JOURNAL OF VIROLOGY, Sept. 1976,p.985-997 Vol. 19,No.3
CopyrightC1976 American Society for Microbiology Printed in U.S.A.
Structural Proteins of La Crosse Virus
JOHN F. OBIJESKI,* DAVID H. L. BISHOP, FREDERICK A. MURPHY, AND ERSKINE L. PALMER
Bureau ofLaboratories, Center for Disease Control, Atlanta, Georgia 30333; and Department of Microbiology, University of Alabama, Birmingham, Alabama 35294*
Received for publication 19April 1976
Preparations of La Crosse virus, a member of the California encephalitis groupofbunyaviruses,werefound to possess three major virion proteins. Twoof the proteins were glycosylated (Gi and G2) and were located on the surface of thevirusparticles. These twoglycoproteinswerepresent inequimolaramounts and possessed apparent molecular weights of 120 x 103 and 34 x 103. Virion
nucleocapsids, isolated by a nonionic detergent and salt treatment, contained
another major protein, N (molecular weight = 23 x 103). A large, but minor,
protein species L (molecular weight = 180 x 103) was also found in virus
preparations. Theapproximatenumber of protein moleculesper virion has been
determined. Electronmicroscopy of purified LaCrossevirusindicated that the
virus particle (mean diameter, 91 nm) is enveloped and possesses irregular
surface projections (length, 10 nm).
LaCrosse viruswasinitially isolatedin 1964
from frozenbrain tissue of a 4-year-old female
child whodiedin1960from
meningoencephali-tis in La Crosse, Wis. (29). Comparative
neu-tralization, complement fixation, and
immuno-diffusion tests have shown thatLaCrosse virus
is serologically related to, but distinguishable
from, other isolates of the California encephali-tisgroupofbunyaviruses (3, 12-14, 22, 24, 31).
Thevirus has been isolated from various
mos-quito speciesthroughout the north-central and
eastern United States, and neutralizing
anti-bodies have been detected in various
forest-dwelling
small mammals inthese same areas(11, 28). Since 1964, several hundred cases of
encephalitis have been attributed to the
Cali-fornia encephalitis group of viruses,
particu-larly inOhio, the southwestern region of
Wis-consin, and othermidwestern states (5).
Apart fromaninitialstudy by McLerran and
Arlinghaus (9),relatively littleisknown about
the chemical composition and structure of La
Crosse virus. We have investigated the
num-ber,
location,
and function of theproteinsofLaCrosse virus and characterized the purified
vi-rusparticle byelectron microscopy. MATERIALS AND METHODS
Reagents. Uniformly labeled L-14C- or 3H-amino acid mixtures, [3H]uridine (32 Ci/mmol),
D-[3H]glucosamine (10 Ci/mmol), D_['4C]glucosamine (0.2Ci/mmol), [32P]orthophosphate, and ['25I]iodine (17 Ci/mg) were obtained from New England Nu-clearCorp., Boston, Mass. Phosphorylase A, a-ga-lactosidase, and pancreatic RNase were purchased from Worthington Biochemicals Corp., Freehold,
N.J. Cytochrome c, myoglobin, chymotrypsinogen, ovalbumin, and bovine serum albumin were ob-tained from Mann Research Laboratories, New York, N.Y.; bromelain and the purified grade of lactoperoxidase were purchased from Calbiochem, La Jolla, Calif.
Growth and purification of viruses. Vesicular stomatitis virus (VSV), Indiana serotype, was grownand purified as described previously (15). La Crosse viruswasobtained from D.Trent, Center for Disease Control, Fort Collins, Colo., and was fur-ther plaque purified in confluent monolayers of BHK-21cells. To prepare working virus stocks, we infected BHK-21 cellsinpetriplates with <10 PFU ofvirus,andat 48hthe virus presentin oneplaque plug was usedtoinfect107confluent BHK-21 cells. This culture was harvested at 48 h postinfection and storedinaliquotsat -70°Cin amedium containing 10%fetal calfserum.Thetiterof virusinthe stock preparationwas 8 x 107PFU/ml.
Roller bottlecultures of confluent BHK-21 cells (1 x 101 to 3 x 10# cells/bottle), grown in reinforced Eagle medium supplemented with 10% fetal calf serum, were infected with the stock virus at an inputmultiplicity of infection (MOI) of0.001 PFU/ cell. Fortheproduction of labeled virus,weincluded in the growth medium either L-'4C-labeled amino acids (0.2 uCi/ml), D_[4C]glucosamine(0.2/,Ci/ml), L_3H-labeled aminoacids (2 ,uCi/ml), D_[3H]glucosa-mine (2
ACi/ml),
[3H]uridine (5tLCi/ml),
or [32P]_ orthophosphate (100 uCi/ml). When labeled amino acids were used, the concentration of unlabeled amino acidsinthegrowth mediumwasreducedto 20%of the normal level.After 48 h ofgrowth at 33°C, the infected cell culture fluids wereclarified by low-speed centrifu-gationat10,000 x gfor 20 min in aBeckman J-21 centrifuge to remove cell debris. The subsequent purification stepswere essentially those described 985
on November 10, 2019 by guest
http://jvi.asm.org/
986 OBIJESKI ET AL.
previously for the purification of other enveloped viruses (1, 15), with certain modifications to im-prove the yield of intact virions and reduce the amountof contaminationby cellular material in the final virus preparation. Virus wasprecipitated from the clarified culture fluids by adding polyethylene glycol 6000 (Carbowax 6000, 70 g/liter)and NaCl (23 g/liter). After the mixture wasstirred 4 h at 4°C, the virusprecipitate was collected by centrifugation at 10,000 x g for 20min, resuspended in 0.01 M Tris-hydrochloride buffer, pH 7.6, containing 0.15 M NaCl and 0.003 MEDTA (TSEbuffer), clarifiedby centrifugation, and loaded over combination equi-librium-viscosity gradients of glycerol and potas-sium tartrate (KT-GLY) (15). After the gradients werecentrifuged 3 h at 4°C (40,000 rpm in a Spinco SW41 rotor), thevisible virusband wascollected by pipette anddialyzed overnight at4°C against TSE buffer. The virus was thenloaded onto a gradient of 20 to 70% (wt/vol)sucrose in 1 M NaCl, 0.01 M Tris-hydrochloride buffer, 0.002 M EDTA, pH 7.4, and centrifuged at 4°C and 35,000 rpm for 3 h. The virus band was harvested, diluted fourfold with TSE buffer, andpelleted through a 1-ml cushion of 30% (wt/vol) sucroseinTSEbuffer by centrifugation for3 hat 35,000 rpm. The final virus pellet was resus-pendedin 0.5 to1.0ml ofTSE buffer and storedat 4°C. Virus protein concentrations were usually be-tween 0.2and 1mg/ml.
Protein determinations and polyacrylamide gel electrophoresis. Protein concentrations were deter-mined by the method of Lowry et al. (7), with bovine serum albumin as aproteinstandard. The separa-tion of sodium dodecyl sulfate (SDS)-dissociated whole virion proteins by electrophoresis hasalready been described (15), using either an 8% (wt/vol) continuous polyacrylamide gel involving a phos-phatebuffer, pH 7.0, and SDS(cont-SDS);a 3 to30% (wt/vol) lineargradientpolyacrylamidegel and the same buffer-SDS system (gradient-SDS); or a dis-continuous gel system involving a 3.6% (wt/vol) stacking gel and a10%(wt/vol)resolving polyacryl-amidegel plus SDS andadiscontinuous buffer sys-tem involving Tris-hydrochloride and glycine buffers,pH 9.0(disc-SDS).Theseparation obtained forlabeled protein specieswasdeterminedbyslicing frozen gels into 1-mm portions and determing their content ofradioactivity after soaking the slices for 18 h in a toluene-based scintillation cocktail con-taining 3% (vol/vol) NCS (Amersham-Searle, Chi-cago, Ill.). The separation ofprotein species was determinedinsomecases by staininggels with Coo-massie brilliantblue, destaining in an acetic acid-methanol mixture, and scanning the gels at 640 nm, asdescribedpreviously (1).
Iodination of viral proteins. Purifiedvirus prepa-rations containing 200
gg
ofproteinin 0.4 mlof0.1 Msodiumphosphate buffer, pH 7.0, wereincubated at20°Cwith 250,uCi of carrier-free['251]iodinein the presence of10,ugoflactoperoxidase and0.01 ml of freshly diluted 0.001 M H202 (26). Five additional 0.01-mlportions ofhydrogenperoxidewereaddedat 5-minintervals (33). After the lastaddition, 2mlof cold (4°C)TSE buffer and0.1ml ofsaturated potas-sium iodide solution were added. Unreactedre-agents were removed from the virus particles by loading the reactionmixture on a20to70%(wt/vol) gradient of sucrose containing 1 M NaCl, 0.01 M Tris-hydrochloride buffer, 0.002 M EDTA, pH 7.6, and centrifuging at 35,000 rpm for 1 hat4°C. The visible virusband wascollected with a pipette and dialyzedagainst TSE buffer. The specific activity of thelabeled virus preparation wasdetermined to be ontheorder of 105 cpm per mg of protein.
RESULTS
Growth curves of La Crosse virus as a func-tion of the input MOI. Confluent monolayers
of3 x 106 BHK-21 cells in 25-cm2 flasks were infected with virus at input MOIs equivalent to 10,
1,
or 0.01 PFU/cell. The monolayers were incubated at 33°C; the release of infectious vi-rus wasmeasured bydetermining the number of PFU presentin thesupernatant fluids. Ina separate experiment, the virus present in asingle plaque plug was used to infect a
con-fluent monolayer of3 x 106BHK-21 cellsat an
inputMOI of0.002 PFU/cell, and the yield of
infectious virus was also determined (Fig. 1).
Thehighestvirusyield wasobtained from the
cellsinfectedatthe lowest
MOI,
but wassup-_-o----o.002PFU/CELL
/ ~~~CLONED
PLAQUE
/ 0
0 .01 PFU/CELL
/0
lo7/ -' -A PFU/CELL
0
Qo0
;,r_
i,
PFU/CELL
I / *v
II
0~~~~~~~-I-I.-1--I,!
0AII12 1/
12 24 36 48 60 72
HOURS POST INFECTION
FIG. 1. Growth curves ofLa Crosse virus as a
function ofthe input multiplicity of infection. Mono-layer culturesofBHK-21 cells, infectedwith 0.002, 0.01, 1, or10PFUlcell, weremonitoredforthe pro-duction of infectious virus by aplaque assay
tech-nique.
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.505.270.463.344.610.2]LA CROSSE VIRUS 987
pressed by even moderate increases of MOI
(Fig. 1). At the calculated input MOI of 0.002
PFU/cell, about 150 infectious units were
pro-ducedpercell. By 48 h postinfection, there were
severe cytopathic changes in all cells and no
further virus production. In light of these
ob-servations, all further virus preparations were
prepared from cultures infected at an MOI of 0.001 PFU/cell.
Electron microscopy of purified virus
prep-arations. A low-magnification electron
micro-graph of gradient-purified La Crosse virus is
presented in Fig. 2A. Large numbers of virus
particles with a mean diameter of91 nm were
observed, and therewas noevidence of
contam-inating cellular debris. Fine, irregular surface
projections, 10 nm in length, were present on
the surface ofall particles (Fig. 2B). No
sym-metric arrayof theseprojections wasobserved,
in contrast to the results described for
Uuku-niemi virus (2, 18, 21, 23). Where negative-contrast medium penetrated the particles, the
envelopecould beseen, andinsomeparticlesa
convoluted, internal structure,
possibly
repre-senting theviralnucleocapsid(s), wasobserved
(Fig. 2C).
Sedimentation coefficient of La Crosse
vi-rus. Purified test samples ofLa Crosse virus
(1.5 mg/ml) were dialyzed against a reference
buffer (0.01 M borate, 0.16 M NaCl, pH 8.0),
and thesedimentation
coefficients
at20°Cweredetermined atspeeds of10, 12, 14, and 17,000
rpm in aBeckman modelE ultracentrifuge
op-erated with Schlieren optics. Sedimentation
coefficients (sapp) werecorrectedto
s,,,8w
valuesby a standard procedure (27). By this
tech-nique, LaCrossevirus wasdeterminedtohave
asedimentation coefficient of 415S.
Identification and characterization of the
structural proteins of La Crosse virus. The
following procedure was used to identify the
structural proteins of La Crosse virus and to
estimate their molecular weights. Asample of
:'H-amino
acid-labeled La Crosse virus wasmixed with a sample of
'4C-amino
acid-labeledVSV Indiana virus, and their proteins, were
dissociated by SDS. The VSV Indiana
struc-tural polypeptides were included as markers
becausethey are known to haveapparent
mo-lecularweights of 160x 10: (L),65 x
10:3
(G), 54x 103 (N), 42 x 10:3 (NS), and 27 x 103 (M) in
cont-SDS-gels (30). The proteins of the two
vi-ruses weresubjected to electrophoresis in
poly-acrylamidegels containing SDS. Three gel
sys-tems, cont-SDS, disc-SDS, and gradient-SDS
(15; Espositoand Obijeski, inpreparation),
de-scribed in Materials and Methods, were used. These particular gelsystems were used because
proteinsof similarmolecularweightscanoften
be separatedby onegel system but notby
an-other(16,25). Also,anestimation of the
appar-entmolecularweightsofthe VSV Indiana
pro-teins, even in the presence of SDS, depends
uponthegel systemused (15).The
electropher-ograms of the proteins of the two viruses are
presented inFig. 3.
The molecularweights of the structural
pro-teinsof La Crossevirus, asdetermined by com-parison with the known major VSV Indiana structural proteins and seven standard
pro-teins, are shown in Table 1. Four structural
polypeptides were identified in the
cont-SDS-gel system, alarge protein (180 x 103daltons,
minorcomponent), a majorpolypeptide (120 x
103 daltons), a third protein present in
appar-ently smalleramounts(34 x 103daltons), anda
fourthcomponent(23 x 103daltons). No
signifi-cant differences in the apparent molecular
weightsor in the number of viral protein
spe-cies were observed by using the other two gel
systems.
Identification oftwovirusglycoproteins in
LaCrosse virus. La Crossevirus wasgrown in
the presence of '4C-labeled amino acids and
[3H]glucosamine,
purified,
and dissociatedwithSDS and mercaptoethanol. The
differentially
labeledvirusproteins wereresolvedby
electro-phoresis in 8% polyacrylamide gels (Fig. 4A).
The largest and smallest viral proteins were
labeled only by the '4C isotope. However, the
twointermediate-sizedproteins werelabeled by
bothisotopes (3Hand 14C), thereby identifying
them as glycoproteins; these proteins were
designated Gl and G2.
Demonstration that the glycoproteins are
atthesurface ofthevirus
particle.
Apurifiedvirus preparation labeled by '4C-amino acids
and [3H]glucosamine wasincubated with 1mg
of the proteolytic enzymebromelain per ml in
TSE buffer for 2 h at 35°C to remove the
pro-teins located on the outersurface ofthe virus
particle (10). The bromelain-treated virions
were reisolated bycentrifugation in agradient
of sucrose anddissociated by SDS. After
poly-acrylamide gel
electrophoresis,
it was foundthat neither ofthe viral glycoproteins (Gl or
G2) had survived the proteolyticenzyme
treat-ment (Fig. 4B). The other two viral proteins remained associated with the protease-treated virions. Thebromelain-treated preparation ex-amined byelectron microscopy contained only
spikeless particles with intact envelopes (Fig.
5). The infectivity ofspikeless particles as de-terminedbyplaquinginBHK-21cell
monolay-ers was about 10- that of untreated control virus preparations (data not shown). It was VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
A
'k
...^ f-fl..
*. I.
s,;*-8.
&4 .. _
A44
)4?7%
I;F-
'1.i
es';A
5A
411"
A~~~~~~A
,,.,.
y{,,~
4 ,, '4jiUs
4' i14'f'
ts- s w* '>>E >
tA v~F~
:':eg >..g
Jr
B
4
4, 5a
C
FIG. 2. Electron micrographs ofpurified La Crosse virus, all negatively stained with 2% sodium phospho-tungstate. (A)Concentratedsuspension ofpurified LaCrosse produced by thedescribedprotocol ( x62,400). (B) Virusparticles showing the characteristic layer of irregular surface projections (x201,500). (C) Virus particles penetrated by the negative-contrast medium showing an electron-lucent membraneand internal structure(x152,400).
988
4
A
;. .1
:: 4"";.
.N, T.
....4 %.:-'A
e
1:1Y '16%
I fl, ., -i
"'.
-t-zf A.
;., M., "E.f
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.505.69.461.55.620.2]LA CROSSE VIRUS 989
T
a-0)
0
I
- 20 40 60 80 +
[image:5.505.44.439.538.615.2]FRACTION NUMBER
FIG. 3. Electropherograms of VSVIndiana and La Crosse viralproteins. Mixtures of'4C-amino acid-labeled VSV and 3H-amino acid-labeled La Crosse viruspreparations were dissociated by SDS and
2-mercaptoethanol,andthe proteinswereresolvedbyelectrophoresisincont-SDS-,disc-SDS-,
orgradient-SDS-gels (see text).After electrophoresis eachgel wassliced into 1-mm segments,and the distribution ofeach
isotopeinthegel wasdetermined byliquid scintillationspectrometry.
TABLE 1. Molecularweights(xlO-3) of La Crosse virus proteins"
Cont-SDS Disc-SDS Gradient-SDS
VSVIndiana LaCrosseb VSV Indiana LaCrosse VSV Indiana La Crosse
L 160 L 180 L 149 L 190 L 160 L 175
G 65 G1 120 G 63 G1 125 G 63 G1 120
N 54 G2 34 N 46 G2 38 N 53 G2 37
NS 42 N 23 NS 54 N 25 NS 42 N 22
M 27 M 27 M 27
aThe molecular weights of the various viral proteins were estimated by comparing theirelectrophoretic mobilities relative to seven marker proteins runinthe same gel system(,8-galactosidase,phosphorylase A, bovineserumalbumin, ovalbumin,chymotrypsinogen, myoglobin, and cytochrome c). The values reported arethemeans offiveseparatedeterminations.
b Thedesignation of La Crosse virus proteins is based on the results giveninthetext. VOL. 19, 1976
on November 10, 2019 by guest
http://jvi.asm.org/
990 OBIJESKI ET AL.
12F
T
x
a-
y-i
a-to C-I
FRACTION NUMBER
FIG. 4. Identification ofLa Crosse virus glycoproteins. (A) Purified La Crosse virus, grown in the presence of '4C-labeledaminoacids (a) andf:3H]glucosamine(0) was dissociated by SDS and 2-mercaptoethanol, and the proteins were resolved by cont-SDS-gel electrophoresis. (B) A similarly labeled virus preparation was treated with bromelain andrepurifiedby centrifugation in a 20 to70% gradient of sucrose containing TSE buffer and 1 MNaCl(35,000 rpm for 45min). The visible band ofparticles was recovered and dissociated by SDSinthe presence of 2-mercaptoethanol; the proteins were resolved by cont-SDS-gel electrophoresis. concluded from these results that the
glycopro-teins constitute thespikesonintact virus
parti-cles.
Additional studiesweredonetoconfirm that theglycoproteins weretheonlyvirion proteins
externaltothe viralenvelope.Apreparationof unlabeled viruswasiodinatedwith '25']iodine, lactoperoxidase, and hydrogen peroxide. After repurification by equilibrium gradient centrifu-gation, the labeled virus particles were
re-covered. An aliquot was dissociated by SDS, and theproteinswereresolved by
electrophore-sis in an 8% cont-SDS-gel. Although some
la-beled material remained at the surface of the
gel,
the majority of the labelmigrated
withelectrophoretic mobilities characteristic of the Gl and G2 glycoproteins (Fig. 6). A small
amount of labeled material was recovered around fraction 40. No label was observed in the regionsofthe gel where thetwo
nonglyco-sylated viral proteins were expected. Another
aliquotof the iodinatedvirusparticleswas
in-cubated with bromelain and repurified by
su-crose gradient centrifugation, and the visible
band ofparticleswasrecovered. After
dissocia-tion of the particles by SDS and resolution of the constituent proteins by electrophoresis in
an 8% polyacrylamide gel, it was found that
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:6.505.124.413.62.455.2]LA CROSSE VIRUS 991
almost all the label
originally
associated withthetwoviralglycoproteins (aswellasthe
frac-tion 40 material) had beenremoved by the en-zyme treatment (Fig. 6).Iodinationof Triton
X-100-disrupted particles, followed by
trichloroa-cetic acid precipitation and dissociation with
SDS, labeled all four structural proteins (data
notshown).
Gradient
centrifugation
ofbromelain-treated La
Crosse
virus.When La Crosse viruswas incubated with
bromelain,
thedensity
ofthespikeless particles was 1.16
g/ml
incombi-nation KT-GLY
gradients
(Fig. 7). Controlpreparations ofvirusincubated intheabsence ofenzyme had a
density
of 1.20g/ml.
Whenvirus preparationsradiolabeled withamixture
ofaminoacidsweretreatedwithbromelain and
centrifuged in KT-GLY
gradients,
approxi-mately40%of thetotal
radioactivity
wasfoundto be associated with the
1.16-g/ml region,
whereas theremainder of the
radioactivity
wasatthetopof the
gradient (Fig.
7C).
Incontrast,when
glucosamine-labeled
viruswasincubatedwith theenzyme, >90%of thetotal
radioactiv-ityremainednearthetopof the gradient(Fig.
7B). Similarly, when '2I-labeled virus was
in-cubated withbromelain, almost all of the radio-activity was released and remained near the origin ofthe gradient (Fig. 7A). Labeled mate-rial from the 1.16-g/ml region wasprecipitated
completely with trichloroacetic acid and could
be recentrifuged in KT-GLY gradients.
Fur-thermore, electron microscopy of the particles
from thisregionofthe gradient showed that the
particles were devoid of their surface
projec-tions (see Fig. 5). Less than 10%ofthe
brome-lain-solubilized radioactive material, which
re-mained at the top of the gradient, could be
precipitated withacid.
Solubilization of La Crosse glycoproteins.
Exposure of many lipid-containing RNA
vi-ruses to nonionic detergents and various salt
concentrationshasbeen showntodifferentially
solubilize viral proteins (4, 6, 8). Purified La
Crosse virus labeled with :H-amino acids
sus-pended in 0.01 M Tris buffer (pH 7.8) was
treated with Triton X-100 (2% vol/vol) and
varying amounts of NaCl. Each preparation
X,;
.ZS~~~~~~~~~~.::i
F"-.-.4.4 '. 'l.
0.4P ,FJ
qu
l1'-.rps r
i-*Ar
'
FIG. 5. Electronmicrographof bromelain-treatedLaCrossevirusparticles.Repurifiedbromelain-treated LaCrossevirus(Fig.4B)wasnegativelystained withsodiumphosphotungstate(x132,300).
VOL. 19, 1976
I
'.t.
e,
.3 #;,
.,!,-"- -4
., 4.,
i4..:,
I,:,.z . '' .-IIlo;., ,.,
i
...-i114'$1
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.505.48.445.331.646.2]992 OBIJESKI ET AL.
2
'10
a-)
-
20
40
60
80
+
FRACTION NUMBER
FIG. 6. Iodination ofintactLa Crosse virions. Unlabeled La Crosse virus, iodinated and repurifiedas describedinthe text,wassolubilizedby SDS and 2-mercaptoethanol, and the proteinswereresolvedby cont-SDS-gel electrophoresis (a). After the iodinated viruswastreated with bromelain (see text), theparticleswere
repurified,dissociated by SDS, and again resolved bycont-SDS-gel electrophoresis(0).
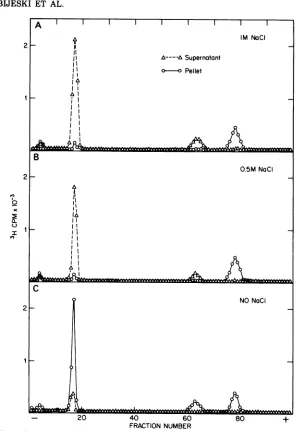
wasseparated intoa soluble and aparticulate
fractionbycentrifugation, and theprotein
con-stituents in each fraction were determined
by
SDS-gel electrophoresis. Figure 8C indicates
that only 5 to 10% of the
largest
La Crosseglycoproteins (Gl and G2)weresolubilized (Fig.
100 treatment alone. However, when the virus
was treated with either 0.5 M or 1.0 M NaCl
togetherwith TritonX-100, almost100%of both
glycoproteins (Gl and
G2)
wassolubilized(Fig.
8A, B). The
particulate
fractions of thesalt-Triton X-100-treated viruscontained
virtually
all of the
largest
and smallest viralproteins.
Identification of the proteins associated
with the nucleocapsids ofLa Crossevirus. In
preliminary attempts to release the viral
nu-cleocapsids
of La Crosse virus with nonionicdetergents, we found that
only
when viruspreparationsweretreated with TritonX-100 in
the presence of NaCl could
nucleocapsids
be obtained essentially free from the viral glyco-proteins (see Fig. 8).Apurified preparation of La Crosse virus, la-beled by L3H]uridine and '4C-amino
acids,
wasdissociated by 2% (vol/vol) Triton X-100 in the presence of 1 M NaCl, and the mixture was
loaded on a KT-GLY gradient containing 1 M NaCl. After 24 h of
centrifugation
at4°C
and 40,000 rpmin aSpincoSW41 rotor,itwasfoundthatessentiallyall the :1H label waspresent in
asingleband (p = 1.32)
together
withapproxi-matelyone-thirdof the '4C label (Fig. 9A). The
rest of the '4C label was recovered as a broad band ata density of 1.20 g/ml. To identify the viral proteins associated with the viral RNA, material from both regions of the gradient was
dialyzed free from the glycerol and potassium
tartrate and precipitated by 5 volumes of 5% (wt/vol) trichloroacetic acid at 4°C. The precipi-tates were recovered and dissociated by SDS,
andthe proteins were resolved by electrophore-sis in 8% cont-SDS-gels. The two proteins asso-ciated withtheviral RNA (fraction I material) were the smallest and the largest species (Fig.
9B). Since the smallestprotein is a majorviral
constituent (based on the number of molecules per virion Lsee below]) and is associated with the viralRNA, it can be tentatively designated asthe majornucleocapsidprotein (N). Because of its size, the large protein, also associated with the nucleocapsid, is designated L until a
function can be ascribed to it. The fraction II
material was found to consist of the two viral
glycoproteins (Fig. 9C).
Number of protein molecules in La Crosse virus particles. The approximate number of the various viral proteins was determined based upon the following assumptions: (i) that the estimatesof the apparent molecular weights of the individual viral proteins given in Table 1 are correct; (ii) that virions contain only one molecule of each size class of viral RNA; and
(iii)that the combined gram molecular weight
of the viral genome is 5.1 x 10' (unpublished
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.505.123.414.56.297.2]VOL. 19, 1976 LA CROSSE VIRUS 993
A
Xobservation).
Indetermining
thevirion ratio ofP
=1.20
RNAtoprotein,
a5-ml
suspension
of La Crossevirus labeled by [3H]uridine containing 0.260
8
mg of protein perml, andatotal of1.56 x10"
cpm was used. After extraction, the purified
RNA had a
specific
activity
of3.53 x 107 cpmO 1- per mg of RNA. It was calculated, therefore,
that therewas 1.3mgof protein to 0.044mgof
1
l viral RNAinthe
original
virussuspension,
i.e.,
a.. Ithat therewas a
protein-to-RNA
weightratioofo e 30:1.
~- 4 l Another
sample
of LaCrosseviruswasdisso-C9
[ ciated by SDSand precipitatedwith 5% (wt/vol)trichloroacetic acid. The
precipitate
wasre-covered bycentrifugation and washed by
alco-P4.1611
hol, andanaliquotwastreated with SDS before^^A electrophoresis in an 8% (wt/vol)
polyacryl-amide gel. After electrophoresis, the gel was
stained
by
Coomassie brilliant blue andscannedat 640 nm. Theareaunder eachpeak of
thescan wascomputed, and themassratio was
A 6 determined for the four viral proteins. From
10 three such
determinations,
the meanpropor-tions of virion proteinrepresented by the L, Gl,
G2, and N species were 3, 51, 14, and 32%,
ll0 e
respectively.
Similarresults were obtainedby
b I t
O0
using '4C- or 3H-aminoacid-labeled virusprep-xx arations (Table2).
-,MW2Assuming that the gram molecular weight of
X-
O La Crosse viral RNA is 5.1 x10",
it wascon-? 5_ cluded that the gram molecular
weight
of thei
total virion proteins was 180 x10'.
From the2 percentageofvirionproteininL, Gl,
G2,
andN proteins, the amount of each protein present per virion wasdetermined(Table
2). Theaver-age number of molecules of each protein per
virion was then calculated by dividing these
amountsby their respective molecularweights (Table 2).
C ,' It was apparent from these results that the
9
- numbers of Gl and G2 molecules per virionwereapproximatelyequal (Table2).Itwasalso
4 concluded that the mass ofN protein
by
com-i
I,f, parison to the mass ofvirion RNA was ofthe
o
o0
orderof10:1.
x 6 Absence of phosphoproteins in La Crosse
A virus
preparations.
A concentratedprepara-a- : 0 tion of La Crosse virus, labeled by
[32PIphos-Il
1- 2 t FIG. 7. KT-GLY gradient centrifugation of bro-l bro-l melain-treated La Crosse virus. Virus labeled with3_
iodine, aminoacids,orglucosamine was incubatedwith (A) orwithout(A)bromelain(1 mg/ml)at35°C for 2 h. Each sample was rebanded in KT-GLY gradientsat40,000 rpm for8 h in an SW41 rotor. Fractions (0.4 ml) were collected from each gradient
A, andcounted. Densitiesweredeterminedbyweighing
0.10-mlsamplesat roomtemperature.(A) '25I-labeled 40 20 TOP La Crosse. (B)[14C]-or
P'Hglucosamine-labeled
LaFRACTION
NUMBER
Crosse.(C) 14C-or:H-aminoacid-labeled La Crosse.. Il.- _-w _.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.505.62.232.52.690.2]994 OBIJESKI ET AL.
2
2
x
I
2
-20 40 60 80 +
[image:10.505.113.409.55.486.2]FRACTION NUMBER
FIG. 8. Electropherogramsofproteins inthesupernatantand pellet fractions of La Crossevirus treated with(A)2%(vollvol) TritonX-100in 1MNaCI, (B) 2% (vollvol) TritonX-100in 0.5MNaCI,or(C) 2%(voll
vol) TritonX-100. Purified La Crosse virus labeledwith :H-amino acids and suspendedin 0.01 M Tris-hydrochloride buffer,pH 7.8,wasincubated withTritonX-100inthepresence orabsence of NaCl for20min
at20°C.Each preparationwasseparatedintosolubleand particulate fractions bycentrifugationat100,000 x
gfor2 h.Theproteins inthesupernatantorresuspended pelletfractionswereprecipitated withtrichloroacetic
acid (5% [vollvol] final concentration) after 100 jAg ofbovine serum albumin was added. The protein
precipitateswerewashedwithethanol, dissociated by SDS, andresolvedbycont-SDS-gelelectrophoresis. phate (1 x 107 cpm in 0.1 ml of TSE buffer),
was adjusted to 1% (wt/vol) SDS to solubilize the viral membrane and release the virion RNA. It was then diluted 50-fold with TSE
bufferand incubated with 0.5mgofpancreatic
RNase at 37°C for 1 h in order to digest the virion RNA. The viral proteins were
precip-itated at 4°C with 5 volumes of 5% (wt/vol) trichloroacetic acid. The precipitate was then
washed twice with5% acidto removeresidual
acid-soluble nucleotides and three times with ethanol to remove residual phospholipids and
acid. After dissociation by SDS, the
proteins
wereresolved by electrophoresisin an8% (wt/
vol) polyacrylamide gel, stained by Coomassie brilliantblue, destained, and then sliced into
1-mm sections. Although all four viral proteins were identified in the stained gels, no 32P was
IM NaCI
II -~ASupernatant
o-I-o Pellet
1A
I' II
NO5 NaCI
J. VIROL.
l
on November 10, 2019 by guest
http://jvi.asm.org/
LA CROSSE VIRUS 995
FRACTION NUMBEC FRACTIONn
12-0.
1 12
(.20.20 2 TPI0 40 6 8
FIG. 9. Isolation ofLaCrosse viral nucleocapsids. Apurified preparationof [3H]uridine (O)- and
14C-aminoacid WA-labeledLa Crossevirus was treatedfor20 min at20°C with2%(vollvol) TritonX-100in thepresence of0.01 MTris-hydrochloridebuffer,pH7.8,and1 MNaCl.Themixturewascentrifuged for
24hat4°C and40,000rpminacombinationKT-GLYgradientcontainingIMNaCl(A). The distribution oflabelineachof thirty0.4-mlfractionswasdeterminedaswellasthedensityofselectedsamples.Pooled
firactions
(IorII)ofthegradient weredialyzedfreefrom potassium tartrate; theproteinswererecoveredbyprecipitationwith5%(wtlvol) trichloroaceticacid,and theprecipitateswere washed with acid and ethanol.
Theproteins weredissociatedbySDS and2-mercaptoethanolandresolvedbycont-SDS-gelelectrophoresis
(B,fractionI; C,fractionII).
recovered in any part of the gel (data not shown).
DISCUSSION
Four proteinshavebeen identified in
prepa-rations ofLa Crossevirus. Two of theproteins
areglycoproteins (GlandG2). Bromelain treat-ment and iodination studies have shown that these glycoproteins are present in the outer
surface of the virus particles. Stoichiometric
considerationsindicatethattheGlandG2 pro-teinsarepresentinequimolaramounts (Table
2). Ofthe two other virionproteins, oneis the
majorcomponentof the viral nucleocapsids (N protein); the other (a large protein, L), also associated with the nucleocapsid, is present in limitedquantities.Themolecular weightsof La Crosse virus proteins (Table 1)wereestimated on the basis ofcomparative electrophoresis in
which three different gel systems with VSV Indiana structural proteins (30) and seven
other nonviral proteins ofknown size (Fig. 3)
wereused.
In investigations of La Crosse virus, Mc-Lerranand Arlinghaus(9) identifiedthree
ma-jorstructural proteins (molecular weight = 85
X 103, 45 x 103, and 26 x 103). Two glycopro-teins (molecular weight = 84 x 10: and 31 x
103) and athird protein of about21 x 103
dal-tonswere identified inOriboca virus (20),
an-othermemberof thebunyavirusgroup. Rosato
and associates (19) showed that each ofseven
bunyaviruses (Bunyamwera, California
en-cephalitis BFS-283, Tahyna, Oriboca,BeAn 17,
BeAn 974, and Murutucu) contained three pro-teins ranging in size from 85,000 to20,000 dal-tons. Morerecently, White (32)identifiedfour
structural proteins in California encephalitis
virusBFS-283. Threeof the proteins (molecular weight = 30 x
103,
38 x 103,and 82 x 103) wereglycoproteins; the fourth (molecular weight =
17.5 x 103) was associated with the virion nu-VOL. 19X 1976
12
on November 10, 2019 by guest
http://jvi.asm.org/
[image:11.505.50.444.53.351.2]TABLE 2. Protein compositioni of La Crosse virus
Total protein('1) Molwt Daltons of protein/ No. ofmolecules/
Proteinspecies - virion virion
Incorporation' Stained (x 10 (x 10)
L ... 5 3 180 4.6 25
GI ... 50 51 120 78 650
G2 ... 14 14 34 21.4 629
N ... 31 32 23 48.9 2,126
Molecular weight estimates are based on the cont-SDS results given inTable1.
Total daltons of virion protein were determined from the 30:1 ratio of virion protein to RNA and the estimatethat the genomecomplexity of La Crossevirus is 5.1 x 10` daltons.
" Numberofprotein molecules per virion was calculated for each protein species bydividingthedaltonsof
protein per virion by their respective molecular weights.
Percentage of total protein fromincorporation studies represents the average of six separate determina-tionsinvolving:H-aminoacid- or'4C-amino acid-labeled virus(minimum3Hpergel = 9 x 104 cpm; mini-mum '4C pergel = 2 x 10'cpm).
Percentage oftotalproteins from a stained gel represents the average ofthreeseparatedeterminations of thearea under each peak.
cleocapsid. Uukuniemi virus, atick-borne
bun-yavirus-like isolate, serologically distinct from the bunyavirus group of viruses, was initially reported (18) to possess an envelope protein (molecular weight = 70 x 101) and another protein (molecular weight = 25 x 10:"). In
fur-ther analyses (2), involving resolution of the virion polypeptides by 15'/k polyacrylamide gels, two envelope proteins were identified.
Our estimate of the size of the largest La Crosse virus, glycoprotein (GI) (Table 1), is some 40,000 daltons larger than the estimate othershavefound for this protein in La Crosse and other related viruses (see above). Its size was determined by comparing its electropho-reticmobility with that of
/13-galactosidase
(mo-lecular weight = 130 x 10:1)and phosphorylase A (molecular 'voight = 92.5 x 10:;) as well asother viral and nonviral proteins. The La Crosse Gl protein migrated in three gel sys-tems slightly faster than /3-galactosidase but slower than phosphorylase A, and was there-fore estimatedto be about 120,000 daltons. We
(J. Gentsch, D. H. L. Bishop, and J. F.
Obi-jeski, unpublished data) have foundone minor
and three major proteins associated with
snowshoe hare, Bunyamwera, and Main Drain viruses, all members ofthe bunyavirus group. Compared withtheresults reported here, rela-tively minor (or no) differences were found in the apparent size ortypes ofproteinsobserved in these three viruses.
Minor quantities ofa very large protein (L) werealways observed in La Crosse virus (and three otherbunyaviruses) inall ofourpurified
virus preparations. Its significance is not known; however, it may represent a virion-associated cellular protein or an uncleaved viralpolypeptide. Alternatively, itcould repre-sent a nondissociable polymeric aggregate of the othervirion proteins(GI, G2, and N). This
isunlikely, however, sinceL is notdissociated to any appreciable extent by boiling purified virions in 2.5% SDS, 8 M urea, and a vast excess of mercaptoethanol (5%). Similarly, re-duction and acetylation of thevirusproteinsdo not causetheamountofL todiminish, suggest-ingthatthis protein is not asimple aggregate. Ithasnotescapedourattention, however, that forrhabdoviruses a large protein is believed to be avirion-associated transcriptase component (30). The function and derivation of the La Crosse Lprotein remains to be determined.
Theestimates ofthe number of protein mole-cules per virion are based on the assumption that the sum molecular weight of the viral RNAis5.1 x 10'. InourstudieswithLaCrosse, snowshoe hare, Bunyamwera, and Main Drain viruses, we have identified three virion RNA components ofapproximate molecular weights of 2.9 x 10, 1.8 x 10', and 0.4 x 10' (manu-script in preparation). Although these RNA speciesareregularly presentinallofourvirus preparations (except when defective interfering particles are present; unpublished observa-tions), we have no evidence to prove that all three species are necessary for La Crosse virus to be infective. Other investigators have also identified segmented RNA genomes for La Crosse virus (9) and Uukuniemi virus (17). If, however, all of the genetic information resides in the largest RNA molecule, our estimates of the numberof protein molecules per virionwill betoo high by almost a factor of2.
ACKNOWLEDGMENTS
We thank Marianne D.Stappand Richard A. Klimas for excellent technical assistance and John Hierholzer for de-termining thesedimentationvalue of La Crosse virus.
Thisstudy wassupportedinpartby Public Health Serv-ice grantAI-13402, from theNationalInstitute ofAllergy and InfectiousDiseases, toD.H.L.B.
996
OBIJESKI ET AL. J. VIROL.on November 10, 2019 by guest
http://jvi.asm.org/
LA CROSSE VIRUS 997
LITERATURE CITED
1. Bishop, D. H. L., and P. Roy. 1971. Dissociation of vesicular stomatitis virusandrelation of the virion proteins to the viraltranscriptase. J. Virol. 10:234-243.
2. vonBonsdorff, C.-H., and R.Pettersson. 1975. Surface structure of Uukuniemi virus. J. Virol.16:1296-1307. 3. Calisher, C. H., and K. S. C. Maness. 1970. Arbovirus identification by an agar-gel diffusion technique. Appl.Microbiol.19:557-564.
4. Hefti, E., P. Roy, and D. H. L. Bishop. 1975. The initiation oftranscription by influenza virion tran-scriptase, p. 307-325. In B. W. J. Mahy and R. D. Barry (ed.), Negative strand viruses, vol. 1. Aca-demic Press Inc., London.
5. Henderson, B.E.,and P. H. Coleman. 1970. The grow-ingimportance of California arboviruses in the etiol-ogyofhuman disease. Prog. Med. Virol. 13:405-461. 6. Kelley, J. M., S. U. Emerson, and R. R. Wagner. 1972. Theglycoprotein of vesicular stomatitis is the anti-genthat gives riseto andreacts withneutralizing antibody.J.Virol. 10:1231-1235.
7. Lowry,0.H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenolreagent. J.Biol. Chem. 193:265-275. 8. McAllister, P. E., and R. R. Wagner. 1975. Structural
proteins of two salmonid rhabdoviruses. J. Virol. 15:733-738.
9. McLerran, C. J., and R. B. Arlinghaus. 1973. Struc-turalcomponents of a virus of the California encepha-litiscomplex: La Crosse virus. Virology 53:247-257. 10. McSharry, J. J., R. W. Compans, and P. W. Choppin.
1971. Proteins ofvesicular stomatitis virus and of phenotypically mixedvesicular stomatitis virus-sim-ian virus 5 virions. J. Virol. 8:722-729.
11. Moulton, D. W., and W. H. Thompson. 1971. California virus infections insmall, forest dwelling mammals in Wisconsin. Am. J.Trop. Med. Hyg. 20:474-482. 12. Murphy, F. A., and P. H. Coleman. 1967. California
group arboviruses: immunodiffusion studies. J. Im-munol. 99:276-284.
13. Murphy, F. A., A. K. Harrison, and S. G. Whitfield. 1973.Bunyaviridae: morphologic and morphogenetic similaritiesofBunyamweraserologicsupergroup vi-rusesandseveralotherarthropod-borne viruses. In-tervirology1:297-316.
14. Murphy, F. A.,S.G. Whitfield, P.H.Coleman, E. R. Rabin,A. B.Jenson, J. L. Melnick, M. R. Edwards, andE.Whitney. California group arboviruses: elec-tronmicroscopicstudies.Exp. Mol.Pathol.9:44-56. 15. Obijeski,J. F., A. T.Marchenko,D. H. L. Bishop,B.
W.Cann,andF. A.Murphy. 1974. Comparative elec-trophoretic analysis of the virus proteins of four rhab-doviruses. J.Gen. Virol. 27:21-33.
16. Obijeski, J. F., E. L. Palmer, L. G. Gafford, and C. C. Randall. 1973. Polyacrylamide gel electrophoresis of fowlpoxand vacciniaproteins. Virology51:512-516.
17. Pettersson, R., and L. Kaariainen. 1973. The ribonu-cleic acids of Uukuniemi virus, a noncubical tick-bornearbovirus. Virology56:608-619.
18. Pettersson, R., L. Kaariainen, C.-H. vonBonsdorff, and N. Oker-Blom. 1971. Structural components of Uukuniemi virus, anoncubical tick-borne arbovirus. Virology 46:721-729.
19. Rosato, R. R., J. M. Dalrymple, W. E. Brandt, R. D. Cardiff, and P. K. Russell. 1974. Biophysical separa-tionofmajor arbovirus serogroups. Acta Virol. 18:25-30.
20. Rosato, R. R., M. L. Robbins, and G. A. Eddy. 1974. Structural components of Oriboca virus. J. Virol. 13:780-787.
21. Saikku, P., and C.-H. von Bonsdorff. 1968. Electron microscopy of the Uukuniemi virus, an ungrouped arbovirus. Virology 34:804-806.
22. Saikku, P., C.-H. von Bonsdorff, M. Brummer-Korven-kontio,and A. Vaheri. 1971. Isolation ofnon-cubical ribonucleoprotein from Inkoo virus, a Bunyamwera supergrouparbovirus. J. Gen. Virol. 13:335-337. 23. Saikku, P., C.-H. von Bonsdorff, and N. Oker-Blom.
1970. The structure ofUukuniemi virus. Acta Virol. 14:103-107.
24. Sather,G. E., and W. McD. Hammon. 1967. Antigenic patterns within the California-encephalitis-virus group. Am. J. Trop. Med. Hyg. 16:548-557. 25. Schlesinger, M. J., S. Schlesinger, and B. W. Burge.
1972. Identification of asecondglycoprotein in Sind-bis virus. Virology 47:539-541.
26. Stanley, P., and E. A. Haslam. 1971. Thepolypeptides ofinfluenza virus. V. Localization ofpolypeptidesin the virionbyiodination techniques. Virology 46:764-771.
27. Stanworth, D. R. 1967.Ultracentrifugation of immuno-globulins,p. 44-111. In D. M. Weir(ed.),Handbook of experimentalimmunology. F. A. Davis Co., Philadel-phia.
28. Thompson, W. H., R.0.Anslow, R. P. Hanson, and G. E. Defoliart. 1972. La Crosse virus isolations from mosquitoes in Wisconsin, 1964-68. Am. J.Trop. Med. Hyg.21:90-96.
29. Thompson, W. H., B. Kalfayan, and R. 0. Anslow. 1965. Isolation ofCaliforniaencephalitis group virus fromafatal human illness. Am. J. Epidemiol. 81:245-253.
30. Wagner, R. R., L. Prevec, F. Brown, D. F. Summers, F.Sokol, and R. MacLeod. Classification of rhabdo-virusproteins: aproposal.J. Virol. 10:1228-1230. 31. Wellings, F. M., G. E. Sather, and W. McD.Hammon.
1970.Immunoelectrophoretic studies of theCalifornia encephalitisvirusgroup. J.Immunol. 107:252. 32. White, A. B. 1975.StructuralpolypeptidesofCalifornia
encephalitis virus: BFS-283. Arch. Virol. 49:281-290. 33. Witte,0.N., I. L. Weissman, and H.S. Kaplan. 1973. Structural characteristics ofsomemurine RNA tu-mor viruses studied bylactoperoxidase iodination. Proc. Natl. Acad.Sci. U.S.A. 70:36-40.
VOL. 19, 1976