A Large Cluster of Highly Expressed Genes Is Dispensable for Growth and
Development in
Aspergillus nidulans
Rodolfo Aramayo, Thomas H. Adams and William
E.
Timberlake
Departments of Genetics and Plant Pathology, University of Georgia, Athens, Georgia 30602
Manuscript received December 19, 1988 Accepted for publication February 7 , 1989
during A. nidulans development.
A
SPERGILLUS nidulans is an ascomycetous funguswhose biological properties make it valuable for studying mechanisms controlling eukaryotic cell me- tabolism, growth and differentiation (PONTECORVO et al. 1953; RAPER, FENNELL and AUSTWICK 1965; CLUTTERBUCK 1974; MACDONALD and HOLT 1976; ARST and BAILEY 1977; COCKER and GREENSHIELDS 1977; KINGHORN and PATEMAN 1977; MCCULLOUGH, PAYTON and ROBERTS 1977; MOSS 1977; PATEMAN and KINGHORN 1977; SMITH et al. 1977; THOMAS
1977; ZONNEVELD 1977; HAWKINS, GILES and KINGH- ORN 1982; ARST and SCAZZOCCHIO 1985; GREEN and SCAZZOCCHIO 1985; HYNES et al. 1985; MAY et al. 1985; OAKLEY, 1985; PATEMAN et al. 1985; BEEVER and LARACY 1986; CADDICK, BROWNLEE and ARST
1986; TIMBERLAKE and HAMER 1986; TIMBERLAKE 1987; TIMBERLAKE and MARSHALL 1988). T h e ge- nome of A. nidulans is compact (2.6 X l o 4 kb), about 5.5 times the size of the Escherichia coli genome (BAIN- BRIDGE 197 1 ; TIMBERLAKE 1978; KOHARA, AKIYAMA and ISONO 1987). It contains only 3% repetitive DNA, most of which codes for rRNA and tRNA precursors (TIMBERLAKE 1978). Laboratory strains appear to lack mobile genetic elements. Structural genes are very closely spaced and either lack or have only a few short introns (ORR and TIMBERLAKE 1982; GWYNNE et al. 1984; HAWKINS, DA SILVA and ROBERTS 1984; MUL-
1987; ADAMS, BOYLAN and TIMBERLAKE 1988). All of these features suggest that despite its metabolic and morphological diversity the A. nidulans genome has been streamlined through evolutionary pressures.
During asexual spore (conidium) production, 6% of the A. nidulans genome, corresponding to 1200 di- verse mRNAs (TIMBERLAKE 1980), is selectively tran- LANEY et UI!. 1985; UPSHALL et al. 1986; BOYLAN et
d .
Genetics 122: 65-7 1 (May, 1989)
ABSTRACT
We investigated the functions of the highly expressed, sporulation-specific SpoCl genes of Asper- gillus nidulans by deleting the entire 38-kb SpoC 1 gene cluster. T h e resultant mutant strain did not differ from the wild type in ( 1 ) growth rate, (2) morphology of specialized reproductive structures
formed during completion of the asexual or sexual life cycles, (3) sporulation efficiency, (4) spore viability or ( 5 ) spore resistance to environmental stress. Thus, deletion of the SpoCl gene cluster,
representing 0.15% of the A. nidulans genome, had no readily detectable phenotypic effects. Impli- cations of this result are discussed in the context of major alterations in gene expression that occur
scribed. Mature conidia contain approximately 200 spore-specific mRNAs that collectively comprise >11% of the mass of mRNA stored in the spore. T h e functions of these mRNAs are unknown. However, based on their abundance in spores, the evolutionary conservation of at least some of the genes encoding them (MULLANEY and KLICH 1987) and the compact- ness of the A. nidulans genome, we have speculated that they code for proteins having important functions in conidium differentiation, maintenance or germi- nation (MILLER et al. 1987).
One group of genes that encodes spore-specific transcripts has been subjected to extensive analysis (TIMBERLAKE and BARNARD 1981; GWYNNE et al.
66 R. Aramayo, T. H. Adams and W. E. Timberlake
MATERIALS A N D METHODS
A. nidulans strains and genetic techniques: T h e geno- types of strains used in this study are given in Table 1 .
Standard A. nidulans genetic techniques (PONTECORVO et al. 1953; CLUTTERRUCK 1974; KAFER 1977) and transforma- tion procedures (YELTON, HAMER and TIMRERLAKE 1984) were employed.
Nucleic acid manipulations: DNA and R N A were iso- lated as described (TIMRERLAKE 1986). Blot hybridizations were carried out as recommended by the membrane man- ufacturer (Hybond-N, Amersham).
Plasmid constructions: The following plasmids were con- structed by using standard procedures (MANIATIS, FRITSCH and SAMBROOK 1982; AUSREL et al. 1987).
1. pDC 1 : A 1.8-kb Sphl-BamHI fragment containing t h e A. nidulans argB+ gene (BERSE et al. 1983; UPSHALL et al.
1986) inserted into the SphI and BamHI sites of pIC20R (MARSH, ERFLE and WYKFS 1984).
2. pDC2: A 0.8-kb SalI-EcoRI fragment from XAn- SpoC1-R2 (GWYNNE et al. 1984) inserted into the Sal1 and GcoRI sites of pIC2OH (MARSH, ERFLE and WYKES 1984).
3. pDC3C: A 2.6-kb BamHI fragment from XAnSpoC1- D5 (GWYNNE et al. 1984) inserted into the BamHI site of pIC2OH.
4. pDC4: A 0.8-kb Xhol-SphI fragment from pDC2 in- serted into the XhoI and SphI sites of pDCl.
5. pDC5R: A 2.6-kb BamHI fragment from pDC3C in- serted into the BamHI site of pDC4.
6. pDC6: A 5.2-kb SmaI-XhoI fragment from pDC5R inserted into the SmaI and XhoI sites of pIC20H.
Spore survival experiments: Conidia were harvested and subjected to various treatments. They were then serially diluted, spread onto agar-solidified minimal medium con- taining appropriate nutritional supplements, incubated in the dark for 4 days at 37" and colonies were counted. Ultraviolet light treatment was done according to BARRON and MACNEILL (1 962). For high temperature' treatment, spores were suspended in H2O at a concentration of 1.5 X
1 O7 ml and incubated at 60" for various times. For extended viability tests, spores were suspended in H20 or maintained in the dry state and incubated at room temperature (22- 25") or at 37" for various times. For exposure to potentially toxic substances, spores were suspended in H z 0 at a concen- tration of 1.5 X lo7 ml and incubated for 2 min at room temperature in the presence of 0.1% (w/v) Nonidet P-40, 0.1 % Tween 80, 0.1 % SDS, 0.1 % Na+ deoxycholate, 0.1 % Triton X-100, 0.15 M NaCI, 5 M NaCI, CHCls-, CsH50HaC2- or CsHeO-saturated H20. Only treatments with the last three compounds significantly decreased spore viability.
Sporulation efficiency and spore germination: Coni- dium production was quantified as described by YACER, KURTZ and CHAMPE (1982), except that 1 X lo5 spores/ Petri dish were incubated at 37" and the number of spores/ cm2 was determined at 30, 32, 34, 36 and 38 h after inoculation. The fraction of spores germinating, the timing of germination and the morphology of germlings were determined by microscopic examination.
RESULTS
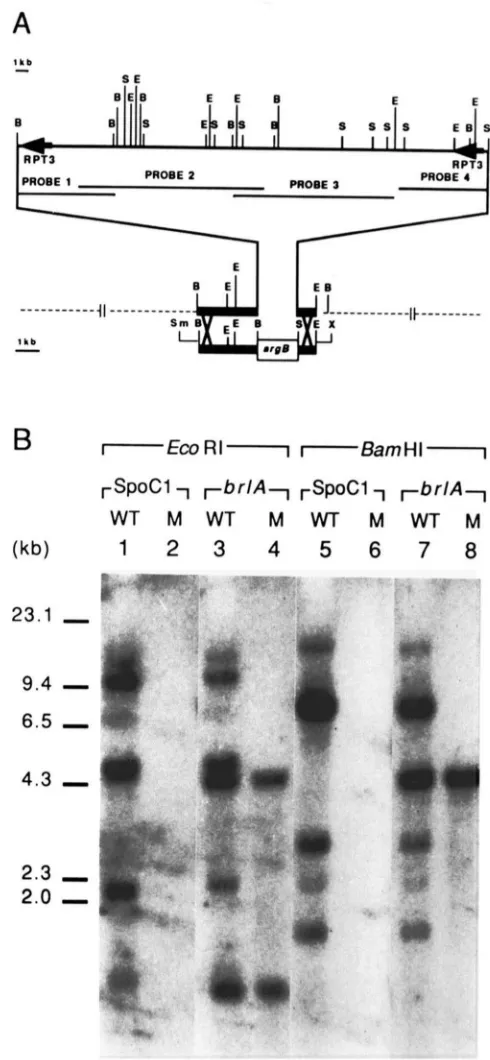
We wished to determine if removal of any SpoCl genes caused a phenotypic alteration. We therefore deleted the entire SpoC 1 region by using a transplace- ment strategy (ROTHSTEIN 1983; Figure 1 A). Plasmid pDC6 containing A. nidulans DNA fragments from
FIGURE 1 .-(A) Diagram of the SpoCl region. A map of the A.
nidulans SpoCl region is given and the sequences used to delete
the cluster are shown below it. The sequences corresponding to the probes are: Probe 1 , 8-kb BamHl fragment from XAnSpoCI-S3; probe 2, XAnSpoC1-L8; probe 3. XAnSpoC1 and probe 4, 7 kb
Sal1 fragment from XAnSpoCl-R2. (GWYNNE el al. 1984). Restric-
tions sites are: B (BamHI), S (Sall). E (EcoRI), Sm (Smal) and X
( X h o l ) . The only X and Sm restriction sites shown are those that
were used to isolate the fragment to delete the cluster. (B) Southern blot ;~nalysis. DNA from the wild-type (WT) and a ASpoC1 (M)
TABLE 1
Aspergillus nidulans strains
~ ~~
Designation Genotype Origin
P" 1 (bioA I '; argB2; mefG I b; velA I ') P. WEGLENSKI, Department
o f Genetics, Warsaw Uni- versity. Pol;md
A.JC 473.14 ( ~ e l A 2 ~ ; a r g B 2 pxnA4'; velA I . JOHN CLUTTERRUCK, Depart-
n i a D l 7 ) ment of Genetics, Glasgow
University, Scotland
FGSC26 (bioA I ; velA I ) Fungal Genetics Stock Center
FGSC237 (yelA2, pabaAl; trpC801, Fungal Genetics Stock Center
FGSC I94 (yelA2, bioA I ; velA I ) Fungal Genetics Stock Center
velA I )
FGSC3.57 (bioA I : wA 3; velA I ) Fungal Genetics Stock Center
TRAOO7 (bioAI; argB2, ASpoC1::argB'; This study; PW 1 transformed
metGI; velAI) w i t h the 5.2-kb Smal-Xhol
fragment from pDC6
RKAOO2 (bioA I ; argB2, ASpoC1 ::argB': Progeny from TRAOO7 X
RRAOO7 (yelA2; metG I ; velA 1 ) Progeny from KRAOO2 X
KKAOOS (yelA2; ASpoC1::argB'; metGI: Same as KRA007
RRAOOS (metG I ; velA I ) Same a s RRA007
RRAOIO (ASpoC 1 ::argB+; mrtG 1 ; Same as KKA007
metG I ; velA I ) AJC 473.14
FGSC237
velA I )
velA I )
' bioAI = biAI; b metGI = methGI; ' velAI = v e A l ; d yelA2 = yA2; ' pxnA4 = pyroA4.
FIGURE 2.-RNA blot :lnal\sis. (;I) Total R N A ( I O pg, lanes 1
; ~ n d 3: 20 pg. lanes 2 and 4) from a ASpoC1 (lanes I and 2) or the wild-type strain (lanes S and 4) w a s fractionated by denaturing agarose gel electrophoresis and blots were hybridized with radiola- bclecl DNA fragments corresponding to a portion of the SpoCl region (XAnSpoC1 and XAnSpoC1-SS). (B) The membrane was \c;lshed and rehybridized w i t h ;In argB probe to confirm loading of the lanes.
chromosomal regions adjacent to the SpoCl cluster fused to a fragment containing the argB+ gene, was constructed. T h e composite SmaI-XhoI A . nidulans D N A fragment was isolated and used to transform an
argB- A . nidulans strain (PW 1, Table 1) to arginine-
independence. T h e chromosomal integration event illustrated in Figure 1A was expected to evict the
entire SpoCl region, including the 1.1-kb direct re- peats (RPT3).
DNA was isolated from 64 transformants and gel blot hybridization analysis was used to determine if both RPT3 sequences had been deleted. A single transformant (TRA007) lacked both repeats and was characterized further. Figure 1 B demonstrates that the entire SpoCl region was deleted from TRA007. T h e hybridization pattern observed for wild-type DNA was as expected (Figure lB, lanes 1, 3, 5 and 7; GWYNNE et al. 1984), while no hybridization to the SpoCl probes (Figure 1A) was observed for TRA007 DNA (Figure lB, lanes 2, 4, 6 and 8). Equal loading of DNA was confirmed by hybridizing the same mem- brane, without removing the previous probes, to ra- diolabeled DNA that corresponds to 6rlA (Figure 1 B,
lanes 3 , 4 , 7 and 8; ADAMS, BOYLAN and TIMRERLAKE 1988). Evidence that the deletion was directed by
integration of the SmaI-XhoI fragment into the SpoCl region was obtained by demonstrating genetic linkage of the argB+ gene to the deletion event (data not shown).
Conidial RNA gel blots were hybridized with two SpoCl internal clones containing most of the SpoCl messages (XAnSpoC1 and XAnSpoCl-S3; GWYNNE et al. 1984) and washed at moderate (0.3 M Na+, 65") or high (0.03 M Na+, 65") stringencies to detect SpoC 1 -related RNAs. As shown in Figure 2, no SpoC 1 complementary transcripts were detected.
6 8 R. Aramayo, T. H. Adams and W. E. Timberlake
Co-isogenic strains
Mixture <;rc.ell-sprcd Yellowspored
I RRAOO9 ( m e t G I " : v e l A I b ) RRA007 (yelA2': metCI: uel.41) 2 UUAO09 ( m e t G I : v e l A I ) RRAOOR CwlAP; ASpoC1::argB +:
3 RRA010 (ASpoCI::argB+: RKA007 (yeIA.2: m e t G I ; v e M I ) metC I ; velA I )
metGI: velA I )
n metC I = methC: b vel,\ I = ueA I : ' yelA2 = ~qA2.
I IIWlb\YYY\
FIGURE 4.-Sensitivity of conidia to UV light. A mixture of green, yellow and white conidia was exposed to UV light as de- scribed under MATERIAU A N D METHODS. Samples taken at the times indicated were serialy diluted, plated and the fraction of green- spored survivors was calculated relative to either yellow- or white- spored srlrvivors. After 90 sec 3.70% of the green, 1.25% of the
yellow and 0.39% of the white conidia remained viable. One stand- ard deviation is indicated (T).
phores, conidia or germlings of strain TRA007 and the strain from which it was derived were detected by light microscopy (Figure 3). Both strains were identi- cal with respect to colonial morphology, radial growth and spore production rates. Conidia from the ASpoCl
FIGURE J.-"orphology of coni- cliophores, spores ; ~ n d gernllings. Kcpresent;ltive s1ructures from the wild-type (panels A, <: ant1 E) and ;I
ASpoCI strain (panels B, D ;Ind F) were photogr;tphecl a t 4 0 0 X by using differential interfercnce contrast op- tics. Conidiophores. panels A and 13:
ungerminated conidia, panels C and D: conidia gernlinatetl for (i 11, p;lnrls E m d F.
FIGURE 5.-Sensitivity of SpoC1+ and ASpoCI conidia to U V light. Mixtures green and yellow of conidia from SpoC1+ and
ASpoC1 strains (Table 2) were exposed to U V light and analyzed a s described in the legend of Figure 4.
strain swelled during early germination and adhered to one another and to plastic Petri dishes, as did the wild type.
Dispensable Genes in
classes changes. If there is a difference in sensitivity due to the tested genotypic variable (e.g., deletion of SpoCl), the difference should be consistent in recip- rocal spore color mixtures.
T h e feasibility of this approach was tested by mixing co-isogenic green (FGSC26), yellow (FGSC 194) and white (FGSC357) (Table 1) spores and exposing the mixtures to UV. Figure 4 shows that with increasing irradiation the fraction of green spores increased in comparison to either yellow or white spores, demon- strating that yellow and white spores are more sensi- tive to UV light than are wild-type green spores. Thus, spore color mixtures provide a convenient, internally controlled method for detecting a differential sensitiv- ity of conidia to environmental factors.
Three spore mixtures were prepared as shown in Table 2 and subjected to potentially detrimental con- ditions (exposure to UV light, high temperature, de- tergents, high salt concentration and organic chemi- cals) or stored for extended periods (2 weeks) either dry or suspended in H 2 0 (see under MATERIALS AND METHODS). Following treatment, the fraction of sur-
viving spores and the proportion of spores in each color class was determined. Figure 5 shows that the fraction of green spores in each of the spore mixtures increases as a function of UV light treatment. There was a similar change in the fraction of green spores in all populations as a function of irradiation, indicating that deletion of the SpoCl cluster had no effect on sensitivity to UV light. Negative results were obtained with all of the other treatments. Thus, these experi- ments failed to detect any phenotypic change associ- ated with deletion of the A. nidulans SpoCl gene cluster.
DISCUSSION
T h e results presented here demonstrate that the A . nidulans SpoC 1 gene cluster is dispensible for growth and development under laboratory conditions. This region represents 0.15% of the genome and codes for numerous poly(A)+ RNAs, that collectively comprise 2 2 % of the mRNA mass produced and stored in conidia. T h e high level of SpoCl gene transcription and the probable translation of SpoCl mRNAs con- stitutes a substantial energy expenditure during coni- dium formation. Because conidiation likely exists as a dispersal mechanism to expand the organism’s popu- lation when conditions are optimal for growth, it seems probable that evolutionary pressures would fa- vor efficient reproductive mechanisms that minimize the energy expended in spore differentiation. T h e strong regulation and high level of SpoCl gene expression would be expected to have been eliminated if the genes provided no biological advantage. In addition, the DNA sequence of at least the central portion of the SpoCl gene cluster is highly conserved
among diverse Aspergillus species (MULLANEY and KLICH 1987), supporting the notion that SpoCl genes have important functions in the Aspergilli.
In view of the preceding considerations, the lack of evidence for SpoCl gene functions during growth and development of A. nidulans might be considered surprising. However, neither the size of this deletion nor the elimination of tightly regulated genes are without precedent. For example, a large (>130 kb) segment of the A. nidulans genome distal to areA is dispensible for growth (CADDICK et al. 1986). Many genes that are specifically expressed by Saccharomyces cerevisiae during ascosporogenesis have been inacti- vated without affecting growth or sporulation (YA- MASHITA and FUKUI 1985; GARBER and SECALL 1986; GOTTLIN-NINFA and KABACK 1986; PERCIVAL-SMITH and SEGALL 1986; PETKO and LINDQUIST 1986). Re- sults from random mutagenesis (GOEBL and PETES 1986) showed that 70% of insertions in the S. cerevisiae genome caused no detectable phenotypic change. Di- rected mutational studies of snRNA genes (PARKER et al. 1988) showed that simultaneous inactivation of 6 snRNA genes had little effect on growth. Thus, a significant number of genes are apparently dispensible in fungi.
Many fungal genes with redundant functions have been described (e.g., MORRIS, LAI and OAKLEY 1979; LINDQUIST 1986). All homologous copies must be mutated before there is a phenotypic effect. We were unable to detect other DNA sequences or transcripts complementary to SpoC 1 in the ASpoC 1 strain by gel blot hybridization. However, some duplicated genes, such as A. nidulans @-tubulin genes, are highly diver- gent (MAY et al. 1985, 1987), indicating that func- tional homology may be difficult or impossible to detect by cross-hybridization.
SpoCl genes may have subtle functions that were not detected in our experiments. For example, A . nidulans grows on substrates as diverse as decaying leaf litter, citrus fruits, and rhinocerous bronchia
(THOMAS 1977; KUTTIN et al. 1984). Conidia may therefore have enzymes in their walls that hydrolyze complex nutrient sources present in their natural en- vironment. Conidia from the phytopathogenic fungus Nectria haematococca, for instance, contain cutinase, an enzyme that digests the waxy cuticle of plant stems and leaves (KOLLER, ALLAN and KOLATTUKUDY 1982). Our experiments were not designed to detect such exotic catalytic activities.
Finally, a large number of fungal genes code for enzymes that catalyze the formation of secondary metabolites such as P-lactam antibiotics and aflatoxins
70 R. Aramayo, T. H. Adams and W. E. Timberlake
Moreover, closely related fungal isolates may have or lack the ability to produce particular secondary me- tabolites without any significant corresponding effect on growth or development (MACDONALD and HOLT
1976; MAKINS, HOLT and MACDONALD 1983). In ad- dition, secondary metabolites may be synthesized by the action of metabolic “networks” rather than by linear metabolic pathways (ZAHNER, ANKE and ANKE
1983). The extensive redundancy of such networks makes them essentially immutable. Some of the 1200 sporulation-specific genes found in A. nidulans, in- cluding SpoCl genes, may be responsible for second- ary metabolite synthesis. Even though such genes may have subtle or redundant functions, and may be of little consequence to the organism itself, their activi- ties in synthesis of beneficial (e.g., antibiotics) and detrimental (e.g., carcinogens) compounds are of great importance to humans.
This work was supported by U.S. Public Health Service grant
GM37886 and U.S. Department of Agriculture grant 88-37262- 3566 to W.E.T. R.A. received support from Empresa Brasikira d e Pesquisa Agropecuiria (Centro Nacional d e Recursos Geneticos e Biotecnologia) and Conselho Nacional d e Desenvolvimento Cienti- fico e Tecnolbgico-Brazil.
We wish to thank S T E V E KARL for his excellent help in statistical analysis of the data, members of the laboratory for their helpful suggestions and CHARLES MIMS and WYATT ANDERSON for their critical reviews of the manuscript.
L I T E R A T U R E CITED
ADAMS, T . H., M. T. BOYLAN and W. E. TIMBERLAKE, 1988 brlA
is necessary and sufficient to direct conidiophore development
in Aspergzllus nidulans. Cell 54: 353-362.
ARST, H. N., JR., and C. R. BAILEY, 1977 Carbon metabolism in
Aspergillus nidulans, pp. 97-129 in Genetics and Physiology of
Aspergzllus, edited by J. E. SMITH and J. A . PATEMAN. Academic
Press, New York.
ARST, H. N., JR., and C. SCAZZOCCHIO, 1985 Formal genetics and molecular biology of the control of gene expression in Asper-
gillus nidulans, pp. 310-343 in Gene Manipulations in Fungz,
edited by J. W. BENNETT and L. L. LASURE. Academic Press, New York.
AUSUBEL, F. M., R. BRENT, R. E. KINGSTON, D. D. MOORE, J. A. SMITH, J. G . SEIDMAN and K. STRUHL, 1987 Current Protocols
in Molecular Biology. John Wi!ey & Sons, New York.
BAINBRIDGE, B. W . , 1971 Macromolecular composition and nu- clear division durng spore germination in Aspergillus nidulans. J. Gen. Microbiol. 6 6 319-325.
BARRON, G . L., and B. H. MACNEILL, 1962 A simplified proce- dure for demonstrating the parasexual cycle in Aspergillus. Can. J . Bot. 40: 1321-1327.
BEEVER, K. E., and E. P. LARACY, 1986 Osmotic adjustment in the fil;unentous fungus Aspergillus nidulans. J. Bacteriol. 168:
1358-1365.
BERSE. B., A. DMOCHOWSKA, M. SKRZYPEK, P. WEGLENSKI, M. A. BATES and R. I,. WEISS, 1983 Clonirig and characterization
of the ornithine carbamoyltransferase gene from Aspergillus
nidulans. Gene 25: 109-1 17.
BOYI.AN, M. T , , P. M. MIRABITO, C. E. WILLET, C. R . ZIMMERMAN and W. E. TIMBERLAKE, 1987 Isolation and physical charac- terimtion of three essential conidiation genes from Aspergillus
nidulans. Mol. Cell. B i d . 7: 31 13-31 18.
CADDICK, M. X., A. G. BROWNLEE and H. N. ARST, JR.,
1986 Regulation of gene expression by p H of the growth medium in Aspergillus nidulans. Mol. Gen. Genet. 203: 346- 353.
CADDICK, M. X . , H . N. ARST, JR., L. H. TAYLOR, R. I . JOHNSON
and A. G. BROWNLEE, 1986 Cloning of the regulatory gene
areA mediating nitrogen metabolite repression in Aspergtllus
nidulans. EMBO J. 5: 1087- 1090.
CLUTTERBUCK, A . J., 1974 Aspergillus nidulans, pp. 447-510 in
Handbook of Genetics, edited by R. C. KING. Plenum, New York.
COCKER, R., and R. N. GREENSHIELDS, 1977 Fermenter cultiva- tion of Aspergillus, pp. 361-390 in Genetics and Physiology of
Aspergzllus, edited by J. E. SMITH and J. A. PATEMAN. Acd-
demic Press, New York.
GARBER, A. T., and J. SEGALL, 1986 T h e SPS gene of Saccharo-
myces cerevisiae encodes a major sporulation-specific mRNA.
Mol. Cell. Biol. 6: 4478-4485.
GOEBL, M. G., and T . D. PETES, 1986 Most of the yeast genomic sequences are not essential for cell growth and division. Cell
GOTTLIN-NINFA, E., and D. B. KABACK, 1986 Isolation and func- tional analysis of sporulation-induced transcribed sequences from Saccharomyces cerevisiae. Mol. Cell. Biol. 6: 2 185-2 197.
GREEN, P. M., and C. SCAZZOCHIO, 1985 A cloning strategy in filamentous fungi, pp. 345-353 in Gene Manipulations in Fungi, edited by J. W. BENNET and L. L. LASURE. Academic Press, New York.
GWYNNE, D. I. , B. L. MILLER, K. Y. MILLER^^^ W. E. TIMBERLAKE,
1984 Structure and regulated expression of the SpoCl gene cluster from Aspergzllus nidulans. J. Mol. Biol. 180: 91-109.
HAWKINS, A. R., A. J. F . DA SILVA and C. F. ROBERTS,
1984 Cloning and characterization of the three enzyme struc- tural genes qutB, 9utC and qutE from the quinic acid utilization cluster in Aspergillus nidulans. Curr. Genet. 9: 305-312.
HAWKINS, A. R., N. G. GILES and J. R. KINGHORN, 1982 Genetical and biochemical aspects of quinate breakdown in the filamen- tous fungus Aspergillus nidulans. Biochem. Genet. 20: 271- 286.
HYNES, M. J., J. M. KELLY, C. M. CORRICK and T. J . LITTLEJOHN,
1985 Structure and expression of the Aspergzllus amdS gene, pp. 157-169 in Molecular Genetics ofFilamentous Fungt, edited by W. E. TIMBERLAKE. Alan R. Liss, New York.
KAFER, E., 1977 Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1 9 AI-A28.
KINGHORN, J. R., and J. A. PATEMAN, 1977 Nitrogen metabolism, pp. 147-202 in Genetics and Physiology of Aspergillus, edited by J. E. SMITH and J. A. PATEMAN. Academic Press, N e w York. KOHARA, Y., K. AKIYAMA and K . ISONO, 1987 T h e physical map
of the whole E . coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50: 495-508.
KOLLER, W., C. R. ALLAN and P. E. KOLATTUKUDY, 1982 Role of cutinase and cell wall degrading enzymes in infection of Pisum satiuurn by Fusarium solani f. sp. pisi. Physiol. Plant Patho]. 20:
47-60.
KUTTIN, E. S . , W. KAPLAN, H . I . SCHOLER, H. BURTSCHER and H. KOHLER, 1984 Sexual and asexual reproduction of Aspergzllus
nidulans in viuo. Mykosen 28: 109- 1 16.
LINDQUIST, S., 1986 T h e heat shock response. Annu. Kev. Biochem. 55: 1 1 5 1 - 1 19 1 .
MACDONALD, K. D., and G . HOLT, 1976 Genetics of biosynthesis and overproduction of penicillin. Sci. Prog. 63: 547-573.
MAKINS, J . F., G. HOLT and K. D. MACDONALD, 1983 T h e genetic location of three mutations impairing penicillin production i n
Aspergillus nidulans. J . Gen. Microbiol. 1 2 9 3027-3033.
MANIATIS, T . , E. R. FRITSCH and J. SAMBROOK, 1982 Molecular
Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory,
Cold Spring Harbor. N.Y.
71
MARSH, J. L., M. ERFLE and E. J. WYKES, 1984 T h e PIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32: 481-485.
MAY, G. S., J. A. WEATHERBEE, J. GAMBINO, M. L. TSANG and R. MORRIS, 1985 Identification and function of beta tubulin
genes in Aspergillus nidulans, pp. 239-25 1 in Molecular Genetics
of Filamentous Fungi, edited by W. E. TIMBERLAKE. Alan R.
Liss, New York.
MAY, G. S., M. L. S. TSANG, H. SMITH, S. FIDEL and R. MORRIS,
1987 Aspergillus nidulans P-tubulin genes a r e unusually di- vergent. Gene 55: 231-243.
MCCULLOUGH, W., M. A. PAYTON and C. F. ROBERTS,
1977 Carbon metdboolism in Aspergillus nidulans, pp. 97-
129 in Genetics and Physiology of Aspergillus, edited by J. E.
SMITH and J. A. PATEMAN. Academic Press, New York. MILLER, B. L., K. Y. MILLER and W. E. TIMBERLAKE, 1985 Direct
and indirect gene replacements in Aspergillus nidulans. Mol. Cell. Biol. 5: 17 14- 172 1.
MILLER, B. L., K. Y. MILLER, K. A. ROBERTI and W. E. TIMBER-
LAKE, 1987 Position-dependent and -independent mecha- nisms regulate cell-specific expression of the SpoCl gene clus- ter of Aspergillus nidulans. Mol. Cell. Biol. 7: 427-434.
MORRIS, R. N., M. H . LAI and C. E. OAKLEY, 1979 Identification of a gene for alpha-tubulin in Aspergillus nidulans. Cell 16:
MOSS, M. 0.. 1977 Aspergillus mycotoxins, pp. 499-524 in Ge-
netics and Physiology of Aspergillus, edited by J. E. SMITH and J.
A. PATEMAN. Academic Press, New York.
MULLANEY, E. J., and M. A. KLICH, 1987 Survey of representative species of Aspergzllus for regions of DNA homology to Asper-
gillus nidulans developmental genes. Appl. Microbiol. Biotech-
nol. 25: 476-479.
MULLANEY, E. J., J. E. HAMER, K. A. ROBERTI, M. M. YELTON and W. E. TIMBERLAKE, 1985 Primary structure of the trpC gene from Aspergillus nidulans. Mol. Gen. Genet. 1 9 9 37-45.
OAKLEY, B. R., 1985 T h e molecular biology of microtubules in
Aspergillus, pp. 225-238 in Molecular Genetics of Filamentous
Fungi, edited by W. E. TIMBERLAKE. Alan R. Liss, New York.
O’DONNELL, C. H., A. UPSHALL and K. D. MACDONALD,
1985 Gene dosage effects and antibiotic synthesis in fungi, pp. 293-307 in Gene Manipulations in Fungi, edited by J. W. BENNETT and L. L. LASURE. Academic Press, New York. ORR, W. C., and W. E. TIMBERLAKE, 1982 Clustering of spore-
specific genes in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 7 9 5976-5980.
PARKER, R., T . SIMMONS, E. 0. SHUSTER, P. G. SILICIANO and C.
GUTHRIE, 1988 Genetic analysis of small nuclear RNAs in
Saccharomyces cerevisiae: viable sextuple mutant. Mol. Cell. Biol.
8: 3 150-3 159.
PATEMAN, J. A,, and J. R. KINGHORN, 1977 T h e regulation of nitrogen metabolism, pp. 203-241 in Genetics and Physiology of
Aspergillus, edited by J. E. SMITH and J. A. PATEMAN. Academic
Press, New York.
PATEMAN, J. A , , C. H . DOY, J. E. OLSEN, H. J. KANE and E. H . CREASER, 1985 Molecular analysis of alcohol metabolism in
Aspergillus, pp. 171-184 in Molecular Genetics of Filamentous
Fungz, edited by W. E. TIMBERLAKE. Alan R. Liss, New York.
PERCIVAL-SMITH, A., and J. SEGALL, 1986 Characterization and mutational analysis of a cluster of three genes expressed pref- erentially during sporulation of Saccharomyces cerevisiae. Mol.
Cell. Biol. 6 2443-245 1.
437-442.
PETKO, L., and S. LINDQUIST, 1986 Hsp26 is not required for growth a t high temperatures, nor for thermotolerdnce, spore development, or germination. Cell 45: 885-894.
PONTECORVO, G., J. A. ROPER, L. M. HEMMONS, K. D. MACWNALD and A. W. J. BUFTON, 1953 T h e genetics of Aspergzllus ni-
dulans. Adv. Genet. 5: 141-238.
RAPER, K. B., D. I . FENNELL and P. K. C. AUSTWICK,
1965 Cultivation, pp. 35-55 in The Genus Aspergillus, edited by K. B. RAPER, D. 1. FENNELL and P. K. C. AUSTWICK. Williams
& Wilkins, Baltimore.
ROTHSTEIN, R. J.. 1983 One-step gene disruption in yeast. Meth- ods Enzymol. 101: 202-21 1 .
SMITH, J. E., J. G. ANDERSON, S. G. DEANS and B. DAVIES,
1977 Asexual development in Aspergtllus, pp. 23-58 in Ge-
netics and Physiology of Aspergillus, edited by J. E. SMITH and J.
A. PATEMAN. Academic Press, New York.
THOMAS, A,, 1977 T h e g e n u s Aspergzllus and biodeterioration, pp. 453-480 in Genetics and Physiology of Aspergillus, edited by J. E. SMITH and J. A. PATEMAN. Academic Press, New York. TIMBERLAKE, W. E., 1978 Low repetitive DNA content in Asper-
gillus nidulans. Science 202: 773-775.
TIMBERLAKE, W. E., 1980 Developmental gene regulation in As-
pergillus nidulans. Dev. Biol. 7 8 497-510.
TIMBERLAKE, W. E., 1986 Isolation of stage- and cell-specific genes from fungi, pp. 343-357 in Biology and Molecular Biology
of Plant-Pathogen Interactions ( N A T O AS1 Series, Vol. H I ) ,
edited by J. BAILEY. Springer-Verlag, Berlin.
TIMBERLAKE, W. E., 1987 Molecular genetic analysis of develop- ment in Aspergillus nidulans, pp. 63-82 in Genetic Regulation of
Development, edited by W. F. LOOMIS. Alan R. Liss, New York.
TIMBERLAKE, W. E., and E. C. BARNARD, 1981 Organization of a gene cluster expressed specifically in the asexual spores of A.
nidulans. Cell 26: 29-37.
TIMBERLAKE, W. E., and J. E. HAMER, 1986 Regulation of gene activity during conidiophore development in Aspergillus nidu-
lans, pp. 1-29 in Genetic Engineering, edited by J. K. SETLOW
and A. HOLLANDER. Plenum, New York.
TIMBERLAKE, W. E., and M. A. MARSHALL, 1986 Genetic regula- tion of development in Aspergillus nidulans. Trends Genet. 4: ,
162-169.
UPSHALL, A., T. GILBERT, G. SAARI, P. J . O’HARA, P. WEGLENSKI, B. BERSE, K. MILLER and W. E. TIMBERLAKE, 1986 Molecular analysis of the argB gene of Aspergillus nidulans. Mol. Gen. Genet. 204: 349-354.
YAGER, L. N., M. B. KURTZ and S. P. CHAMPE, 1982
Temperature-shift analysis of conidial development in Aspergil-
lus nidulans. Dev. Biol. 93: 92-103.
YAMASHITA, I . , and S. FUKUI, 1985 Transcriptional control of the sporulation-specific glucoamylase gene in the yeast Saccharo-
myces cerevisiae. Mol. Cell. Biol. 5: 3069-3073.
YELTON, M. M., J. E. HAMER and W. E. TIMBERLAKE,
1984 Transformation of Aspergzllus nidulans by using a t r p c plasmid. Proc. Natl. Acad. Sci. USA 81: 1470-1474.
ZAHNER, H., H. ANKE and T. ANKE, 1983 Evolution and second- ary pathways, pp. 153-171 in Secondary Metabolism and D q e r -
entiation in Fungz, edited by J. W. BENNET and A. CIEGLER.
Marcel Dekker, New York.
ZONNEVELD, B. J. M., 1977 Biochemistry and ultrastructure of sexual development in Aspergillus, pp. 59-80 in Genetics and
Physiology of Aspergillus, edited by J. E. SMITH and J. A. PATE-
MAN. Academic Press, New York.