JOURNAL OFVIROLOGY, July1979,p.220-230 Vol. 31,No. 1 0022-538X/79/07-0220/11$02.00/0
Biochemical
Characterization of
Temperature-Sensitive
Rabies Virus Mutants
NAIMASAGHIANDANNE FLAMAND*
Groupe des Laboratoires de BiologieExperimentale, Universite de Paris-Sud, 91405 Orsay Cedex, France Received forpublication27February1979
Biochemical characterization of
70temperature-sensitive
(ts)
mutantsof rabies
virus has been done
by
following the
appearanceof
viral proteins and RNA
molecules in infected cells
atboth
permissive and
nonpermissive
temperature.The
presence orabsence
of the
nucleocapsid protein (N)
wasdemonstrated
by
treating infected cells with anti-N fluorescent antibodies.
At33°C,
all the
mutantsinduced
afluorescence
comparable
tothe wild
type. At39.6°C,
the
mutants canbe
classified into three
groups.Three
mutantsinduced
afluorescence
comparable
to
the wild
type(F+ mutants);
54mutantsinduced
afaint
fluorescence which
wasproportional
tothe
multiplicity
of infection and increased with time
(F+-
mu-tants). No fluorescence
could be detected for the
13remaining
mutants(F-mutants). The
synthesis of all viral
proteins
wasshown
tobe normal for
F+
mutants,
indicating that
transcription
and
replication
of the virus
werenormal
and that the
tslesion
waslocated
in aprotein which is
notdirectly required for
those functions. The
synthesis
of all viral
proteins
wassimilarly
decreased for
F+-mutants
and
undetectable for the F-
mutants.This
suggeststhat
the
tslesion
affects the
transcription
and/or
replication
of the virus.
By
annealing
techniques
it
wasdemonstrated that the F+-
mutants wereable
toperform
someamountof
secondary
transcription
atnonpermissive
temperature. Nosecondary
transcrip-tion occurred with
F- mutants.When detectable
(i.e.,
athigher
multiplicity of
infection), primary
transcription of
F- mutants wasnormal.
Bussereau et
al.
(5;manuscript in preparation)
have isolated
117spontaneous orinduced
ther-mosensitive
(ts)
mutantsof the CVS strain of
rabies virus. Those
mutants wereunable
toplaque
at38.6°C,
although
the titer
at33°C
wasnormal.
Seventy of them showed
aresidual
growth
at anonpermissive
temperature(NPT)
less
than 2% of wild
type and wereretained for
further studies. These
mutantsfailed
tocomple-ment
each other under conditions in which
com-plementation
could be obtained for other
rhab-dovirus
ts mutants(see
A.Flamand,
In D. H.L.
Bishop (ed.), Rhabdoviruses,
inpress,
andref-erence 15for areview). The present report de-scribes a
biochemical
characterization of the ra-bies mutants to deternine whether the muta-tions involvethe same function.MATERIALS AND METHODS
The Orsay stockofrabies virus used isaclone of the CVSstrain ofrabies, whichtitratesat 1.5 x 107
PFU/ml. Viral multiplication was done in BHK-21 cells. Biochemical studies ofRNAandprotein synthe-sis were performed with another hamster cell line, CER (a generous gift of T. Wiktor), inwhich rabies formsplaques between30 and38.6°C.
The following procedurewasused to isolate
spon-taneousandchemically inducedmutants.The muta-genized orcontrol stockwastitratedat33°C,a per-missive temperature(PT). Well-isolatedplaqueswere
suspendedinsalinemedium,and thissuspensionwas then titratedat33and38.6°C,thelatterbeingaNPT. Clones that did not yield plaques atthe NPT were retainedasputativetsmutants.Once thetscharacter was confirmed, a stock was created and stored at
-70°C. The majority of mutant stocks titrated at approximately 107 to 2 x 107PFU/ml. Mutants were selectedat38.6°CasNPT sincewild-typeplaques did not formon CER cellsathigher temperatures. The wild-type viral production wasnormal up to 39.6°C (seeFig.6), and this temperaturewaschosenasNPT for biochemical studies. Most of the mutants have been isolated aftermutagenesis with5Fu, i.e.,mutants 01 to0102, 0105, and 0109to 0115. Mutants 0103 and 0104 are spontaneous. Twomutantshave been isolated after mutagenesis with nitrousacid (tsO106 and0107)orethyl methane sulfonate(EMS) (tsOI16 and0117).
Fluorescentantibodylabelingof cells. Pasteur or Wistar anti-rabies nucleocapsid fluorescent anti-bodieswereused.LyophilizedPasteur antibodies were twice diluted in isotonic buffer before use. Wistar antibodies (a generousgiftofT.Wiktor)werediluted 1/50 inisotonic buffer andstored at-70°C until used. Sterile chambers (Labtek)were seededwith BHK-21 cells and incubated overnight at 37°C, after which 220
on November 10, 2019 by guest
http://jvi.asm.org/
ts RABIES VIRUS MUTANTS 221
they were drained and infected with 50 ul of viral suspension. The inoculum was removed after 30min at room temperature and replaced with 0.3 ml of minimal essential medium (MEM) supplemented with 0.1% (wt/vol) bovine serum albumin (0.1% BSA-MEM). After 15, 20, or 30 h of incubation in a 5%C02 atmosphere at 33 or 39.6°C, the cells were washed twice with0.2Mphosphate buffer (pH 7.2), dried with acetone, and incubated with 0.1 ml of the fluorescent antibody solution for 1 h at 37°C. The preparations were then washed several times with phosphate buffer and distilled water, and slides were microscopically observed with an UV light source.
Determination of viral protein synthesis: in-fection and labeling of cells. Growth medium was removed from confluent CER monolayers (3 x 10'
cells) in 60-mm-diameter culture dishes. Cells were then infected with 0.2 ml of either viral suspension (multiplicity of infection [MOI] between 1.2 and 2.3 PFU/cell, see legends to thefigures) or sham infected with 0.2 ml of MEM. Adsorption was for 30 min at room temperature, andcells were then incubated for 15, 20, or 24 h at 33 or 39.6°C in 3 ml of 0.1% BSA-MEM in5%CO2. Cells were then treated with hyper-tonic amino acid-free medium (Earle salt solution supplemented with Earle vitamins, 10% fetal calf se-rum,andan excessofNaCl (600 mosM), as described by Madore and England (14) for 30 min.
Cellswerelabeled for120minin1ml ofhypertonic amino acid-free medium containing 25 yCi of [35S]-methionineat afinalspecific radioactivity of 50 to 100 Ci/mol.
Preparation ofcell extracts. Labeled cells were washed twice withice-cold TD buffer (0.15 M NaCl-0.01MTris-hydrochloride, pH 7.4) and were scraped from the tissueculture dish with 2 ml of the same ice-cold buffer. A 10-mlamountofethanol was added to thesuspension, which was storedovernight at -20°C. A 15-min centrifugation at 9,000 rpm in a Sorvall swinging-bucketrotor wasthenperformed. Thepellets were dried and then dissolved in 150 Il of protein dissociation buffer (62.5 mMTris-hydrochloride [pH 6.8], 2% sodium dodecyl sulfate, 10% glycerol, 5% 2-mercaptoethanol, 0.001% bromophenol blue). Samples were kept at roomtemperature for 1 h, boiled for1 min, and stored at -70°C until used. The protein concentrationwasdeterminedasdescribedby
Bram-hail
et al. (4). Appropriate amounts ofbuffer were addedso that all samples were atthe sameprotein concentration.Polyacrylamide gel electrophoresis. Samples
containing 20tIofcelllysatewereelectrophoresedin 10% discontinuous slabs containing sodium dodecyl sulfate (13). After electrophoresis for 4 h at 80 V, thegelswerefixed inmethanol-acetic acid-water (3: 1:6, vol/vol/vol), dehydratedindimethylsulfoxide, in-filtrated with 20% 2,5-diphenyloxazole in dimethyl-sulfoxide, dried,andfinallyexposedtoRPRoyal "X-Omat" film at -70°C, as described by Bonner and Laskey (3).
Preparation of unlabeled RNA from infected cells. Cellswere pretreated 30min before infection with 3 ml of 0.1% BSA-MEM containing 100 ,ug of cycloheximide per mlormock treated with 0.1%
BSA-MEM. They were then infected with 0.2 ml of the wild type or mutant viral suspension as described above and incubated in 0.1%BSA-MEM in the presence or absence ofcycloheximide (100 ,ug/ml). Efficiency of cycloheximide treatment wascontrolled before use: at adose of 100ug/ml, a 96% inhibition of cellular protein synthesis was found. Reversibility of the action of the drug was controlled as well to see whether a 24-h treatmentwith 100 ,ug ofcycloheximide per ml was not toxicfor the host cell. Itwasfound that 30 min after removal of the drug, protein synthesis in 24-h-treated cells increasedto66% ofsynthesis in the control non-treatedcells.
Aftervarying periodsof timeat 33 or39.6°C, cells weredrained, washed in Eagle medium, and solubilized with 5 ml of 0.01 MTris-hydrochloride (pH 7.4), 0.4 MNaCl, 1% sodiumdodecylsulfate,0.1mlof diethyl-pyrocarbonate (as RNase inhibitor), and 5 ml of phenol-cresol (500 g of redistilled phenol, 70 ml of redistilled m-cresol,0.5g of8-hydroxyquinoline satu-rated with200ml of0.01 M Tris-hydrochloride [pH 7.4], and0.15MNaCl)wasthen addedtothe suspen-sion. The mixture was then sonicated to fragment DNA, reduce the viscosity of the solution, and aid recoveryof RNA from thephenol-water interface. An MSE ultrasonic disintegratorwasused atfull power for40 s. After centrifugation, the phenol phase and theinterfacewerereextracted with1ml ofbuffer, and thecombined aqueousphaseswerereextracted with5 ml of phenol-cresol beforeaprecipitation with2 vol-umesof ethanol inasiliconized Corexcentrifugetube (Corning Glass, Corning, N.Y.). After an overnight
storage at -20°C, nucleic acids were recovered by
centrifuging for 30 min in a Sorvall HB4 swinging-bucketrotor at9,000 rpm. The nucleic acidpelletwas dissolved in 1 ml of0.01 M Tris-hydrochloride (pH 7.4)-0.4 MNaCl andwasreprecipitated with3ml of cold ethanol to removeresidual phenol and sodium dodecyl sulfate. After3h at-20°C,nucleic acidswere recoveredby centrifugationasabove, drained, dried,
dissolved in0.2 ml of0.01 MTris-hydrochloride (pH
7.4)-0.4 M NaCl, and finally frozen at -20°C until used.
Preparation of labeled viral RNA:preparation
of labeled virus. Three bottles of confluent BHK-21 cells (3x 107 cells)wereinfected with theOrsaystock of rabies virus at anMOI of0.1 PFU/ml.After ad-sorptionat roomtemperaturefor30min, infected cells wereincubated in 0.1% BSA-MEM (80ml/bottle)for 24hat370C in 5%CO2.The mediumwasthenreplaced
withanequal volume of 0.1% BSA-MEM containing
4mCi of[3H]uridine (Commissariat al'Energie Ato-mique,Saclay, France),and incubationwascontinued foranadditional48h.The supernatantwasremoved andclarifiedbycentrifugationat1,500 rpm for15min. Virus particles were concentrated by precipitation
withpolyethylene glycol(70 gofpolyethylene
glycol-6000per liter+23g ofNaCl perliter).Thesuspension
was kept at 4°C for 3 h and wascentrifuged
in aSorvall HB4rotorfor30minat5,000 rpm.The
pellets
werethen dried anddissolvedin 2ml of isotonic buffer (0.01 M Tris-hydrochloride [pH 7.4]-0.15 M NaCl).Theviralsuspensionwasthen loadedontoa10to60% sucrosevelocitygradientand
centrifuged
inaSW25.1 VOL. 31,1979on November 10, 2019 by guest
http://jvi.asm.org/
222
SAGHI AND FLAMANDrotorat23,000 rpmfor30min. The material in the gradient above the viral bandwascarefullyremoved,
andthe viral materialwasthen removed witha
pi-pette. No bands corresponding to defective particles
could be observed in thegradients.
Extraction of labeled RNA. A 4-ml amountof thephenol-cresolmixturewasaddedtothe viral sus-pension, and RNAwasextractedasabove except that thesonication stepwasomitted. The final RNA prep-arationwasdissolved in0.5mlof 0.01 M
Tris-hydro-chloride(pH 7.4)-0.4 MNaCl,and50-,ilportionswere storedat-20°Cuntilused. Thispreparationcontained 48 ugof RNA per ml withaspecific radioactivityof
1.5x 108 cpm/mgof RNA.
Annealingof RNA and RNase
digestion.
Unla-beled RNA from infected cells was annealed with labeled viral RNAaspreviouslydescribed(7).Briefly, 20, 5, or1IlI
of unlabeled RNA extracted from infected cells was mixed with 730, 1,460,or3,650 cpmof labeled viral RNA. Thevolume of the reactionwasadjustedto 25,ulwith 0.01 MTris-hydrochloride (pH 7.4)-0.4 MNaCl. The mixture wasincubated insealed tubes
for36hat
600C.
Eachsamplewasthen transferredto0.6ml of 0.01 M Tris-hydrochloride (pH 7.4)-0.4 M NaCl, anda
0.3-mi
portionwasprecipitatedwith tri-chloroacetic acid before orafterdigestion with pan-creatic RNase A at 20 ug/ml at 37°C for 30 min. Controls consisted of labeled viral RNA annealed with extractsfromuninfectedcells under thesame condi-tions.Each determinationwasmade induplicate,and a mean valuewascalculated from the results. Viral RNAdemonstrateda12% resistancetoRNase beforeannealing, and this figure was not significantly in-creased after incubation (17.5%). A relatively high RNaseresistance of RNA frompurified rabies virions hasbeen observedonseveral occasions (2; Flamand,
unpublished data). Postannealing RNase resistance was considered to be significant when it was more than20%(andless than80%) of the total radioactivity. When RNase resistance after hybridization was in-cluded between thesetwovalues, the total radioactiv-ityhybridizedbyunlabeled RNA extracted from one petri dish (3x 106 cells) was calculated and corrected for the background RNase resistance of viral RNA
(17.5%).
RESULTS
Initially, it
wasverified whether the
70 tsmutants
could induce the synthesis of rabies
nucleocapsid protein ininfected
cells
at both PT and NPT. The presence or absence of N proteinwas
demonstrated
by treating infectedcells
withanti-N
protein fluorescent
antibodies.Subse-J. VIROL.
quently, mutant-induced
protein
and RNA
syntheses
wereexamined with
electrophoresis
and
RNA-RNA
hybridization.
Fluorescence
induction in
infected cells.
When
cells infected with
wild-type
rabies
weretreated with anti-rabies
nucleocapsid
fluorescent
antibodies, fluorescence
began
to appearafter
11
h
at 37 or39.60C. Fluorescence became visible
after
15h
at33°C. This
early
fluorescence
wasa
function of the MOI.
Itwasdecided
toclassify
the mutants on
the basis of
fluorescence
induc-tion
ininfected
cellsafter
15,20,
or30h at PTand NPT
at anMOI lower than
1PFU/cell.
Fluorescence intensity
wasestimnated
onBHK-21
monolayers in which less than 70% of the
cells
had been infected.
Inthis
case, mostcells
wereinfected with
only
1PFU.
At
330C,
all the
mutantsinduced
afluores-cence
which
wascomparable
tothat induced by
the wild
type(Fig. 1).
At a temperatureof
39.60C,
three
tsmutants,ts022, ts034 and ts094,
exhibited
afluorescence similar
tothat
of the
wild
typeand
wereclassified
asfluorescent-pos-itive
mutants(F+). The remainder of the
70mutants
tested exhibited either
aweak
orun-detectable
fluorescence after
15or 20hof
incu-bation
at NPTand
weretermed F+-
orF-,
respectively.
When incubation
wasfor
longer
periods, i.e.,
30h,
or whencells
wereinfected
with
agreaterMOI,
mostofthese
mutants(F+-)
exhibited fluorescence. Thirteen
F- mutantsex-hibited
abarely detectable fluorescence after
30h
of incubation
or at anMOI of
10:tsO6,
tsO12,
tsO16,
ts018, tsO3M,
ts042, ts048, tsO55, ts083,
tsO9W, tsO96, tsOlOl,
and
ts0104.
When
detect-able, the fluorescence of
NPT was notdue
toleakiness of the
mutantssince residual
matura-tion
at39.6°C
wasless than 2%.
Conclusions based
on theresults of
fluores-cence
induction lack
a certainprecision,
sincethey depend
onincubation
time,
MOI, and other
factors, such
asthe
quality of the UV light
source
and
the
microscope and the
concentra-tion of
antibody preparations. Thus, with
amorepowerful
light
source orwith
moreconcentrated
preparations of fluorescent antibodies, it was
possible
to detect somefluorescence,
whereasthe test wasnegative in other conditions.
Viral
protein
synthesis
incells
infected
FIG. 1. Fluorescence of cells infected by wild-type and ts mutants of rabies virus. After 20 or 30 h of incubationat 33or
39.60C,
infected cells were treated with fluorescent antibodies directed against the rabies nucleocapsid protein as explained in the text. This figure shows successively: fluorescence induced bytsO55 (F- mutant) after 20 (a) and 30 h (b) of incubation at 33°C (MOI=0.7); fluorescence induced bytsO55 (F-mutant)after20(c) and 30 h (d) of incubation at39.60C
(MOI=0.7);3°Cfluorescence induced bytsO22 (F+ mutant) after 20 (e) and 30 h(D
of incubation at39.60C
(MOI=0.7); fluorescence induced by tsO23 (F+-mutant)after20(g) and 30 h (h) of incubation at39.60C
(MOI=0.7). Fluorescence induced by the wildtype,F+, andF+ mutants at 33°C or the wild type at
39.60C
resembles that of F mutants at33°C and is not shown inthisfigure.on November 10, 2019 by guest
http://jvi.asm.org/
20
h
30
h
on November 10, 2019 by guest
http://jvi.asm.org/
224 SAGHI AND FLAMAND
with
F+,
F',
and F- mutants. Slabgel
elec-trophoresis
wasusedtostudy
viralprotein
syn-thesis
in CERcells infected with the threemu-tantclasses and with the wildtypeat anMOI of 1 to 2
PFU/cell
after15,
20,
and 24 h of incu-bation at PT and NPT. Cellularprotein
synthe-sis was
selectively
depressed by
high-salt
treat-ment
(14).
Under theseconditions, proteins N,
Ml,
and
M2
areclearly
visible,
whereasproteins
L
and G
areless
easily
detectable.Autoradi-ographs
of thesegels
appear inFig.
2through
5.The
residual
growth of
mutants was measuredin
the
sameexperiment
as acontrol,
and itwasfound
thatthis
parameterwasless
than 0.1% ofU\
--v\17
\I
-9--
s-
-_
-000_
-W
a_~~ _
_4
_
_
Fr
10-_ "
_
W_
Ww m
-*..* S..e
..
:
tr
n._
:9
'';
\ -v
\l
-
1-
v
_
_
-
w
,
_
_
w
.
_
_. _
... ;..
\1
2 .- -11-,
low,X,
,9.,_~A"M
0
hPNP
151-H 20H
p NP
24 H FIG. 2. Intracellularprotein synthesis after infec-tion withwild-typerabies virus.Autoradiographs of 10%o slabgel electrophoresis. Cellswereinfectedwith
the wildtype ataMOIof1.2 PFU/cell.After 15, 20,
or 24h of incubation at 33 (PT) or39.60C (NPT),
infected cells were treated with hypertonic amino
acid-freemediumasexplainedin thetext.This treat-mentselectively depressescellularprotein synthesis andallowsthe detectionofviralproteins L,G,N,Ml, and M2. Labeling was for 2 h with 25 ,aCi of
[35S]methionine. A20-plamountof celllysate(from 3x105cells)wasloadedontoeach well and electro-phoresed for4hat80 mA.Positionsof the fiveviral proteinsdetermined by comigration ofproteins from purified labeled virus mixed with unlabeledcellular extractsareindicatedbyarrows. C, Residual intra-cellularprotein synthesis in uninfected cells (hyper-tonictreatmentandlabelingasforinfected cells).
'15
20 } 24FIG. 3. Intracellularproteinsynthesisafter
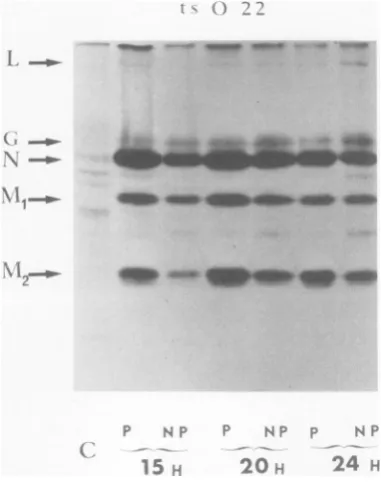
infec-tionwithanF+ts mutantof rabies virus. Autoradi-ographof10%oslab gelelectrophoresis. Labeling and electrophoresisasexplained in thetextand the leg-end toFig.2. The MOIwasequalto2.1PFU/cell.
that
of the wild
type atNPT. Mutant
multipli-cation at PT was normal
(cf.
Fig.
6for
atypical
result).
Protein
synthesis induced by
ts022 and ts094(F+) under these conditions
wascompared
toeach other at PT and NPT
and
wascomparable
to
that of the
wild
type.The
results
concerning
ts022 and thewild typeareshown in
Fig.
2and 3.For
F+
mutantsprotein
synthesis
seems tobe
normal
atNPT.
At
NPT,
protein
synthesis
wasdetectable
fortwo
F+-
mutants, ts023(Fig. 4) and tsO21 (data
not
shown),
although
it wasdepressed
incom-parison
to that at33°C
or tothat of
thewild
type. These results are consistent with the
hy-pothesis
that the syntheses ofall
viralproteinsare
similarly depressed.
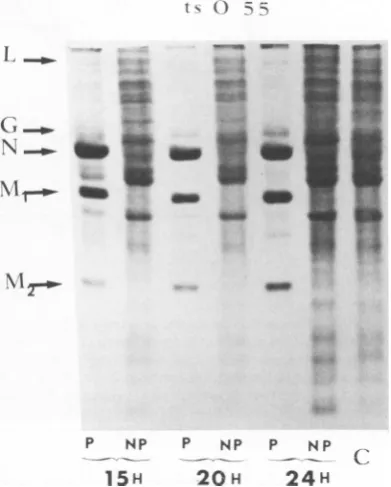
Although
protein synthesis in cells infectedwith five F- mutants,
ts055,
tsO31, tsO16,
tsOlOl and
tsOI04,
was normal at PT, it was undetectable at NPT. Results forts055
are shown inFig.
5.Since the
synthesis
ofall
viral proteins wassimilarly affected
in theF+-
and F- mutants,this suggests that the replication and/or
tran-scription
of the virus is affected atNPT.RNA synthesis
induced
byF`
andF-J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.508.267.458.79.323.2]ts RABIES VIRUS MUTANTS 225
1 v. ... _ _
x~
q-
S. U
4h,wIw.
P NP P NP P NP
[image:6.508.55.247.66.345.2]15
H2
0H
24
HFIG. 4. Intracellularprotein synthesis after
infec-tion withanF'- tsmutantofrabies virus. Labeling
andelectrophoresisasexplainedin thetextandthe
legendtoFig.2.The MOIwasequalto1.7PFU/cell. mutants. To
determine
at whichstep
of theviral
cycle,
i.e.,
atthelevel of
primary
transcrip-tion
orsecondary
transcription,
F'- and
F-mu-tants were
blocked,
viral RNA
synthesis
wasstudied
at PT and NPT inthe
presence orabsence of
cycloheximide.
Itis
well documented
that the
primary transcription
of
vesicular
sto-matitis virus
(VSV) (the
transcription
ofthe
infecting
genomeby
the
structural
enzyme),
thefirt step
ofthe viral
cycle,
occursnormally
in the presence ofcycloheximide. Replication
andsecondary
transcription,
however,
are blockedby
the
drug
when added
early
as aresult
of theinhibition
ofprotein synthesis.
It has beenshown
that
rabies,
asotherrhabdoviruses,
con-tainsa
particle-associated transcriptase
(10, 12)
and that the
primary
transcription
of this virus takesplace
in infectedcells
in the presence ofcycloheximide (1).
The levels of RNA
synthesis
arelow,
espe-cially
in the presence ofcycloheximide. Thus,
the more sensitive
technique
ofhybridization
was used to demonstrate it
(7, 8).
Unlabeled RNA from infected CERcels
incubated at PT and NPT forincreasing
periods
(0
to 18h)
wasextracted and hybridized with labeled RNA of
known
specific activity extracted from purified
rabies
virions
(seeMaterials and Methods
andthe
legend
toFig.
7for
details).
Thistechnique
enabled
us to determine thelevel
ofsynthesis
ofmolecules which
werecomplementary
tothe
viral
genome,i.e.,
primarily
viral
messengers,which represent themajority
of
RNAmolecules
synthesized during the
cycle of viral infection
(6).
These
biochemical studies involved
twoF+-mutants
(ts023 and tsO2), three
F- mutants(tsO55,
ts042, and
ts031)
and the wild
type.Results obtained with
wild-type
virus
areshown
in
Fig.
7. Inthe
presenceof
0.1mgof
cyclohex-imide
perml,
primary transcription
wasunde-tectable
at anMOI of
approximately
1.2PFU/
cell.
At anMOI of
10,it
wasclearly detectable
and
proceeded for
atleast
18h.
In
the absence
of
thedrug considerable
am-plification of RNA
synthesis occurred.
Forin-stance,
RNA
extracted
from 3 x106
cyclohexi-mide-treated
cells after
18h of infection
could
hybridize
4.3 x103
cpm,and
RNAextracted
from
the
samenumber of
nontreated
cellscould
hybridize
7 x104
cpm,although
the
starting
MOI
was 10times lower
(Fig.
7).
O%
()N-:1:~~~~~~~~~~~~
tI
&
Ae
N
--o.UP
ti
N
I
-40 _g_am__
_0 _
-tos*
r,*t
__
4ms
a"7
_
S~~~~
NiT0
p NP
1 5H
P NP
20H
P NP
24H
CI
FIG. 5. Intracellularprotein
synthesis
after
infec-tion with anF- ts mutantofrabies virus.
Labeling
andelectrophoresisasexplained
in thetextand the legendtoFig.2. TheMOIwasequal
to2.3PFU/cell.VOL. 31,1979
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.508.259.455.364.608.2]226 SAGHI AND FLAMAND
ppu.&l
7
10
04
id'
le
a 12 s 24 30
Time
(h)
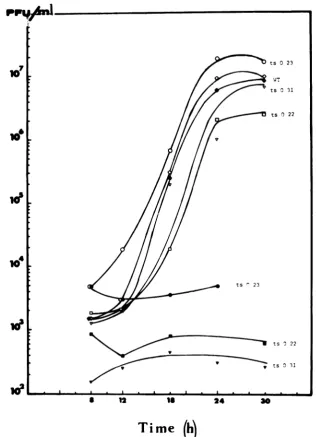
FIG. 6. Productionof infectious virus in cellsinfectedby thewildtypeandts mutantsofrabiesvirusat33 and39.6°C. Confluent CERmonolayers(3x 106cells) wereinfected with0.2ml of the viral suspension (MOI
wasbetween1and5PFU/ml, dependingonthestrain). Adsorptionwasfor30minatroomtemperature;cells
werewashed twice andincubatedat33or39.60C in3mlof 0.1%BSA-MEMin5%CO2. Mediumwaschanged
after1htoremovedesorbed virus.Aliquots(0.2 ml)weretakenatvarying periodsoftime and titratedas
explained in Flamandetal.(11). Open symbols:mutantsorwildtype atPT;closedsymbols:mutantsorwild
typeatNPT.
Since the specific activity ofthe probe was
known (1.5 x 108 cpm/mgof RNAor 1cpm =
8.6x 105molecules), the quantity of+ strands
present in the cytoplasm was calculated from
the number of counts per minute hybridized.
Resultswereexpressedin"genome
equivalent"
mass copies per infected cell (Table 1). In thepresenceofcycloheximide, synthesiswaslinear
foratleast 5h, givinganaverageof120genome
equivalents per infected cell per h. Since the
7t
ts 0 23
WT
ts 0 3I
ts 3 22
ts 3 22
Y ~ ~ ~ -~~ ts 3I1
. I I . I AL A
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.508.104.421.76.514.2]ts RABIES VIRUS MUTANTS 227
10
WT
m< P / 39.6'c(-C)
W.
0 _ 10
39.6',c (.C)
10
1 2 a 12 Time (h'
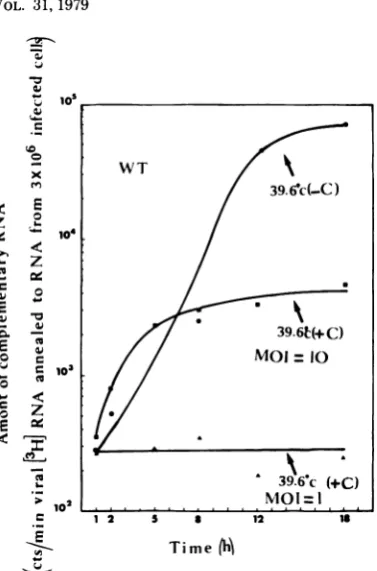
FIG. 7. Production of viral complementary RNA incellsinfected by wild-typerabies virusat39.60C in thepresence orabsence of cycloheximide. Total
un-labeled RNAs were extractedfrom rabies-infected
CER cellsatvarious periods oftimeafterinfection. TheMOIwas eitherequalto1.2orto10PFUper
cell. In this lattercase the virus has been
concen-trated by centrifugation (30,000rpmfor90minina
Beckman42 rotor). The pelletwasdissolved in 0.1%
BSA-MEM and titrated as usual. Since pelleting
couldcauseaggregation of the virus, theevaluation
of the MOI basedonthe titer in PFUswas aminimal
figure.Theproduction of rabies complementary RNA
wasmeasured inthepresenceand absence of
cyclo-heximideby annealing theunlabeled RNAs extracted from infected cellsto3H-labeledRNA extracted from purified rabies virions, asexplained in thetext.
Spe-cific activity of the RNA was 1.5x 108 cpm/mgof
RNA (1 cpm = 8.6 x 105 molecules). Symbols: 0,
39.60C inthe absence ofcycloheximideatanMOI of
1.2PFUpercell; A,39.60C in thepresenceof
cyclo-heximideatanMOI of1.2PFUper cell; U,39.60C in
thepresenceof cycloheximideatanMOI of10PFU
percell.
MOI was equal to 10 PFU/cell, the rate of synthesis was equal to 12 genome equivalents
per cell perinfectiousparticleperh. Therateis
therefore in the order ofonegenomeequivalent per5min.
In the absence ofcycloheximide, an average
of 9,860 and 20,880 genome equivalents were
present inthe cytoplasm 12 and 18 h after
infec-tion, respectively.
Clearly,
this numeration gives no indicationabout the
relative
amount of mRNA species. Itis
probable that
some are moreabundant thanthe
others, since the five viral
cistrons may notbe transcribed with the
sameefficiency,
depend-ing on their position on the viral genome (9, 14).
The
results
obtained with the two F+-mu-tants
(Fig.
8Aand
B)
werecomparable.
RNAsynthesis
atNPT islower than that
at33°C
orthan that of wild
type.Nevertheless, RNA
syn-thesis is
greaterin
the absence of
cycloheximide
than in its presence,
indicating
that atleast
somesecondary transcription occurred
atNPT. Theamount of mRNA present in the cytoplasm rep-resents
around
1/10 of what isnormally
foundwith
thewild
type at NPT.The
results
concerning the three
F- mutantsare
shown in
Fig. 8C and
Dand
9.RNA
synthesis
induced
by
mutantsts055and
tsO31
atNPTis
considerably
lower than that of wild
type orof
the
mutants atPT. Inaddition, it
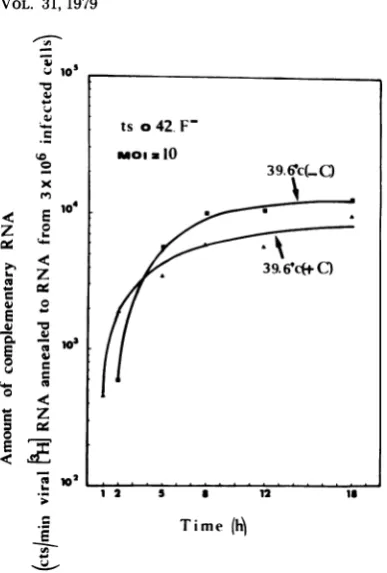
islower
thanthat
induced
by F+-
mutantsatNPT.When it
is
detectable, RNA synthesis
is nothigher
in the
absence than in the
presenceof
cycloheximide.
At
higher
MOIs, where primary transcription
atNPT is
clearly detectable,
RNAsynthesis
inthe presence orabsence
ofcycloheximide
iscompa-rable, confirming
thatonly
primary
transcrip-tion occurred
atthis
temperature(Fig. 9).
Itis
also
comparable
tothe
wild typeatsimilar MOI
(Fig.
7;wild
type+cycloheximide).
DISCUSSION
Numerous reports
confirm that the molecular
biology
of rabies virus is similar
tothat of other
rhabdoviruses (6,
9-12).
Although
modality
of
the
transcription resembles that of
VSV,
the
rate
is
different.
Ittakes
5min
withrabies and
only
90 swith
VSV
(8)
toobtain
a setofmessen-gers
equivalent
inlength
to the genome. Thenumber of
messengersregularly
increasesduring
TABLE 1. Numberof+strands inrabies-infected
cells, in the presence and absenceof cycloheximide'
Time after infec- +
Cyclohexi-tion(h) mideb -
Cycloheximide'
1 101 78
2 232 145
5 638
8 870 638
12 957 9,860
18 1,334 20,880
aExpressedin genomeequivalentsper infected cell.
bMOI = 10.
cMOI
=1.2.VOL. 31,1979
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.508.60.249.65.351.2]228 SAGHI AND FLAMAND
i@'
10^
0
le
u
v
E
WI
z
8 0
S
0 4. E
.9
a
102
Time
M
10'
104
-r-w.1
x m
< E
0
,va
_ t
E
a 'n
cL-to
C
)
I
o3
104
0 L
12 s a a
Time (h)
J. VIROL.
Time (Ih Inilh
FIG. 8. ProductionofviralcomplementaryRNA in cellsinfectedbyF+-orF-mutants at 33and39.6°Cin thepresenceandabsenceofcycloheximide. Totalunlabeled RNAswereextractedfrom rabies-infectedcells atvariousperiods oftimeafter infection and annealed to3H-labeled RNA extractedfrompurified rabies
virions, asexplainedin thetextand in thelegendtoFig. 7.MOIswererespectively equalto1. 7(tsO23),4.2
(tsO2),1.3(tsO31) and 2.3 PFUpercell(ts055). Symbols:0,mutants at33°C (leftpanels),wildtypeat39.60C
(rightpanels)withoutcycloheximide; ,mutants at39.60Cwithoutcycloheximide;A,mutants at39.60C with cycloheximide (100 pugml).
morethan18h in thecaseofrabies andduring
6 hforVSV.At maximumproductionthereare
2 x
104
genome equivalents of messengers perrabies-infected celland 2x105perVSV-infected cell (8). This indicates that transcription (and
probablyreplication) is slower and lessefficient
forrabiesthanforVSV.
On the basis of fluorescence induction in
in-fected cells at NPT, the ts mutants of rabies
virus studiedwere classified into threegroups,
F+,
F',
and F-. Approximately 80% of the ts mutants were F+-, although the borderlinebe-tween the most affected F+- and F- was not
precise since the classificationdependedonthe
levels of viralproteins experimentally detected.
Ourbiochemical studies showed thatsynthesis
B ts a2. F
*-D tso55
F-3Ut(-C)
4*(-C)
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.508.89.437.73.490.2]ts RABIES VIRUS MUTANTS 229
a
10
ts o42. F-moia10
x
< °E 104
Za
3I| /4 ~
~~~~~~~
39.6P*c C)E
102 .... .. .... ...
*- 1 2 5 a 12 1
E T ime
(h)
-~
FIG. 9. Production
of
viralcomplementary
RNA incellsinfected
byanF-mutant at39.60CatanMOIof10PFUpercell, in thepresence and absenceof
cycloheximide.
Total unlabeled RNAswereextractedfrom rabies-infected cells at various periods after infectionand annealedto3H-labeled RNA extracted
from purified
rabiesvirions, asexplained
in thetext.Since the viruswasconcentrated by
centrifugation,
the evaluationof
the MOIbasedonthe titer in PFU isaminimalfigure
asexplained
inFig.
7.Treatmentby
cycloheximide
(100tLglml)
was asexplainedin thelegend
toFig.
7of the five viral
proteins
was either normal forF+
mutants,
similarly
inhibited
forF+-
mutants,
or
undetectable
for F- mutants. Aunique
mu-tational event could therefore
depress
the syn-thesis of all viralproteins, indicating
that
this eventwasatthelevel of
viraltranscription
and/
or
replication.
Our
hybridization
experimnents
indeed dem-onstrated thatF+-
werecapable
ofperforming
at least somesecondary transcription (and
thereforesome
replication)
atNPT. F-mutants,
however,
couldperform
primary transcription
butnot
replication and/or secondary
transcrip-tionatNPT.
Since
F+
mutantprotein synthesis
is normalat
NPT,
thetslesionmustbelocated inaprotein
which is not
directly
required
fortranscription
and
replication, i.e., probably
inproteins Ml, M2,
or
G. In the absence of
complementation, it isnot
possible
todetermine
whether F+ mutantsare
affected
in the same function. Thecharac-terization of
the mutated protein iscurrently
under study.
Ithas been
well
documented that 87% of VSV ts mutantsbelong
tocomplementation
group Iand
aremutated in
the
transcriptase (for reviewssee 15;
Flamand, in
press; andC.
R. Pringle andJ.
Szilagyi,
InD.
H. L.Bishop,
Rhabdoviruses,in press). In most cases VSV mutants of
group
I are
able
toperform primary
transcription, but replication and secondary transcription arecom-pletely inhibited
at NPT. In this respect theyresemble F- mutants of rabies virus.
Inthe case of rabies, the majority of ts mu-tants are
of
theF` phenotype. Secondary
tran-scription of
F`
mutantsis
depressed
at NPTcompared
with thewild
type, although someamplification of RNA synthesis could be clearly
detected. The
transcribing
structureof
therhab-dovirus is the
nucleocapsid. Rabies nucleocapsid
is composed of
twoviral proteins:
L,which
islikely
tobe the
transcriptase, and
N (16).The
hypothesis that
F`
and
F- mutantscould be
affected
inthe
N or Lprotein
isunderinvesti-gation.
We have shown that all
our ts mutantsof
rabies virus do
nothave the
samephenotype.
Presumably,
they are notmutated
inthe samefunction. Why it
wasimpossible
todemonstrate
complementation
in testsinvolving F+, F+-, and
F- mutants is
still
anintriguing question.ACKNOWLEDGMENTS
We thank J. Benejean, B. Jaillard, and F. Aguero for excellent technical assistance. WearegratefultoPh.Vigierin whose laboratory this work was done and to Dr. Andral, Directorof "Centre d'EtudesurlaRage deNancy"for their interest and encouragement. We thank Tadeusz Wiktor and WilliamWunner for criticalreading of the manuscript.Figure 1and finaltyping has been doneatthe WistarInstitute where A. F. isonleavefor ayear.
This researchwassupported bytheCentreNational de la RechercheScientifique throughthe L. A.40086,bythe Com-missariata l'Energie Atomique,Saclay, Franceandby the Institut National de la Santeetde la RechercheMedicale (contractno.77.I.160.I).
LITERATURE CITED
1. Bishop,D.H.L.,and A. Flamand.1975.Transcription process of animal RNA viruses, p. 95-152. In 0. C. Burke and W. C. Russell(ed.),Controlprocessin virus multiplication. Cambridge University Press, Cam-bridge.
2. Bishop,D.H.L., andM.Smith. 1976.Rhabdoviruses, p.167-225. InK. D.Maya(ed.),Themolecularbiology of animalviruses.Dekker,New York.
3. Bonner, M., andR.A.Laskey.1974. Afilm detection methodfortritium-labelled proteinsand nucleicacids inpolyacrylamidegels.Eur.J. Biochem. 46:83-88. 4. Bramhall,S.,N.Noack,M. Wu,and J. R.
Loewen-VOL. 31,1979
on November 10, 2019 by guest
http://jvi.asm.org/
[image:10.508.55.249.60.347.2]230 SAGHI AND FLAMAND
berg.1969.Asimplecolorimetric method for determi-nation of protein. Anal. Biochem. 31:146-148. 5. Bussereau, F., and A. Flamand. 1978. Isolation and
preliminary characterization of ts mutants ofrabies virus, p.701-708.InB. W. J. MahyandR. D. Barry (ed.),Negativestrand viruses and thehostcells. Aca-demic PressInc.,New York.
6. Ermine,A., and A. Flamand. 1977. RNAsynthesisin BHK21cells infected by rabies virus.Ann.Microbiol. (Paris)128 A:477-488.
7. Flamand,A., andD. H. L. Bishop. 1973.Primaryin vivotranscription ofvesicular stomatitis virus and
tem-perature-sensitivemutantsof fivevesicularstomatitis viruscomplementationgroups. J. Virol.12:1238-1252. 8. Flamand,A.,and D. H.L.Bishop. 1974. Invivo
syn-thesis of RNA by vesicular stomatitis virus and its
mutants.J.Mol.Biol. 87:31-53.
9. Flamand,A., and J. F. Delagneau. 1978. Transcrip-tionalmappingof rabies virus in vivo.J.Virol. 28:518-523.
10. Flamand, A., J. F. Delagneau, and F. Bussereau.
1978. AnRNApolymerase activity in purified rabies virions. J. Gen. Virol. 40:233-238.
11. Flamand, A., D. Pese,and F.Bussereau. 1977.Effect ofactinomycin D andcytosine arabinoside onrabies and VSVmultiplication. Virology 78:323-327. 12.Kawai, A. 1977.Transcriptase activity associated with
rabies virion. J. Virol.24:826-835.
13. Laemmli, U. K. 1970. Cleavage ofstructural proteins during the assembly of the head of bacteriophage T4. Nature(London)227:680-685.
14.Madore,H.P.,and J. M.England.1977.Rabiesvirus protein synthesisininfected BHK-21cells.J.Virol. 22: 102-112.
15. Pringle, C. R.1977. Genetics of rhabdoviruses,p. 239-290. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 9. Plenum Publishing Corp., New York.
16.Sokol, F. 1975. Chemical composition and structureof rabiesvirus. In G.M.Baer(ed.), The naturalhistory of rabies. Academic Press Inc., New York.
J. VIROL.