary structure. Fewor noantibodiesweremade to the 30-kDa bandinjected into rabbits and were therefore not efficient in any of the
assays performed. No conclusions can be made about the role of the 30-kDa protein in virus binding. High levels of
REFERENCES
1. Acuto, O., R. E. Hussey, K. A. Fitzgerald, J. P. Protentis, S. C. Meuer, S. F.Schlossman,and E. L. Reinherz.1983.Thehuman Tcellreceptor: appearance inontogeny andbiochemical rela-tionship ofalpha and beta subunits on IL-2 dependent clones andTcelltumors. Cell 34:717-726.
2. Boyle, J. F., D. G. Weismiller, and K. V. Holmes. 1987.Genetic resistance tomouse hepatitis viruscorrelates withabsence of virus-binding activity on target tissues. J. Virol. 61:185-189. 3. Choppin, P. W., and A.Scheid. 1980. The role of viral
glyco-proteinsinadsorption, penetrationandpathogenicity of viruses. Rev. Infect. Dis.2:40-61.
4. Cork,L. C., W. J. Hadlow, J. R. Gorham, R. C. Piper, and T. B. Crawford. 1974. Infectious leukoencephalomyelitis and arthritisin goats. J. Infect. Dis. 129:134-141.
5. Crane,S.E., J.E. Clements, and0. Narayan. 1988. Separate epitopesintheenvelope of visna virusareresponsible for fusion andneutralization: biological implications for fusion anti-bodies inlimiting virus replication. J. Virol. 62:2680-2685. 6. Creutz, C. E., W. J. Zaks, H. C. Hamman, S. Crane, W. H.
Martin, K. L. Gould, K. M. Oddie, and S. J. Parsons. 1987. Identification of chromaffingranule-binding proteins. Relation-ship of the chromobindins to calelectrin, synhibin and the tyrosine kinase substrates p35 and p36. J. Biol. Chem. 262: 1860-1868.
7. De Duve, C., and J. Berthet. 1981. The use of differential centrifugation in the study of tissue enzymes. J. Cell Biol. 91:225-275.
8. Frade, R.,M. Barel, B.Ehlin-Henriksson,and G. Klein. 1985. gpl40,the C3d receptorofhuman Blymphocytes, isalso the Epstein-Barr virus receptor. Proc. Natl. Acad. Sci. USA 82: 1490-1493.
9. Greve, J. M.,G. Davis, A.M. Neyer, C. P. Forte, S.C.Yost, C. W.Marlor,M.E.Kammarck,andA.McClelland. 1989.The majorhumanrhinovirusreceptoris ICAM-1. Cell 56:839-847. 10. Jolly, P. E., D. L. Huso, D. Sheffer, and 0. Narayan. 1989.
Modulationof lentivirusreplicationbyantibodies: Fcportionof immunoglobulin moleculeis essentialforenhancementof bind-ing,internalization, andneutralization of visnavirusin
macro-phages.J. Virol.63:1811-1813.
11. Jolly, P. E.,and0. Narayan. 1989. Evidence forinterference, coinfections, andintertypicvirus enhancementof infectionby
ovine-caprine lentiviruses.J. Virol.63:4682-4688.
12. Kennedy-Stoskopf, S.,and0.Narayan. 1986.Neutralizing anti-bodiestovisnalentivirus: mechanism andpossibleroleinvirus persistence. J.Virol. 59:37-44.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.612.127.244.80.241.2]13. Lifson, J. D., K. M. Hwang, P. L. Nara, B. Fraser, M. Padgett, N. M. Dunlop, and L. E. Eiden. 1988. Synthetic CD4 peptide derivatives that inhibitHIVinfectionandcytopathicity. Science 241:712-716.
14. Maddon, P.J., A. G. Daigleish, J. S. McDougal, P. K. Clapham, R. A. Weiss,and R. Axel. 1986. The T4 gene encodes the AIDS virusreceptor and is expressed in the immune system and the brain. Cell 47:333-348.
15. Maddon, P.J., J. S. McDougal, P. R. Clapham, A. G. Daigleish, S.Jamal, R.A. Weiss, and R. Axel. 1988.HIV infection does not require endocytosis of its receptor, CD4. Cell 54:865-874. 16. Markwell, M. K., S. M. Haas, L. L. Bieber, and N. E. Tolbert.
1978.Amodification of theLowryprocedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210.
17. McDougal, J. S., M. S. Kennedy, J. M. Sligh, S. P. Cort, A. Mawle, and J. K. A. Nicholson.1986.BindingofHTLV-III/LAV to T4+ Tcellsby a complex of the 110K viral protein and the T4 molecule. Science 231:382-385.
18. Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellularreceptorfor poliovirus: molecular cloning, nucleotide sequence,andexpression ofa newmemberof the immunoglob-ulin superfamily. Cell 56:855-865.
19. Narayan, O.,and J. E. Clements. 1989. Biology and pathogen-esis oflentiviruses. J. Gen. Virol.70:1617-1639.
20. Narayan,O., J. E. Clements, D. E. Griffin, and J. S. Wolinsky. 1981. Neutralizing antibodyspectrum determines theantigenic profiles of emerging mutants of visna virus. Infect. lmmun. 32:1045-1050.
21. Narayan, O., J. E. Clements, J. D. Strandberg, L. C. Cork, and D. E.Griffin. 1980.Biologic characterization oftheviruscausing
leukoencephalitisarthritis in goats. J. Gen. Virol. 50:69-79. 22. Narayan,O.,D. E. Griffin, and J. Chase. 1977. Antigenic shift of
visna virus inpersistently infected sheep. Science197:376-378. 23. Narayan, O., D. E. Griffin, and J. E. Clements. 1978. Virus mutation during "slow infection." Temporal development and characterization of mutants of visna virus recovered from sheep. J. Gen. Virol. 41:343-352.
24. Pyper, J. M., J. E.Clements, S. M. Molineaux, and 0. Narayan. 1984. Genetic variation among lentiviruses: homology between visna virus and caprinearthritis-encephalitis virus is confined to the 5' gag-pol region and a small portion of the env gene. J. Virol. 51:713-721.
25. Sattentau, Q. J., and R. A. Weiss. 1988. The CD4 antigen: physiological ligand and HIV receptor. Cell 52:631-633. 26. Sigurdsson, B., P. A.Paisson, and H. Grimsson. 1957.Visna, a
demyelinating transmissible disease of sheep. J. Neuropathol. Exp. Neurol. 16:389-403.
27. Smith, D. H., R. A. Bryn, S. A. Marsters, T. Gregory, J. E. Groopman, and D. J.Capon. 1987. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science 238: 1704-1707.
28. Stanley, J., L. M. Bhaduri, 0. Narayan, and J. E. Clements. 1987. Topographical rearrangements of visna virus envelope glycoprotein during antigenicdrift. J. Virol. 61:1019-1028. 29. Staunton, D. E., V.J. Merluzzi, R.Rothlein, R.Barton, S. D.
Marlin, and T. A. Springer. 1989. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell 56:849-853.
30. Thormar,H.1963.Thegrowth cycleof visna virus inmonolayer culturesofsheepcells. Virology19:273-278.