Vol. 64, No. 2 JOURNALOFVIROLOGY, Feb.1990, p.950-956
0022-538X/90/020950-07$02.00/0
CopyrightC) 1990,AmericanSociety forMicrobiology
Phenotypic Analysis
of Bovine
Papillomavirus Type 1
E2
Repressor Mutants
PAUL F. LAMBERT,t* BRAD C. MONK, ANDPETERM. HOWLEY
Laboratory of Tumor Virus Biology, National CancerInstitute, Bethesda, Maryland 20892 Received 1August 1989/Accepted 17 October 1989
The bovine papillomavirus type 1 (BPV-1) E2 open reading frame encodes three proteins: the E2
transcriptional transactivator,the E2transcriptionalrepressor(E2-TR), and theE8/E2fusionpeptide.Inthis
study,wedescribethephenotypesofBPV-1mutantswhicharedisruptedin theircapacityto encode eitherthe
E2transcriptionalrepressororthe E8/E2 fusionpeptide.We also describeexperiments which demonstratethat
the E8/E2geneproductfunctionssimilarlytoE2-TR. In the context of the entire viral genome,disruptionof
E8/E2 expression had little effectonviral processes, whereas disruption of E2-TRexpression resulted in a complexphenotypemarkedbya10-to20-fold increaseinviral DNAplasmidcopynumberaswellasincreased transformationpotential.A doublemutant, defective in theexpressionof bothE8/E2and E2-TRproteins, had highlevelsofE2 transactivationactivityyet had reduced plasmid replication capacityandadelayed capacity totransform rodent cells.
Bovine papillomavirus type 1 (BPV-1) transformation of mousecells has servedas a
model
forgeneticstudiesofthepapillomaviruses (19a). The viral genomepersists as a
mul-ticopyDNAplasmidinBPV-1-transformedmousecells,and
asubset oftheviral genes, theearlygenes,areexpressedat
low levels (14, 26). Viral gene expression appears to be at
least partially under the control of the viral E2
transcrip-tional regulatory circuit, which is comprised of the E2
transactivator, the E2 repressor, the E2 transcriptional
re-pressor(E2-TR), and the E8/E2fusionprotein (5,22,42, 48; this study). All three proteins are expressed in
BPV-1-transformed cells (18, 21). TheE2 transactivator, the
prod-uctof thefull-lengthE2 openreadingframe(ORF),activates viral transcription through E2-responsive elements (10, 11, 13, 41), which are cis-acting enhancer elements located
predominantly within the long control region (LCR) of the
viral genome. The E2 transactivator binds to a partial
palindromic sequence (1, 31), multiple copies of which are
located within the E2-responsive elements (10, 13, 40, 41)
and elsewhere in the viral genome (27). The E2 repressor
was shown tobe encoded by the 3' half of the same ORF, and its translation was predicted to initiate at an internal ATG codon (22) which is located just downstream of the P3080 promoter (2). A second E2 gene product that was truncated at the N terminus was previously predicted (15, 23)tobe thetranslation product of a viral RNA spliced from nucleotide (nt) 1234 to nt 3225 (5, 44) which fuses the upstreamE8ORF to the 3' end of the E2 ORF. Both the E2 repressor and the E8/E2 fusion protein contain the DNA-binding domain of the E2 transactivator, which maps to the
C-terminal100amino acids of the E2 ORF(30),and all three
E2 proteins share the capacity to dimerize with themselves andeach other (7, 29). Both competitive DNA binding and subunitmixinghave been proposed as potential mechanisms
bywhich the repressor molecules inhibit E2 transactivation. The purpose ofthis study was to identify the transcriptional
regulatory activity of the E8/E2 gene product and to
char-* Correspondingauthor.
tPresentaddress:DepartmentofOncology,McArdleLaboratory for Cancer Research, University of Wisconsin-Madison, 450 N. Randall Ave., Madison,WI 53706.
acterize the biological consequences ofdisrupting the tran-scriptional repressor activities encoded by BPV-1.
AnE8/E2 cDNA was constructed (Fig. 1A) and tested for its ability to function as a repressor in the two assays
originallyutilized in the identification of E2 repressor
activ-ity: inhibition of BPV-1 transformation and repression of E2 transactivation (22). In both assays, the E8/E2 cDNA was functionally identical toE2-TR. Cotransfection of the
plas-mid pE8/E2withan equal amountof the full-length BPV-1
plasmid p142-6 resulted inagreater-than-10-fold decrease in the number of transformed foci ofmouse C127 cells (Fig. 1B). Intransient cotransfections ofmonkey CV-1cells, the E8/E2 cDNAplasmid repressed the ability of the E2 trans-activator to enhance chloramphenicol acetyltransferase
(CAT)expression from the reporter plasmid by 5-to10-fold
(Fig. 1C). Not surprisingly,the E8/E2 geneproduct did not function as an E2 transactivator (data not shown), since it doesnotcontain the N-terminal E2 domainwhichis required fortransactivation(8, 12, 29). Thus, the E8/E2 gene encodes
a transcriptional repressor which is functionally
indistin-guishable from the previously identified E2 repressor E2-TR. In agreement withour data, Choe et al. (5) have isolated a
partialcDNA from BPV-1-transformed C127 cells encoding
theE8/E2 gene and have also concluded that theE8/E2 gene functionstoinhibit E2 transcriptionalactivation.
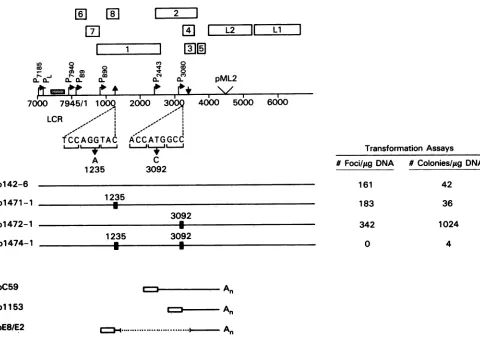
To examine the role of the repressor genes E2-TR and E8/E2 in thetranscriptional regulation of BPV-1, base sub-stitution mutations predicted to disrupt these genes were
engineeredinto p142-6, aplasmid containing the full-length
viral genomecloned in the bacterial vector pML2D (36). To
disrupttheE2-TRgene,amissense mutation was generated
alteringT toC at nt3092,givingrise to the plasmidp1472-1
(Fig. 2).Multiple syntheticoligonucleotides were utilized to
generate adouble-stranded DNA fragment with the BPV-1 sequencefrom the FspI site at nt 3023 to the HgaI site at nt 3113andcontaining the T-to-C base substitution at nt 3092. This DNAfragment was then cloned intop142-6 cut at the
samerestrictionsites,FspIand HgaI, by partial digestion to
give the clone p1472-1. The T-to-C base substitution in
p1472-1 caused a Met-to-Thr amino acid change in the E2
ORF. When engineered into the E2-TR cDNA p1153 (Fig. 2), this mutation eliminated the ability of this cDNA to
950
on November 10, 2019 by guest
http://jvi.asm.org/
NOTES 951
PSV40e a C'cN
CNC
, 'r
_
.
p1
42-6
I,,.
e*V. % * ' 1
p142-6
S::.
/0 ,,#.
v J-irJ
S _
Jt rd w
i'
f fa ,# sw
T: Xi.
'
.
\ z
:: *
S
a.
5 '-
:...
..p142-6 + pE8/E2
I
o
U'-CL +
)UJ
LO 00
Uw
+L FIG. 1. (A) Line drawing of plasmidpE8/E2,whichwasconstructed by cleaving the plasmidpCW1-28 (22) with the restrictionenzymes
XmaIandTthI and inserting the double-strandedDNAfragmentsynthesized with oligonucleotides withthe BPV-1coding strandsequence,
5'-CCGGGTTGCTGAAAATGAAGCTAACCGTGTTCTTACGCCCCTCCAGAGATCGCCCAGACG-3'. This sequence corresponds to
BPV-1 nt1193 to 1234 andis contiguous withnt 3225to3238. The sequence ofpE8/E2wasconfirmedbydirect sequence analysis. (B)
Inhibition of BPV-1 transformation. SubconfluentC127 cellsin6-cmdishesweretransfected by the calcium phosphate precipitationprotocol,
aspreviously described (22), with 1 ,ug of the wild-typeBPV-1 plasmid p142-6 withorwithout 1 pgof pE8/E2 plasmidDNA. Plateswere
stained withmethylene blue 2 weeks posttransfection.Darkly stainedareas aretransformedfoci. (C) Thin-layer chromatography of acetylated (fast-migrating formsattop)and unacetylated(originatbottom)forms of14C-chloramphenicoltaken from reactions in whichthesubstrate
was incubated for 30min with50 pug ofprotein extracted from CV-1 cells transiently transfected, as previouslydescribed(22),with the indicatedamountsof thefollowing plasmids: p407-1,5 ,ug; pC59, 1,ug; pE8/E2,3 ,ug;p1151 (cDNA-vectorcontrol plasmid [22]), 3,ug.
repress E2 transactivation (data not shown), in agreement with the finding that this ATG codon is utilized for
transla-tion initiation of E2-TR(21). The capacity for this mutation to disrupt E2-TR expression is further described in the accompanying paperby Riese etal. (35).
Todisrupt the E8/E2gene, aG-to-A basesubstitutionwas engineeredat nt1235,givingrisetotheplasmid p1471-1 (Fig. 2). This mutation was generatedby the protocol of Kunkel (19), using a single-stranded recombinant M13mpl8 clone containing the SmaI (nt 945)-to-EcoRI (nt 2113) BPV-1
subgenomic fragment and the antisense oligonucleotide primer with the sequence 5'-CTCCAGATACAGG-3'. The
sequenced clonemutatedat nt1235wasusedto reconstruct thefull-lengthmutantclonep1471-1 by exchangingthe XmaI
(nt 943)-to-BspMII (nt 1582) fragment of the mutated M13
recombinant intop142-6. TheG-to-A base substitutionat nt 1235 is a silent mutation in the E8 ORF and causes a
Val-to-Ile amino acidchange in the El ORF. This mutation altered the 5' splice sequence from 5'-AGGT-3' to 5'-AGAT-3'.Thecapacityof this mutationtodisruptutilization ofthent1234splicesitewasconfirmedby analysisofRNA isolatedfrom cells transfectedbyp1471-1 (datanotshown).
Adouble repressormutant,p1474-1, was alsogeneratedby introducing both of the individual mutations into the full-lengthBPV-1 genome (Fig. 2);the EcoRI(nt2113)-to-KpnI (nt 3455) fragmentfromp1472-1wasexchangedintop1471-1.
Hubbert et al. (18) identified the presence of three E2 peptidesinBPV-1-transformedrodentcells,with sizes of48, 31, and 28 kilodaltons (kDa). The E2-TR (in pl472-1) and E8/E2 (pl471-1) mutant plasmids were used in identifying
the E2 genes responsible forencoding the three E2
immu-nospecific peptides (21); the most abundant 31-kDa E2-specific proteinmaps tothe E2-TRgene;the less abundant 28-kDaE2-specific proteinmapstotheE8/E2gene;and the largestand least abundant48-kDaE2-specific protein maps
tothe E2 transactivatorgene.Theanalysis byLambertetal.
(21)confirmed thegenetic identityof the E2immunospecific peptides and provided proof that, in addition to the E2
transactivator, both the E8/E2 and E2 repressor genes are expressed in BPV-1-transformed cells. Furthermore, it
es-tablished that the repressor gene products werein greater abundance than the transactivator and that E2-TRwasthe predominantrepressorprotein (Table 1).
The disruption of the E2repressorgeneswasalso shown
(21)toalter the relative abundance of E2 transactivatortoE2 repressor gene products in cells harboring the repressor mutants p1471-1, p1472-1, and p1474-1 (Table 1). We were
therefore interested inmeasuringthetransactivation
poten-tial of these mutants. Levels of E2 transactivation were
measured by usingthereporter plasmidp407-1, which
con-tains the BPV-1 LCRasanenhancerclonedupstreamof the simian virus40earlypromoter. Expressionof the CATgene in this plasmid is dependent upon the presence of E2 transactivatoractivity (42)and isproportionaltothe relative levels of the E2 transactivator andrepressor proteins (22).
CATactivity was measured inmonkeyCV-1 cells cotrans-fected with thereporterplasmidand eithermutant or wild-type BPV-1plasmids (Table 1).Disruptionof the E8/E2gene (in p1471-1) led to no observable change in levels of E2 transactivationcomparedwith the levelofwild-typeBPV-1,
A.
pE8/E2
(W)
0
04
N9
E2
B.
C.
VOL.64, 1990
on November 10, 2019 by guest
http://jvi.asm.org/
952 NOTES
E
El
EZII
I
LO
co * 0
0O~O O~
CY
J m CD
arQ -i r-c o c
CL CL X1 X- X
7000 7945/1 1009
LCR i
TCCAGGTAC
A
1235
r
2-
L[E
I
L2I[
Ll
I
1
1
cs
cN
0 0
0-f"*
2000 3009 4000
--ACCATGGCC
C
3092
pML2
5000 6000
Transformation Assays
#
Foci/ag
DNA #Colonies/4g
DNA1235
m
3092
1235 3092
m
.~~~
FIG. 2. Location ofpointmutationsonthefull-lengthBPV-1genomicmapandtransformationpotentialofwild-typeandmutantgenomes. AtthetopisamapoftheBPV-1circulargenomelinearizedatnt7000 foreaseofpresentation.Thenumbered boxesindicate thelocations ofboththeearlyand late(Li andL2)ORFs.Thepositionandidentity of the viraltranscriptionalpromotersaregiven bythehorizontalarrows
atright angles.Thestipledbox indicates thepositionofE2-responsiveelement 1 within the LCR. The locationof thebacterial plasmidvector pML2DsequenceattheuniqueBamHIsite is indicated(36).The verticalarrowsindicatethepositionofnt1234and3225splicesignals.The nucleotidesequencessurroundingnt1235 and 3092aregivenbelow the linedrawing, andarrowsiadicatethebasechangeintroducedbythe respectivemutationsateach site;Horizontal brackets indicatethecodons in the E8 and E2 ORFs. Below thenucleotidesequences arethe linedrawings for the wild-type (p142-6)andthemutantplasmids,withsolidboxesindicatingthepresenceofpointmutations. Transformation assaydata foreach plasmidaregiventotherightofeach linedrawing.Theleft-hand column presents the number of focigeneratedon a6-cm dish ofsubconfluent C127cells transfectedaspreviouslydescribed(22)with 1 p.geach of theBPV-1plasmids and stainedat2weekswith
methyleneblue. Theright-handcolumnpresentsthe number of soft-agar coloniesgeneratedpermicrogram ofplasmid DNAtransfectedinto C127cells, as previouslydescribed (20). Briefly, 6-cm dishesof subconfluent C127 cells weretransfected, and 48 hlater the cellswere
trypsinized, suspended in 0.3%agar,and fed for4weeks,atwhichtime thenumber of colonieswasdetermined.Atthe bottomareindicated
the BPV-1 codingregions ofpC59 (48), which encodes the E2 transactivator; p1153 (22),which encodes E2-TR; andpE8/E2, which is described inFig. 1A.
as measured by CAT activity. A 10-fold increase in CAT activity, however, was observed with the ATG missense
mutant p1472-1, in which E2-TR is disrupted. A 30-fold
increase in CAT activity was observed with the double
mutant p1474-1. The transactivation potential of the
mu-tants, therefore, appears to correlate with their coding capacity; the double mutant, which expresses only the transactivator, had the greatest capacity to activate the
E2-responsive promoter.
To furtherexamine the biological effects of these
muta-tions,mouseC127 cellsweretransfected withthemutantor
wild-type BPV-1 genomes and analyzedfor viral
transcrip-tion, viral DNA replication, and cellular transformation.
Levels of viral transcription and DNA replication were
measured in pooled G418-resistant cell populations. These cell lines were derived by cotransfection of each viral plasmid with the plasmid pMMTneo, which encodes the
bacterial neomycin resistance gene transcribed from the mouse metallothionein promoter (25). Cellular
transforma-tion was measured by focus formation and by soft-agar colony formation of transfected C127 cells as well as by analysis ofpooled G418-resistant, cotransfected cell popu-lations.
Cellular transformation by BPV-1 involves the direct action oftwo independent viral genes, the ES and the E6 oncogenes (38, 47), andpossibly the E7 gene (32). The E2 genes are believed to indirectly affect the transforming capacityof BPV-1throughtheirmodulationof viral
promot-erswhich direct theexpression of theseoncogenes, primar-ilytheE2-responsive promoters: P89 (11, 41) and P2443 (16,
33). A recent study (4), though, has proposed a direct
involvement of the E2 genes in the immortalization of primaryrodentfibroblasts.Totestthetransformingcapacity
of the E2repressormutants, mouse C127 cells were trans-p142-6
p1471-1
p1472-1
p1474-1
161
pC59
p 153
183 342
0
42
36
1024
4
pE8/E2
=- An
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.73.553.72.411.2]NOTES 953 TABLE 1. Relative abundance of E2 proteins and levels of
transactivation in BPV-1repressormutants Level of E2 BPV-1 Protein withmolecular
plasmid mass(kDa)a:
Trans-DNA activationb
48 31 28
p142-6 1 21 6 1.0
p1471-1 0.4 13 1.3
p1472-1 3 15 9.7
p1474-1 0.2 27.0
aRelative abundance of E2 proteins present in C127 cellsharboringthe indicated BPV-1 plasmid DNAs. The abundance of the 48-kDa E2 species present inwild-type-transfected cells was arbitrarily set at 1.0. The data were taken fromLambert et al. (21) with the exception of that forp1474-1 (P. Lambert, N. Hubbert, J. Schiller and P.Howley, unpublished data).
b Relative rates ofchloramphenicol acetylation for extracts of CV-1 cells transiently transfected with 5 ,ug ofp407-1and 3 ,g of theindicated BPV-1 plasmid. The rate of acetylation inp142-6-transfected cells was arbitrarily set at 1.0. All rates were corrected for background CAT activity in cells transfected with p407-1 only.
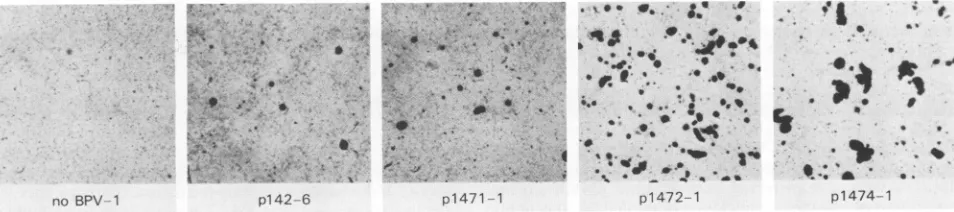
fected with the wild-type or the repressor mutant BPV-1
DNAs, and the efficiency of focus formation or the efficiency of colonyformation in soft agar was measured (Fig. 2). The
plasmid p1471-1was similar towild-type (p142-6) BPV-1 in
its abilitytoinduce formation of foci and soft-agar colonies.
The plasmid p1472-1 displayed a reproducible twofold
in-crease inits efficiencyof focus formation (Fig. 2). The foci
observedwithp1472-1 appearedsoonerand grewlargerthan
thoseresulting fromwild-type BPV-1transfection; cell
pop-ulations derived from individualclonedp1472-1 focigrew to
higher cell densities andcharacteristicallyturned themedia
acidic more quickly than did wild-type BPV-1-transformed
cells(data notshown). Inthe soft-agarassay, p1472-1gave
riseto atleast20-foldmorecolonies, whichappearedwithin
7daysposttransfection, thandidp142-6 (wild-type BPV-1),
whose colonies appeared in 14 to 18 days. In addition,
p1472-1 coloniesgrew tolarger diametersthandidcolonies
appearinginwild-type-transfected cells.
Thedoublerepressormutant(pl474-1)was
impaired
initstransformation capacity in each of these transformation
assays(Fig.2).Some fociwereobservedinamodified focus
assay, however, in which the cells were
trypsinized
48 hposttransfection
andplated onto two6-cmdishes. Thetime ofappearanceofthesefociwassignificantly delayed,
and theefficiency offocusinductionwasless than5%thatobserved
with thewild-typeplasmid in this assay. Experimentswere
performed to determine whether the double mutant was
cytotoxic, given the absence of transformation in the
stan-dardtransformationassays.The double mutant was
cotrans-fected with severaldifferent plasmids which provided
dom-inant selection, such as G418 resistance by using pMMTneo and BPV-1-independent transformation by using pEJRAS and pHARAS, which encode activated forms of the RAS oncogene.There was nosignificant decrease in the efficiency
of selection foreither of these dominant markers (data not
shown),indicatingthat the double repressor mutant was not
cytotoxic.
The cells generated by cotransfection of the neomycin resistance geneplasmid pMMTneo with each of the BPV-1
plasmids were analyzed for their capacity to grow as
colo-niesin soft agarafter continued cell passage (Fig. 3 [exper-iment performed at cell passage 17]). Cells harboring the
E8/E2mutantp1471-1exhibited a similar capacity to grow as
colonies in soft agar as did the wild-type BPV-1
transfor-mants, whereas cells harboring the E2-TR mutant p1472-1
grew moreaggressively in soft agar, with approximately59c
ofthe input cells forming colonies as compared with less
than 0.5% for the wild-type BPV-1 transformants. Cells
harboringthedoublemutantp1474-1acquired atransformed
phenotypeafter continuedpassage, as clearlyevidenced by theircapacity toefficiently form large colonies in soft agar.
Todeterminewhether the increasedtransformation capacity
seen with the double mutant in expanded populations of
G418-resistant cellswasreflective of levelsofviral
oncopro-teins, E5 immunoprecipitations were performed (Fig. 4A).
CellsharboringtheE8/E2repressormutantp1471-1had E5 levelswhichwerecomparabletothoseof cellsharboringthe wild-type BPV-1plasmidp142-6. Cellsharboring the E2-TR
mutant
p1472-1
had approximately 10-fold-higher levels ofE5comparedwith the levels inwild-type BPV-1-transfected
cells. This corresponded both to the highly transformed
phenotype of these cells (Fig. 2 and 3) and to elevated
expression ofan E5-CAT fusion gene (35). Cells harboring
thedoublemutant
p1474-1
hadE5 levels which were up tothreefoldhigherthanthose of the wild type. Thishighlevel
ofE5
expression
wasreproducible
indifferentpopulations
ofcells harboring
p1474-1, including
cell lines at an earlierpassage (passage5; datanot
shown).
Levelsofviral transcriptionweremeasured byNorthern
analysis (Fig. 4B) oftotal cellularRNA extracted from the
G418-resistant C127 cell
populations.
RNA was extractedfrom early (passage 5) and late (passage 17) passage cells.
The smalldifference in the
hybridization
patternat passage17 likely resulted from
slight
RNAdegradation
in thesesamples, which was reflected also in
hybridization
to thecontrol
,-actin
probe (data
notshown).
The levels ofviral#4
*oP -.p142 *6
* /
a
_.e'.: 1
a.
*
' ~. . .a,
'4
l
,
*47*2
p1472-1
"I I
.
0 .
i
;
*
*
4
. 4 1
p1474- 1
FIG. 3. Growth in softagarofG418-selectedpopulationsof C127cells.Poolsof G418-resistant coloniesweregenerated by cotransfection
of1,ugofpMMTneowith1,gof the indicatedBPV-1plasmidsandselection
by
growth
in mediumcontaining 420 ,ug of G418perml for 2weeks beforeresulting colonieswerepooled.Atotal of105cellswasplacedinto0.3%agaroseandfedfor 4 weeks beforestaining with 0.05%
iodonitrotetrazolium.TheG418-resistantpooledcellswereatpassage 17attimeofassay. VOL.64, 1990
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.612.77.554.582.688.2]954 NOTES
B.
c( N 4
r-
r-_ It
(3 a CL {1 a
Passage 5 Passage 17
,,.,.
. ...FtoF
0. 0.
t (9 .L4~
rN r cN r- rN rN
Nt St .t Mt Str
am a a 0.C.a
C.
mr' a a) a a
m
(N0.0.0.0X0.0.0
7.5 Kb-43Kd-|
29
Kd-11
4.4
Kb-18
14
Kd-6 Kd- _
3
Kd-FIG. 4. Levels of E5protein,total viralRNA,and DNAcopynumber inpooledG418-resistant cellpopulations.Cells usedin these assays
aredescribed in the legendto Fig. 3. (A) Cells (at passage 17) harboring the indicated BPV-1 plasmid were metabolically labeled with [35S]methionine for 3 h, and E5 was immunoprecipitated as previously described (39 [with modifications by Goldstein and Schlegel, manuscript in preparation]).Thearrowindicates thepositionof theE5peptide.Molecularmassmarkersareshownontheleft. Thecontrol (c)cell linewaspooledG418-resistant colonies ofC127 cellstransfectedwithpMMtneo only. (B)Northernanalysisof totalcellularRNAfrom pooledG418-resistant colonies harboringtheindicated BPV-1plasmidsextractedattheindicatedpassage.Total cellularRNA(2.5 ,ug)was electrophoresedon a1.4%agarosegel, transferred toa nylonmembrane(GeneScreen), andhybridized accordingto the protocol ofthe
manufacturer by using32P-labeled BPV-1 DNAastheprobe.Mobilities ofRNAstandardsareindicated atleft(Kb, kilobases).Control(c) RNAwasfrom thesame cells as inpanel A. (C)Southernanalysis of total cellularDNA extracted from pooled G418-resistantcolonies harboring theindicated BPV-1plasmidsextractedatpassage17. DNA(2.5 ,ug)wasshearedbypassagethrougha22-gaugeneedle fivetimes, electrophoresedon a0.7%agarosegel,andtransferredtonitrocellulosepaper.Hybridizationwasperformed by using 32P-labeledBPV-1DNA
astheprobe.The first lanecontained25pgofp142-6 plasmid.Thepositionsof form Iand formIIDNAsareindicatedatleft.Alsoincluded
areDNAsamplesobtained from cellsharboringeitherof thereplication-defectiveBPV-1genomes(p745-1andp743-23)and theBPV-1 mutant alteredatthe 5' splicesiteat nt3225(plO39-1) (15, 34). Control(c)DNAwasobtained fromsamecontrol cellsasinpanelA.
RNA in the cells harboringp1471-1wereequaltoorlessthan twofold higher than thoseinthecells harboring the wild-type viralgenomep142-6. Thelevel of virus-specific RNA in cells harboringp1472-1 was twofold higher than that of the wild
type atpassage5 butincreasedtoeightfold higheratpassage 17, which is thesamepassageusedfor the analysis of colony
formation insoftagar(Fig. 3), E5expression (Fig. 4A),and
viral DNAcontent(Fig. 4C). The level of viral RNAseenin
the double mutant p1474-1-transfected cells was at least fivefold lower than that in the wild-type-transfected cells even at the later passage, when aggressive growth in soft
agarandhigh levels ofE5 expressionweremeasured.
The BPV-1 double-stranded circulargenome persistsas a multicopyplasmid in virally transformed rodentcells, which may correspond to the viral replicative state in the basal epithelial cells and thedermal fibroblasts ofafibropapilloma (26). Thevirus encodes bothcis andtransgenetic elements
required for plasmid replication(19a). The E2 transcriptional
regulatory circuit is involved inthe maintenance of the viral
plasmid state,since disruption of the E2 transactivatorgene
product leadstothe loss ofplasmid replication(6, 9, 34). The E2-TR and E8/E2 mutations were therefore predicted to affecttheviral plasmidDNAreplicativecapacity. To
exam-ine the state of the viral DNA, wild-type and mutant viral
plasmids were cotransfected with plasmid pMMTneo and drug-resistant colonieswereclonedorpooled; total genomic
DNAwasisolated and probed forBPV-1-specificDNA(Fig.
3C). Cells harboring the mutant p1471-1, which was dis-ruptedintheexpression ofE8/E2, hadaplasmidDNAcopy number similar to that of wild-type transformant cells,
rangingfrom50to 100copiespercell in the differentpooled populations analyzed. Cells containing the E2-TR mutant p1472-1 hadaplasmid copy number of 500to 1,000, which
was 10- to 20-fold higher than that of wild-type transfor-mants. Significantrearrangementoftheinputp1472-1DNA
was evidenced byheterogeneity of the form IDNA. These
rearrangements appeared tobe confined principally to the
bacterial plasmidvector sequences, since restriction
diges-tion withBamHI,which releasesthe viral DNAinsert,gave risepredominantly to full-length BPV-1 genomic fragments (data not shown). The high-copy-number phenotype of p1472-1wasalso observedwhencellswere selected directly for their transformed phenotype by cloning foci (data not shown). The double mutant p1473-1 was also replication competent but hadareducedcopy numberof 5to25,in the rangeof 10to30% thatof thewild-type viral plasmid. The
lowplasmidcopynumber andthe lack of cytotoxicity of this
double mutantindicatethat this isnota runawayreplicon.
Inthisstudy, the E8/E2geneproduct is showntobeanE2 transcriptional repressor which, like E2-TR, inhibits the activity of the E2 transactivator. Disruption of the E8/E2 genebyamutationatthent1234 5' splicesignal in the viral genome had no observable effect on E2 transactivation capacity, viral transcription, viral replication, or levels of virally induced cellular transformation. Perhaps this is
re-flective of the fact that E8/E2 protein is in low abundance compared with the alternate repressor E2-TR (20). The absence of a phenotype for p1471-1 is of added interest
because thepointmutation ofthesplice signalat nt1235was predictedtoaffect theexpression ofatleastoneother viral
A.
J. VIROL.
1A 1VW_
W-f'
Md.:
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.612.63.559.84.289.2]NOTES 955 gene,
E1-M,
which is apparently required for stable plasmidreplication (3, 28). The El-M gene product is a 23-kDa phosphoprotein encoded by a spliced message which utilizes the nt 1234 5' splice signal (46). Yet the mutant
p1471-1,
which is defective in its
utilization
of the nt 1234 5' splice signal, is replication competent. M. Botchan (unpublished observation) has observed that the 23-kDaEl phosphopro-tein is replaced by an apparent full-length El ORF phos-phoprotein in cells harboring an identical nt 1235 point mutation mutant. It is possible that this larger El protein retains the El-Mactivity, permitting the nt 1235 mutant to remain replication competent.Disruption of the major E2 repressor gene E2-TR had a pleiotropic effect on viral processes, including increases in E2 transactivation activity, viral RNA, viral plasmid copy number, transforming potential, and abundance of the viral
E5
oncoprotein. Increases in these processes were pre-dicted, given that there was a large change in the relative abundance of the transactivator to repressor proteins result-ing from the lack of E2-TR expression in this mutant (21). This change in relative abundance was significant, despite an increased level of E8/E2 expression by this mutant (Table 1). A similar study was performed on an identical ATG mis-sense mutant at nt 3092 and arrived at a similar conclusion that disruption of E2-TR resulted in an increase in the potential of the virus to transactivate, replicate, and trans-form (35). It will be of interest to understand the conse-quences of disrupting E2-TR expression in dermal fibro-blasts and in basal epithelial cells of a bovine fibropapilloma. The behavior of the double repressor mutant, disrupted in the expression of both E2-TR and E8/E2, is complex and enigmatic. As predicted, transactivation activity was 30-fold higher than that of the wild-type viral genome in CV-1 cells (Table 1). Yet the steady state level of viral RNA in stably transfected C127 cells was 5- to 10-fold lower than that ofthe wild-type virus; correspondingly, the mutant viral plasmid copy number was low, and there was a delay in the obser-vance of a transformed phenotype. One possibleexplanation for the delayed onset of transformation is that, in the absence of E2-TR and E8/E2 expression, there is a pertur-bation in the temporal sequence of events occurring subse-quent to the introduction of the viral genome into the recipientcell.This could cause a delay in the time neededto obtain the threshold level of viral oncoprotein expression sufficient for cellular transformation. In the context of this argument, coselection of transfected cells by using G418 could provide the necessary expansion of recipient cells, allowing them to outgrow the nonrecipient cells, which are thought to inhibit cellular proliferation (24).It remains to be understood why the absence of bothE2 repressors, E2-TR and E8/E2, would retard the transforma-tion process whereas the absence of only the major repres-sor, E2-TR, causes a more predictable increase in viral processes. The complex phenotype of the double mutant raises the possibility that either E8/E2 or E2-TR encodes functions in addition to the repressionof E2transactivation and that the loss of such functions is phenotypically mani-fested only in the complete absence ofrepressor activity. It is also possible that the behavior of the double mutant supports the proposal that the full-length BPV-1 E2 gene product, the E2 transactivator, can under certain circum-stances repress BPV-1 transcription, as describedpreviously
for the BPV-1 P7185 promoter (43) and, similar to the capacity of thefull-length E2 gene product, can repress the HPV-18 P97 promoter (45). Alternatively, itispossible that BPV-1 encodes additional transcriptional regulators which
might override the E2transcriptional circuit under certain
conditions, as had been proposed in the context ofDNA
replication-defective BPV-1 mutants disrupted in the El
ORF (20, 37).
This study provides evidence that BPV-1 utilizes two mechanisms toexpress E2 repressor activities and that these repressor activities play important rolesin
modulating
viralprocesses. It is curious that BPV-1 encodes two repressor genes. There are multiple stages in the life
cycle
of thepapillomavirus. Perhaps E2-TR and E8/E2
play
differentroles inthistemporalprocess. E2-TRalsocontains moreof
the E2codingregionthandoes
E8/E2,
raising
thepossibility
that the two proteins may have different activities which haveeluded detection with our current assays.
Irrespective,
the datagenerated in this study leadtothe conclusionthat the high excess in the levels of the E2 repressor gene products over the E2 transactivatorgene
product
inmouse cellstransformedbywild-typeBPV-1isrequired
toestablishandmaintain the tightregulation of viral processes in these cells. Thecodingpotentialfor
multiple
E2geneproducts
in some human papillomaviruses (17) suggests that this may provide a general mechanism forregulating
thetransactiva-tionpotential of the papillomaviruses.
Weacknowledge theexpert technical assistance ofJ. Kim. We aregratefultoA.Griep, B. Spalholz, and F. Thierry for
critically
reading the manuscript and to D. DiMaio and M. Botchan for communication of resultspriortopublication.
LITERATURE CITED
1. Androphy,E. J., D. R. Lowy, andJ. T. Schilier. 1987. Bovine papillomavirus E2 trans-acting gene product binds to
specific
sites in papillomavirusDNA. Nature(London) 325:70-73. 2. Baker, C. C., and P. M. Howley. 1987. Differential promoterutilizationbythebovinepapillomavirusintransformed cells and productively infectedwarttissues. EMBOJ. 6:1027-1035. 3. Berg, L., M. Lusky, A. Stenlund, and M. Botchan. 1986.
Repressionof bovinepapillomavirusreplication ismediated
by
avirally encodedtrans-acting factor. Cell 46:753-762. 4. Cerni, C.,B.Binetruy,J.T.Schiller,D. R.Lowy,G.
Meneguzzi,
and F. Cuzin. 1989. Successive steps in theprocess of immor-talizationidentifiedby transfer ofseparate bovine
papillomavi-rus genes into rat fibroblasts. Proc. Natl. Acad. Sci. USA 86:3266-3270.
5. Choe, J., P. Vaillancourt, A. Stenlund, and M. Botchan. 1989. Bovinepapillomavirustype 1encodestwoforms ofa
transcrip-tionalrepressor: structural and functional
analysis
ofnewviral cDNAs. J. Virol. 63:1743-1755.6. DiMaio, D., and J. Settleman. 1988. Bovine
papillomavirus
mutant temperature defective for transformation,
replication
andtransactivation. EMBOJ.7:1197-1204.
7. Dostatni,N.,F.Thierry,and M. Yaniv.1998.Adimer ofBPV-1 E2proteincontainingaproteaseresistantcoreinteractswithits DNAtarget.EMBO J. 7:3807-3816.
8. Giri,I., and M. Yaniv. 1988.Studyofthe E2 gene
product
ofthe cottontail rabbit papillomavirus reveals a common mechanismof transactivation among the
papillomaviruses.
J. Virol. 62: 1573-1581.9. Groff, D.E.,and W. D. Lancaster.1986. Genetic
analysis
ofthe 3' early transformation andreplication
functions of bovine papillomavirustype 1. Virology 150:221-231.10. Harrison,S.M.,K. L.Gearing,S.-Y.Kim,A.J.Kingsman,and S. M. Kingsman. 1987. Multiple cisactive elementsin thelong control regionof bovine
papillomavirus
type 1. Nucleic Acids Res. 15:10267-10284.11. Haugen, T. H., T.P. Cripe,G. D.Ginder, M.Karin,and L. P. Turek. 1987. Trans-activation of an upstream
early
gene pro-moterof bovinepapillomavirustype 1byaproduct
of the viral E2 gene. EMBO J.6:145-152.12. Haugen, T.H.,L. P.Turek,F. M.Mercurio, T. P.
Cripe,
B. J. Olson, R. D. Anderson, D. Siedl, M. Karin, andJ. C. Schiller. VOL.64, 1990on November 10, 2019 by guest
http://jvi.asm.org/
956 NOTES
1988. Sequence specific and general transactivation by the BPV-1 E2 transactivator require an N-terminal amphipathic helix-containingE2domain. EMBO J. 7:4245-4253.
13. Hawley-Nelson, P., E. J. Androphy, D. R. Lowy, and J. T. Schiller. 1988. The specific DNArecognition sequence of the bovinepapillomavirusE2proteinisanE2-dependentenhancer. EMBOJ. 7:525-531.
14. Heilman, C.A.,L.Engel,D. R.Lowy,and P. M.Howley.1982. Virus-specific transcription in bovine papillomavirus trans-formedmousecells. Virology 119:22-34.
15. Hermonat, P. L., and P. M.Howley. 1987. Mutationalanalysis
ofthe 3' openreadingframes and the splice junctionat nucle-otide 3225 ofbovine papillomavirus type 1. J. Virol. 61:3889-3895.
16. Hermonat, P. L., B. A.Spalholz,and P. M.Howley. 1988. The bovine papillomavirus P2443 promoteris E2 trans-responsive: evidence forE2autoregulation. EMBO J. 7:2815-2822. 17. Hirochika, H.,T.R.Broker, and L. T. Chow. 1987. Enhancers
andtrans-actingE2transcriptional factors ofpapillomaviruses. J.Virol. 61:2599-2606.
18. Hubbert, N. L., J. T.Schiller,D. R.Lowy,and E.J.Androphy. 1988.Bovinepapillomavirustransformedcells containmultiple E2proteins. Proc.Natl. Acad.Sci. USA 85:5864-5868. 19. Kunkel, T. A.1985.Rapid and efficientsite-specific mutagenesis
without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492.
19a.Lambert, P. F., C. C. Baker, and P. M. Howley. 1988. The genetics ofbovine papillomavirus type 1. Annu. Rev. Genet. 22:235-258.
20. Lambert,P.F., and P.M.Howley.1988. Bovinepapillomavirus type 1 El replication-defective mutants are altered in their transcriptional regulation.J. Virol. 62:4009-4015.
21. Lambert, P. F., N. L. Hubbert, P. M. Howley, and J. T. Schiller. 1989. Genetic assignment of multiple E2 gene products in bovine papillomavirus-transformed cells. J. Virol. 63:3151-3154.
22. Lambert, P. F., B. A. Spalholz, and P. M. Howley. 1987. A transcriptional repressorencodedby BPV-1 shares acommon carboxy terminal domain with the E2 transactivator. Cell 50: 69-78.
23. Lambert, P. F., B. A. Spalholz, and P. M. Howley. 1987. Evidence that bovine papillomavirus type 1 may encode a negative transcriptional regulatory factor. Cancer Cells5:15-22. 24. Land, H., A. C. Chen, J. P. Morgenstern, L. F. Parada, and R. A. Weinberg. 1986. Behaviorofmycand ras oncogenes in transformation ofrat embryo fibroblasts. Mol. Cell. Biol. 6: 1917-1925.
25. Law, M. F., J. C. Byrne, and P. M. Howley. 1983. A stable bovinepapillomavirushybrid plasmid thatexpresses adominant trait. Mol. Cell. Biol.3:2110-2116.
26. Law, M.F.,D.R. Lowy,I.Dvoretzky, and P. M. Howley. 1981. Mouse cellstransformedby bovinepapillomavirus contain only extrachromosomal viralDNA sequences.Proc.Natl. Acad. Sci. USA 78:2727-2731.
27. Li, R., J. Knight, G. Bream, A.Stenlund,and M. Botchan. 1989. Specific recognition nucleotides and their DNA context deter-mine theaffinity ofE2protein for17binding sites in the BPV-1 genome.GenesDev. 3:510-526.
28. Lusky, M., and M. R. Botchan. 1985. Genetic analysis of bovine papillomavirustype 1trans-acting replication factors. J. Virol. 53:955-965.
29. McBride, A. A., J. C. Byrne, and P. M. Howley. 1989. E2 polypeptides encoded by bovine papillomavirus type I form dimers throughthe common carboxyl-terminal domain: trans-activationismediatedby the conserved amino-terminal domain.
Proc.Natl. Acad. Sci. USA 86:510-514.
30. McBride, A. A., R. Schlegel, and P. M. Howley. 1988. The carboxyterminal domain sharedbythe bovine papillomavirus E2 transactivator and repressor proteins contains a specific DNAbinding activity. EMBO J. 7:533-539.
31. Moskaluk, C., and D. Bastia. 1987. The E2 gene of bovine papillomavirus encodes an enhancer-binding protein. Proc. Natl. Acad. Sci. USA 84:1215-1218.
32. Neary, K.,and D. DiMaio. 1989.Open readingframes E6 and E7 of bovine papillomavirus type 1 are both required for full transformationofmouseC127 cells. J. Virol. 63:259-266. 33. Prakash, S.S.,B. H.Horwitz,T.Zibello,J. Settleman,and D.
DiMaio. 1988.BovinepapillomavirusE2geneexpression regu-lates expression of the viral E5 transforming gene. J. Virol. 62:3608-3613.
34. Rabson,M. S., C. Yee, Y.C. Yang, and P. M. Howley. 1986. Bovinepapillomavirustype 1 3'early regiontransformation and plasmidmaintenance functions. J. Virol. 60:626-634.
35. Riese,D.J., II,J. Settleman,K. Neary, and D. DiMaio. 1990. Bovine papillomavirus E2 repressor mutant displays a high-copy-numberphenotypeand enhancedtransforming activity.J. Virol.64:944-949.
36. Sarver, N.,J.C. Byrne,and P. M. Howley.1982. Transforma-tion andreplicationinmouse cellsofabovine
papillomavirus-pML2 plasmid vectorthat can be rescued in bacteria. Proc. Natl. Acad. Sci. USA 79:7147-7151.
37. Schiller, J. T.,E.Kleiner,E.J.Androphy,D. R.Lowy,and H. Pfister.1989.Identification of bovinepapillomavirusElmutants with increased transforming and transcriptional activity. J. Virol. 63:1775-1782.
38. Schilier,J. T.,W.C.Vass,and D. R.Lowy. 1984. Identification ofasecondtransforming regioninbovinepapillomavirusDNA. Proc. Natl. Acad. Sci. USA 81:7880-7884.
39. Schlegel, R., M. Wade-Glass,M. S. Rabson, andY.-C. Yang. 1986. The E5 transforminggene of bovine papillomavirus en-codes synthesis ofa smallhydrophobic protein. Science 233: 464-466.
40. Spalholz,B.A., J.C.Byrne,and P. M.Howley. 1988. Evidence forcooperativity between E2bindingsites inE2
trans-regula-tionof bovinepapillomavirustype 1. J. Virol. 62:3143-3150. 41. Spalholz,B. A.,P. F. Lambert,C. L. Yee, and P. M.Howley.
1987. Bovinepapillomavirus transcriptional regulation: localiza-tion of the E2responsiveelements of thelongcontrolregion.J. Virol. 61:2128-2137.
42. Spalholz,B.A.,Y.C.Yang,and P. M.Howley. 1985. Transac-tivation ofa bovine papillomavirus transcriptional regulatory elementbythe E2 geneproduct.Cell42:183-191.
43. Stenlund,A.,G.Bream,and M.Botchan.1987. A promoter with aninternalregulatorydomain inpartof theoriginofreplication in BPV-1. Science236:1666-1671.
44. Stenlund, A., J. Zabielski,H. Ahola,J. Moreno-Lopez, and U. Petterson.1985.MessengerRNAsfrom thetransforming region of bovinepapillomavirustype 1. J. Mol. Biol. 182:541-554. 45. Thierry, F., andM. Yaniv. 1987. The BPV-1 E2trans-acting
proteincan be eitheranactivatororrepressorofthe HPV-18 regulatory region. EMBOJ.6:3391-3397.
46. Thorner, L., N. Bacay, J. Choe, and M. Botchan. 1988. The
productofthebovinepapillomavirustype 1 modulator geneisa phosphoprotein.J. Virol. 62:2474-2482.
47. Yang, Y. C., H. Okayama, and P. M. Howley. 1985. Bovine
papillomavirus contains multiple transforming genes. Proc. Natl.Acad. Sci. USA82:1030-1034.
48. Yang,Y.C.,B. A.Spalholz,M.S. Rabson, and P. M. Howley. 1985.Dissociationoftransformingandtransactivatingfunctions forbovinepapillomavirustype 1. Nature(London)318:575-577. J. VIROL.
![FIG. 4.arepooledelectrophoresedRNAmanuscript(c)manufacturerharboringelectrophoresedarealtered[35S]methionineas the Levels of E5 protein, total viral RNA, and DNA copy number in pooled G418-resistant cell populations](https://thumb-us.123doks.com/thumbv2/123dok_us/1322562.85996/5.612.63.559.84.289/arepooledelectrophoresedrnamanuscript-manufacturerharboringelectrophoresedarealtered-methionineas-levels-protein-pooled-resistant-populations.webp)