Molecular Organization
and
Germinal
Instability
of
R-sti#@led
Maize
William B.
Eggleston, Mary Alleman'
and
Jerry L. Kermicle
Laboratory of Genetics, University of Wisconsin, Madison, Wisconsin 53706
Manuscript received October 24, 1994 Accepted for publication June 5 , 1995
ABSTRACT
The spotted seed allele R-stippled (R-st) is comprised of the following genetic components: strong seed color (Sc), inhibitor-ofR (I-R) and near-colorless seed (Nc). I-R is a mobile element that represses (Sc) expression irregularly. Germinal I-R losses produce progeny with fully colored seed. Southern blot analysis revealed four r-hybridizing segments in R-st and three, two or one in two sets of unequal crossover deletion products. By comparison to published reports of r gene structure, we maintain that each segment contains at least one r gene. The proximal r gene, Sc, confers strong seed color; the three distal r genes together produce near-colorless seed. R-st's seed spotting phenotype is correlated with the presence of a 3.3-kb insert in Sc identified as I - R The level of the near-colorless phenotype is inversely correlated with the number of r genes present, suggesting involvement of a multiple copy silencing mechanism in their regulation. Phenotypic changes in R-st occurred primarily by unequal exchange between r genes. The locations of exchange positions showed Y a strong polarity, nearly all occurring in the 3' portions of the identified r genes.
"
T
HE
red ( r ) color family of maize genes encodes transcriptional activators of the m y class of helix- loophelix DNA-binding proteins (LUDWIG et al. 1989;RADICELLA et al. 1991). In the absence of r product, expression of genes encoding enzymes of the %hydroxy anthocyanin pathway are at a low level or absent. Alleles of r and its cognate (homologous) locus b confer pig- mentation in particular vegetative, flower and seed parts (STADLER and FOGEL 1945; STYLES et al. 1973). Whereas alleles of b are simplex, in some r alleles pig- mentation of different plant parts is regulated by muta- tionally independent and recombinationally separable components (STADLER 1948a; STADLER and NUEFFER
1953). These components behave as separate comple- mentation groups, i.e., as separate genes.
r alleles manifest three types of instability: somatic variegation, germinal mutation and heritable interac- tion between alleles in certain heterozygotes (paramu- tation) (BRINK 1958). Somatic variegation is observed as spots of color on otherwise colorless or faintly colored tissue. In alleles such as R-stippled and R-marbled, these pigmented spots occur in the aleurone layer of the ker- nel. Spotting alleles such as R-st and R-mb mutate germi- nally to produce solid expression in the aleurone (ASH- MAN 1960; BRINK and WEVERS 1957). In contrast, nonvariegating alleles such as R-r:standard produce col- ored aleurone and certain plant and floral tissues, and mutate to lose either aleurone or plant/floral expres- sion ( STADLER 1948a). R-r:standard is comprised of two
genetic components in proximal to distal order on the
C m a p m d i n g author; William B. Eggleston, Department of Biology,
'
Cuffat address:Department of Biological Sciences, Duquesne Uni- Virginia Commonwealth University, Richmond, VA 23284.versity, Pittsburgh, PA 15282.
Genetics 141: 347-360 (September, 1995)
long arm of chromosome 10: (P), which conditions anthocyanin expression in plant and floral tissues (root, coleoptile and anthers), and (S), which allows pigmen- tation in the aleurone layer of the kernel (STADLER 1948b). Molecular analyses have shown that R-r:standard is comprised of four separate r genes in a combination of direct and inverted orientation (ROBBINS et nl. 1991;
WALKER et al. 1995). Phenotypic instability of the com- plex has been shown to be due primarily to unequal recombination events between the various r genes in the complex and to insertions and deletions in specific r genes (ROBBINS et al. 1991).
Paramutation involves a heritable change in expres- sion of one allele following heterozygosity with another allele of the locus, which is itself unchanged by the heterozygosity. Alleles that induce the change are para- mutagenic and alleles that are sensitive to be being changed are paramutable. Following paramutation, sensitive alleles are referred to as paramutant or para- mutated. Several alleles of the r locus participate in paramutation. The two that have been studied most extensively are the paramutable allele R-r:standard and its derivatives and the paramutagenic allele R-stippled (R-st) and its derivatives (reviewed in BRINK et al. 1968;
BRINK 1973).
The purpose of the present study is to explore the molecular organization of pigmentation components and the basis for genetic instability of R-stippled. An
348 W. B. Eggleston, M. Alleman and J. L. Kermicle
viewed in BRINK 1973). The self-colored (Sc) compo- nent is capable of solid pigmentation of the aleurone layer in seeds. However, inhibitor of R (Z-R) inhibits (Sc) expression unstably, producing colored aleurone sec- tors. Occasionally, fully colored, germinal revertant ker- nels or self-colored (R-sc) mutations arise. R-st’s somatic spotting and germinal reversion to R-sc parallels obser- vations on the effects of mobile elements in maize and other organisms (MCWHIRTER and BRINK 1962). This suggestion is supported by the observation that certain classes of R-SC’S arise in conjunction with recovery of new modifiers that increase the intensity of R-st’s so- matic spotting, as would be expected if these R-SC’S arose by transposition of a mobile element from r to a new site in the genome (WILLIAMS et al. 1984). The near- colorless (Nc) component produces lightly mottled, near-colorless pigmentation of the aleurone superim- posed on the fully colored spots resulting from the in- teraction of I-R with (Sc).
Notwithstanding evidence that the somatic and some germinal instability of R-st is attributable to the action of a mobile element, KERMICLE (1970) showed that nearly half of R-SC’S recovered from R-st homozygotes were associated with recombination between genetic markers flanking r. These results suggested unequal recombination between genetic components within the R-st complex as one basis for R-st’s germinal instability. This possibility has been supported by molecular analy- ses showing that the self-color derivative R-sc:124 is com- prised of a single rgene (ALLEMAN and KERMICLE 1993). If deletion by unequal exchange frequently accompan- ies R-st mutation, an analysis of the products of muta- tion could be used to characterize the molecular organi- zation of the complex, as has been done for R-?-:standard
(ROBBINS et al. 1991; WALKER et al. 1995).
In the present study, molecular clones of the r locus were used to probe the organization of the R-st complex and the molecular basis for its germinal instability. TWO sets of derivatives, distal deletions of the locus created by unequal exchange with the displaced r gene leaf color (IC) and internal deletions resulting from unequal ex- changes within the complex that produced R-sc’s, were used to determine the number, order, orientation and interactions between pigmenting components in the complex. These results also were used to identify the mobile element I-R and to determine the patterns and molecular bases for instability in the complex.
MATERIALS AND METHODS
Genetic stocks: All alleles studied were in the W22 inbred
background. Many alleles of r are designated by a two-letter
designation indicating independent expression in seed and plant parts. In this nomenclature, the first letter indicates seed color status and the second indicates plant color status.
For alleles of the R-r class, R- indicates colored seed and -r
indicates colored plant parts. Similarly, R-g indicates colored
seed and green plant parts, r-r indicates colorless seed and
colored plant parts and r-g indicates colorless seed and green
plant parts. In the case ofvariegated alleles like R-st, the letters
after the hyphen indicate the pattern of pigmentation distri- bution within the seed.
Intralocus recombination analysis between R-st and LC: h a . color ( L C ) is a displaced r gene located 2 map units distal to r
on the long arm of chromosome 10 (DOONER and KERMICLE
1976; LUDWIG et aL. 1989). The dominant LC allele used here,
LC, permits anthocyanin expression in leafblade, auricle, peri-
carp, coleoptile, roots and nodes. Recessive LC alleles behave
as the absence of an r gene at this location (DOONER and
KERMICLE 1976). LC was incorporated into the R-st chromo-
some by recombination from the accession R-r:Ecuadorll72
LC. Losses of LC phenotype were selected among the progeny of R-st Lc/R-st LC homozygotes. Testcross populations of ker-
nels needed for screening seedlings were produced in an
isolation plot. Detasseled ear parents were hand pollinated
with a strain carrying a distinguishable allele of r and the
recessive mutation waxy. These markers were verified in puta-
tive recombinant offspring to confirm the intended paternity.
Loss of LC phenotype was identified among seedlings germi- nated in shallow pans under continuous illumination. The scutellar node of Le-containing seedlings pigments intensely when kernels are oriented embryo face up and the tempera-
ture reduced from 27 to 21” upon incipient germination. For
estimating the frequency of LC loss, the number of seedlings
screened is reduced in proportion to the fraction of selections that were not tested successfully.
Evaluation of inhibitor of striate: Alleles of the inhibitor of
striate (isr) locus inhibit expression of certain chlorophyll-
striping mutations such as striate2 in a dosage-dependent
manner. This locus maps just distal to r (KERMICLE and AX
TELIA 1981). R-st is associated with a strongly acting allele (Isr-
s t ) . The isr gene is present at a corresponding location distal to LC (Isr-Le). This allele inhibits albino striping less strongly
than Isr-st. isr alleles were distinguished as described pre-
viously (KERMICLE and AXTELI. 1981).
Evaluation of nearcolorless (Nc) phenotype: The kernel
spotting pattern from which R-st gets its name consists of
intensely pigmented clonal sectors resulting from interaction
between (Sc) and the transposable element I-R. Superim-
posed on this pattern is a faint near-colorless phenotype con- sisting of patches of weakly pigmented cells interspersed with
colorless in a mottled pattern. ASHMAN (1965) associated this
phenotype with a recombinationally separable component in
R-st, designated (Nc). The near-colorless phenotype is incom-
pletely penetrant on a single kernel basis. To evaluate the
(Nc) composition among LC-loss derivatives of R-st LC, kernels
of aleurone genotype R-st/R-st/r-r or R-st/R-st/r-g were exam-
ined under 20X magnification using a dissecting microscope.
Material for this purpose was produced in a winter nursery
located near Homestead, FL, where, for unknown reasons,
the (Nc) phenotype is more strongly expressed than in the summer crop produced at Madison, WI.
Origin of R-sc alleles: The 80 R-sc alleles tested here had been
characterized previously for paramutagenicity (MCWHIRTER and
BRINK 1962). They derive from a set of 83 single kernel R-sc
revertants established by ASHMAN (1960) from R-st/R-st ear par-
ents (frequency of 18.61 X
Nucleic acid purification and analysis: Genomic DNA was isolated from seedlings using the procedure described by
SURE et aL. (1983) as modified by CHEN et al. (1992). DNA from two or more seedlings was tested for each allele and/or
a single sample was tested two or more times. Approximately 5
Mix [final concentrations 0.09% sodium dodecyl sulfate
(SDS), 3.5% sucrose, 8.7 mM EDTA pH 8.0, 0.009% Bromo- phenol blue, 0.009% Xylene cyano11 (T. HSIEH, personal com- munication), fragments were size fractionated by electropho- resis through 1 % agarose slab gels in 1 X TBE (89 mM Tris- borate, 89 mM Boric acid, 2 mM EDTA pH 8.0). The 1-kb ladder (BRL) was used as molecular weight standard in all gels. Separated fragments were photographed under W illu- mination after staining with ethidium bromide and trans- ferred to nylon-reinforced nitrocellulose (Schleicher and Schuell) or Duralon-UV (Stratagene) by the method of SOUTHERN (1975). In some cases, gels were deprotonated by a 10-min wash in 0.25 M HCl before denaturation. DNA was fixed to nitrocellulose by baking for 2 hr under vacuum and to Duralon-UV by UV crosslinking with a Stratalinker (Stra- tagene) at 1200 pJ/cm2 or by crosslinking and baking. DNA fragments used as probes were released by restriction endonu-
clease digestion, separated by electrophoresis through 1%
(w/v) low melting point agarose gels in 1 X TAE (40 mM Tris-
acetate, 1 mM EDTA pH 8.0) and excised from ethidium bromide-stained gels with a scalpel. Approximately 25-50 ng of DNA in agarose fragments was labeled with 32PdCTP ( A m - ersham) using a random primer labeling kit (USB) and the method of FEINBERG and VOGELSTEIN (1983). Membranes were prehybridized at 42" for 1-4 hr in hybridization solution 50% formamide (v/v), 5X SSCP (1 X = 150 mM sodium chlo- ride, 15 mM sodium citrate, 10 mM sodium phosphate), 0.2% SDS, 2 X Denhardt's solution [l X = 0.02% (w/v) Ficoll, poly- vinylpyrrolidone and BSA] and 50 pg/ml sheared herring sperm DNA before addition of heatdenatured labeled DNA fragments. Hybridizations were carried out overnight at 42". Labeled fragments were used at a concentration of
-lo6
cpm/ml of hybridization solution. Filters were posthybridized by a room temperature rinse in 2 X SSCP followed by two 10- min washes in 2 X SSCP, 0.2% SDS at 55" and two 10-min washes in 0.1X SSCP, 0.2% SDS at 55". Membranes were aut@ radiographed for 3-7 days at -70", using a Quanta I11 intensi- fymg screen (Dupont) and XAR-5 X-ray film (Kodak).Probes used in this analysis contained regions from the 5' and 3' ends of the r transcription unit. pR-nj:l, which was cloned from R-nj, contains a region of the rpromoter (DELLA-
PORTA et al. 1988). The clones pSc323:114 and pSc323:J20 were recovered from R-sc:m323 and contain adjacent frag- ments from the 3' end of the r transcription unit (KERMICLE
et al. 1989; ALLEMAN and KERMICLE 1993).
RESULTS
blosses among progeny of R-st
LC
homozygotes: Forty-one instances of the absence of Lcphenotype were identified among seedlings produced from the cross R- st Lc/R-st LC X r-g k/rgIC.
Of these, 34 were recovered among 22,100 seedlings produced from spotted ker- nels; a frequency of 15.38 X These alleles are denoted as R-stAIc's. Seven instances of Le-loss were recovered among the selfcolored revertants present in a total testcross population of 24,530, a frequency of2.85 X
lop4.
These seven are denoted as R-scAlc's. Such Le-losses from r LC homozygotes involving r al- leles other than R-st have been shown to originate via unequal crossing over between homologous sequences at or near r and LC (DOONER and KERMICLE 1976). In the case of R-st, itself a duplication allele, it was antici- pated that more or less of its distal region might be lost depending on the exact position of the crossover. Thatthe 41 Le-loss isolates originated as segmental deletions rather than mutations of LC is shown by coincident change in inhibition of albino striping associated with isr. Because effects of isr genes are cumulative, parental R-st LC, which contains both Isr-st and Isr-le, inhibits striping strongly. All 41 of the Le-loss derivatives inhib- ited striping less strongly than R-st LC, either resembling the effect of R-st (Isr-st) alone or the even weaker LC (Isr-le) alone (Figure 1 ) .
Classification of the Le-loss variants for retention of the R-st components I-R and (Nc), in addition to isr constitution, identified four genetic classes. Table 1
gives frequencies of the four classes and indicates the extent of the genetic region deleted in each. Most con- spicuous is the change in the seven R-scAlc derivatives from spotted to uniform (self-colored) pigmentation. Recombinant R-sc's originate through separation of the Sc promoter region from the transposable element I-R (ASHMAN 1970; KERMICLE 1970; ALLEMAN and KERMICLE
1993). With R-st LC, such crossovers involving R-st and homologous sequences in LC would account for the simultaneous loss of I-R, Isr-st and LC in origination of the R-scale class.
The remaining derivatives retained I-R (R-stAlc classes). One derivative (R-stAkg327) appeared to have lost nearcolorless expression. To confirm the visual de- termination of (Nc) loss in this derivative, Sc was re- placed by the P gene from R-rxtandard through intralo- cus recombination. Upon removal of SC expression from R-st, any kernel pigmentation is attributable to (Nc) (ASHMAN 1965) since P does not pigment seed parts. All such derivatives involving g327were colorless, whereas those from the three other R-stale isolates tested (g301, g309, and g347) had (Nc) expression. De- rivative g327 is designated as a R-stAlc ( I ) class allele. The remaining 33 R-stale isolates were divided into two groups based on isr composition (Figure 1 ) . Twenty- nine retained Isr-lc, designated R-stAlc (11) 's; four re- tained Isr-st, designated R-stAk (111) 's.
Southern blot hybridization to rlocus clones was used to analyze the molecular complexity and structure of four parental R-st LC sublines and the 41 Le-loss deriva- tives. If Le-losses resulted from unequal crossing over between homologous sequences in or near LC and the R-st complex, as inferred from genetic analysis, a reduc- tion in molecular complexity of r-hybridizing fragments was expected.
350 W. B. Eggleston, M. Alleman and J. L. Kermicle
A R-St LC
e R-st IC
8 r-g:e LC
4 0 R - S t A l C
X R - S C A l C
".i
u) b
0 0
e o
0
0
0
X
I I I
I I I I
I
3 0 4 0 5 0 6 0 70 8 0
Leaf width (mm)
FIGURE 1.-Modifymg effect of R-st LC and its derivatives on striate-2 phenotype. R-st LC strongly inhibits albino striping and
narrows the leaves of sr2/sr2 plants due to the presence of Isr-st and Isr-lc. R-st (Zsr-st) alone inhibits striping more weakly and
LC (Isr-LC) alone (i.e., r-g:e LC), still weaker. Outlying entries are classified using striping as the primary variable.
based on available restriction map data, each region/ segment will be referred to as an r gene (see below and DISCUSSION). These results suggest that R-st LC contains a total of five r genes, four associated with R-st and one with LC. For all four categories of LC-loss alleles to have originated by unequal crossing over, the four r genes in R-st should lie in the same orientation as LC. The 5'
to 3' orientation of Sc was shown to be centromere 5'-
3' telomere (ALLEMAN and KERMICLE 1993). Combin- ing the present information concerning r gene number with the available genetic evidence suggests the overall structure of the R-st LC chromosome pictured in Figure 2A. Identification of the various r genes derives from further analysis of the Southern blot evidence as illus- trated below for BstXI digests.
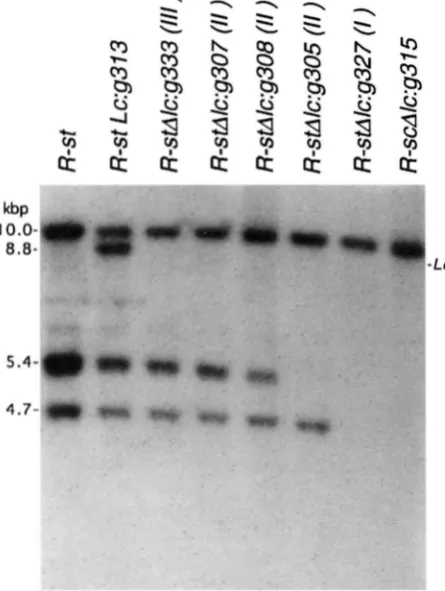
Figures 3 and 4 illustrate the molecular structures encountered among R-st, R-st LC and the LC-loss deriva- tives when digested with BstXI and hybridized with the promoter region probe pR-nj:l and the coding region probe pSc323320 (positions shown in Figure 5). Hy- bridizing bands associated with LC can be inferred from their presence in R-st LC but absence in R-st. Conse- quently, the 8.8-kb fragment present in R-st Lc:g313 but not R-st in Figure 3 and the 3.0-kb fragment in Figure 4 can be identified as released from the 5' and 3' ends of LC, respectively. All 41 LC-loss derivatives lack the 8.8- kb pR-nj:l fragment specific for the 5' end of LC. This finding is consistent with the crossover origin of these derivatives indicated by the isr data and the fact that the 5' ends of r genes confer the tissues-specific expres-
TABLE 1
Phenotypes, number and extent of genetic deletions produced by unequal crossing over between R-st and LC in R-st LC homozygotes
Genetic components retained (+) or lost (-) Isolates
Class (SC) I-R (Nc) Isr-st LC zsr-lc No. Frequency ( X W 4 )
R-SCA IC
~~
+
- NT" - -+
7 2.85R-stA IC ( I )
+
+
+
1 0.45R-stA IC (ZI)
+
+
+
+
29 13.12R-stA IC (IZI)
+
+
+
+
4 1.81- - -
- -
- -
a Not evaluated phenotypically. Absence of (Nc) inferred from loss of flanking components I-R and Zsr-st coincident with the
R-st
2-
NcI--N&“ Nc3;3
LC3
CENTROMERE
-
- . Z : TELOMEREA
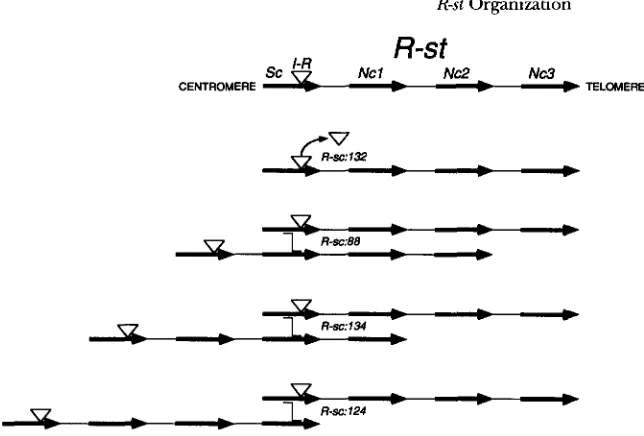
sc FIGURE 2.-Representation of the
v -
-
-
-
, _ - , crossing over (offset lines). The molecu-overall structure of R-st Lr, and origin
of various derivative classes by unequal
v -
-
-
-
-
-
R-&:g346(/g1.1- .
R-s10/&333(//0B
lar analysis for representative I A o s s al-leles indicated are shown in Figures 3
Y7-
-
-
-
-
- .
-
and 4. Arrows represent r-hybGdizing0 -
-
-
-
-
-
-
- .-
, .I
R~sCA/c:g307(j0 - ,c
segments/genes, with 1.c shaded. Thin
lines remesent undetermined distances
v -
between rgenes and proximal and distalD
regions. V represents an insertion at the3’ end of the S C transcription unit corre-
sponding to be the mobile element I-R.
I<-.ytAk (I ) , ( I I ) and (ILI) class alleles are
-
-
- .
-
-
-
cv -
-
-
-
-
-
- .
-
. -1
f k 9 g 3 0 ; ( / 00 -
-
-
-
- .
, .-
-
0 -
-
-
-
-
-
- .
-
. -
1
R~srd/c:g30?(/0E
defined in Table I and in text.sion of alleles studied (LUDWIG et al. 1990). The distal exchange distal to R-stand to
LC:
(Figure 2B). Each allele crossover point (shown as the lower chromosome in also lost either I.sr-.st or Isr-1: (Figure 1). R-stAlcg333Figure 2, B-F) could occur either within L C or distal and three other R-stAlc alleles ( ~ 3 0 9 , g326 and g350)
to it. Both types are represented among the Lc-loss de- designated as class (ZII) retained Zsr-st. The remaining rivatives. allele, R-stAkg346, retained Isr-kc rather than Isr-st, de- Five IdC-lo~s alleles, represented in Figure 4 by R- fining it as an R-StAk (II) allele. Rased on their molecu-
stAlcg333, lack the 3.0-kb fragment specific for the 3’
end of LC. Loss of this fragment is attributed to unequal ”
s s s
? ”z
\z
5.4-
4.7-
h
0 cr)
9
Y
3
(?
a:
0 u) rc)
9
,o
3
(?
a:
I
-LC
kbP
6.4
3.c
c
?
a:
cr)
R
r?,
s
A
9
Q
“ . -
h 0 (3
9
s
A
Q
9
3
(3r?,
Y
A
9
cc
Ir,8
r?,
Y
A
9
cc
LC
FIGURE 4.-Hybridization of pSc323:J20 to RslXI-digested
genomic DNA from R-st, R-st LC and I,c-loss derivatives. See
text and Figure 5 for description and location of probe. The
two high molecular weight fragments present in all alleles
FIGURE 3.-Hybridization of pR-nj:l to BstXIdigested ge- also were present in the r locus deletion allele r-A902 (not
nomic DNA from R-st, R-st L C and LC-loss derivatives. See text shown) indicating that the fragments originate outside the R-
and Figure 5 for description and location of probe. R-stAlc st complex from other myc homologues in the genome. R-
352 W. B. Eggleston, M. Alleman and J. L. Kermicle
V D D X V B
I-R
sc 1 I
3’
3’
3
3’
I I I I I I
B D X X X B
I I / I I I I I I
V DBX D X X X B
I I / I \ I I l l I 1
V DBXB D B B X X B
I I / I \ I I l l I I
V
D B X B D B B X X B
:Iv
Ncl
Nc2
5’
Nc3
5’
LC 5’ I
3’
I I -111 I\ I
D BV D B B B X X
+
pR-nJ:l mX D B
pSc323:’14 I pSc323:JZO
FIGURE 5.-Restriction maps of r genes identified in R-st
LC. Genes are stacked from top (proximal gene) to bottom (distal gene) to highlight monomorphic and polymorphic restriction sites. See text for details. The structure of LC is
based on the LC cDNA (LUDWIG et al. 1989), sequence analysis
of LC genomic DNA (S. LUDWIG, L. HABERA and S. WESSLER,
personal communication) and Southern blot restriction map
analysis of genomic DNA as described in the text. Boxed area, Ftranscribed region; 0, transcribed 5’ leader and 3’ tail se- quences; B, protein coding regions. The insertion denoted
as I-R is a 3.3-kb mobile element-like insertion in exon 7 of Sc
(W. EGGLESTON and J. KERMICLE, unpublished observations).
Locations of probes used in Southern blot hybridizations are shown at the bottom. Restriction sites are as follows: B, BamHI; D, HindIII; R, EcoRI; S, SucI; V, EcoRV, X, BstXI. Sites based
on Southern blot analysis of genomic DNA are 50.1 kb.
lar genetic compositions, the above alleles result from unequal exchanges distal to LC and to r genes in R-st and either distal to or proximal to the imloci flanking R-st and LC.
The remaining types of LC-loss derivatives involve re- combination within R-st and LC rather than distal to the two loci. The R-stAk (Il) alleles g307, g308 and g305 lack the 8.8-kb Lc-specific fragment in Figure 3 but re- tain the 3.0-kb LC-specific fragment in Figure
4.
Since no novel size fragments are observed, all alleles that lost the 5’ but retained the 3’ end Lc-specific fragments are inferred to have resulted from an exchange between a site in the LC gene and a site within an r gene in the R-st complex. R-stAkg307 also retained all pR-nj:l fragments associated with R-st, suggesting an exchange between the 3’ end of LC and the 3‘ end of the most distal r gene in R-st, which is arbitrarily defined as Nc3 (Figure 2C). Such hybrid r genes are designated by a capitalized 5’ end separated from a lowercase 3’ end by double vertical lines. For example, the designation of the hybrid gene in g307 is NcjlZc. R-stAk:g308 likewise retains all R-stspecific pR-nj:l fragments but has a re- duction in the intensity of hybridization to the 5.4kb fragment (Figure 3). This reduction, rather than com- plete loss, indicates that more than one pR-nj:l-hy- bridizing 5.4kb BstXI fragment is released from R-st, with loss of hybridization intensity in R-stAkg308 being due to exchange betweenLC
and the second most distalr gene in R-st, which is defined as Nc2 (Figure 2D). R- stAZc:g305 has lost all homology to the 5.4kb fragment
as expected from exchange between LC and the third most distal r gene in R-st, defined as Ncl (Figure 2E). Therefore, indistinguishable 5.4kb fragments with ho- mology to pR-nj:l are released from Nc3 and Nc2, the distal and second most distal r genes in R-st.
The sole R-stAk
(l)
allele, R-stAkg327, and the R-scAlc allele class represented by R-scAZc:g315 have lost all but one fragment in each hybridization: the 10-kb R-st-specific pR-nj:l-hybridizing fragment (Figure 3) and the 3.0-kb LC-specific pSc323:J20-hybridizing frag- ment (Figure
4).
These alleles result from exchanges between L C and the most proximal r gene in the R-stcomplex, Sc (Figure 2F). Consequently, the 4.7-and 10- kb fragments with homology to pR-nj:l (Figure 3) are assigned as being released respectively from the third most distal (Ncl) and the fourth most distal (i.e., Sc) r
genes in R-st.
Since R-scAkg315 has fullcolor expression in the seed, it should have resulted from an exchange between LC and the region of Sc proximal to I-R Because R- stAkg327 retains the spotting associated with I-R, it
should have resulted from an exchange distal to I-R
All R-scAkderivatives had lost and all R-stAZcderivatives had retained a 3.3kb insertion present in the 3’ end
of Sc but not in any other r gene in R-st. Because this insertion is located at the 5’ end of the pSc323g20 region (Figure 5) and flanked closely by BstXI sites, its
presence/absence is not revealed in Figures 3 and 4.
By
comparison to the restriction map of genomic DNA and sequence analysis of cDNA from LC (LUDWIG et al.1989; S. LUDWIG, L. HABERA and S. WESSLER, personal communication), these results indicate that I-R is a 3 . 3 kb insert at the 5‘ end of SC’S seventh exon. Further support for this conclusion is presented below. The Sc
plus I-R combination will be designated as Sc::I-R to
denote insertion of the mobile element I-R into Sc.
Restriction maps of the five r genes in R-st L C are
shown in Figure 5. Enzymes were mapped singly and in combination with HindIII, which recognizes a centrally located site in all five genes. The maps were constructed based on differences between R-st, R-st LC and LC-loss
derivatives deduced to have retained one, two, three or
R-st Organization
TABLE
2Component structures of the progenitor R-st LC stock and 41 Loloss derivatives
Class"/allele Molecular genetic structureh
R-.Tt L C
g312, g313, g336, g35I Sc::I-R Ncl Nc2 Nc3 Isr-st LC Isr-lc
g309 Sc::I-R Ncl NcAlnc3 Isr-st or Sc::I-R Nclllnc2 Nc3 Isr-st
g326, g333, g350 SC::I-R NcI N c ~ N c ~ Isr-St
g346 &::I-R Ncl Nc2 Nc3 Isr-k
g303, g307, g310, g311, g319, g323, Sc::I-R NcI N c ~ N c j l k I . - k g324, g329, g331, g334, g348
g302, g304, g306, g308, g314, g318, SC::I-R NcI NcAllc Isr-1~ g320, g321, g325, g328, g332, g335, g349
g301, g305, g330, g347 SC::I-R N ~ l l l k Isr-1~
(132 7 Sc::I-Rllk Isr-1~
g315, g316, g317, g338, g339, g353, g355 Sc(llc Isr-lc
" Classes defined in Table 1 and in text. R-stA k ( I I I )
R-SlAk ( I I )
R-stAk ( I )
R-SCALC
By convention, the insertion of I-R into Sc is denoted by a double colon Sc::I-R Hybrid r genes detected by Southern blot analyses are denoted by capitalized 5' region, double vertical lines, lowercase 3' region. See text for details.
The molecular genetic structures of the four R-st LC
alleles and the 41 LC-loss derivatives tested are summa- rized in Table 2. The 41 LC-loss derivatives are of the seven types shown in Figure 2, explicable, with one exception, as arising by single exchanges between LC
and different positions in or distal to the R-st complex. For R-stAkg309 it is necessary to propose more than a single exchange. Molecular analysis of this allele was unable to distinguish between the two possibilities listed in Table 2. For recombination events between sites within r genes, exchanges evidently occurred at corre- sponding regions, i e . , although unequal, each involved homologous sequences.
The overall pattern of exchange is summarized in Figure 6. Analyses of the gains and losses of isr alleles, restriction site polymorphisms and I-R were used to de- termine which r genes and which region of the genes participated in recombination. The large majority of
exchanges, 36 out of 41, involved sites within the F
hybridizing regions of R-st and LC. The remaining five exchanges occurred at more distal sites. LC recombined with each r gene in R-st between four and 13 times at frequencies ranging from 1.81 to 5.88 X A chi- square analysis detected no significant differences in the frequency of exchange between L C and each of the
four r gene in R-st (d.f. = 3,
P
= 0.16). However, a significant dichotomy was observed for the location of exchange within the r genes. All 36 recombination events between sites in r genes occurred in the 3' por- tions of the transcription units, 3' to the centrally lo-cated BumHI site in LC (Figure 5 ) .
Penetrance of the nearcolorless phenotype among R-stAk isolates: Table 3 reports penetrance data for
the nearcolorless phenotype among 34 R-stAk isolates. The sole R-stAlc (I) allele, g327, produced the lowest score, 0.30%. Because g327 was shown by recombina-
'I...
..I.I..
II
I D
'I.
I.. L I-lLJW. IVA N B V
.
..
I-+
, I , I V 111 W
- 1
I.. I 111 !I
r 1
IA IB
lAIBB x
X 0 B B B B BBE XB? BB XB B X X B B B B X B
\ " L \ V < \
T
"Ncl
Nc2
-
Nc3
"
r
r
2kbP
1sr:st 1sr:lc
I-R
354 W. B. Eggleston, M. Alleman and J. L. Kermicle
TABLE 3
Range and average frequencies of R-stAIC/R-stAIC/r IC
kernels showing nearcolorless phenotype
Kernels of nearcolorless
phenotype (%)
r genes No. of
in R-stAk isolates" Rangeb Average
1 1
-
0.302 4 1.9-25.2 9.42 3 14 2.0-13.7 7.77
4 15 1.1-8.8 4.57
An average of 15.5 ears were examined per R-stAlc isolate. Ranges are for average values of isolates within the group.
tion and molecular analyses to lack nearcolorless genes, the value of 0.30% is attributable to misclassifi- cation of finely spotted regions for nearcolorless and provides a baseline for comparison with the remaining isolates. Penetrance of the nearcolorless phenotype in the remaining 33 R-stAlc isolates varied from ear to ear, from test year to test year, and between isolates having the same number of r genes. The ranges and averages nevertheless show a regular trend.
As
r gene number decreased, penetrance of the nearcolorless phenotype increased, Le., penetrance was related inversely to gene number in the order 4<
3<
2.R-sc revertants among progeny of R-st homozy-
gotes: To determine the basis for reversions of R-st to
R-sc, the molecular structures of R-sc derivatives from homozygous R-st females (ASHMAN 1960) were deter- mined as described above for the Lc-loss derivatives of
R-st LC. DNA prepared from 80 R-sc alleles and from R-st
were digested with EcoRVplus BumHI, Southern blotted and hybridized with pR-nj:l, pSc323:114 and pSc323J20
as above. A subset of alleles also was characterized after HindIII, Ssp1 and XbuI digestion as above.
Figure 7 shows a Southern blot of R-st and representa- tives of the four structural classes present among the
R-sc alleles when digested with EcoRV plus BumHI and hybridized with pR-nj:l. The only change in alleles of the R-sc:132 type is replacement of the 5.9-kb fragment by an 8.2-kb one. Reversion from R-st to R-sc resulted in this case from loss of I-R from Sc::I-R without change in other r genes. Consequently, the 5.9-kb fragment is interpreted as being released from Sc::Z-R and the 8.2- kb fragment from Sc in the absence of I-R. In the three remaining revertant allele types, not only is the 5.9-kb
Sc::I-R fragment replaced by the 8.2-kb fragment, there also is a reduction in the number/intensity of r-hybridiz- ing fragments relative to R-st. Coordinate loss of Z-R and reduced complexity of hybridization to r locus clones is explained by unequal exchange between Sc::I-R and the more distal r genes in the complex.
As
shown in Figure 5, the 3.7-kb fragment is released from the 5' end of Sc, the 9.1-kb fragment from Ncl and the 4.1-kb fragment from Nc2 and Nc3. That all R-sc derivatives2
0, d
c
kbP 9.1
-
8.2-5.9-
4.9-
4.1
-
3.7-
FIGURE 7.-Hybridization of pR-nj:l to BamHI plus EcoRV- digested DNA from R-st and four representative R-sc deriva- tives. The weakly hybridizing large (>9.1 kb) fragment in R- st and the R-sc alleles is interpreted as arising from partial
methylation of the BamHI sites in r promoter regions.
with reduced complexity have lost the 9.1-kb fragment released from Ncl is consistent with their origin by un- equal exchange. Alleles resulting from exchange be- tween Sc::I-Rand Ncl, such as R - s c : ~ ~ , lose only the 9.1- kb fragment. In alleles of the R-sc:l34 type, the 4.1- and 4.9-kb fragments are reduced in intensity relative to R- st, whereas in alleles of the R-.w:124 type both fragments are absent. Based on these data, both Nc2 and Nc3 re- lease 4.1- and 4.9-kb fragments. R-sc:l34 results from exchange between Sc::I-R and Nc2 and R-sc:124 from exchange between Sc::I-R and Nc3. The basis for two fragments being released from both Nc2 and Nc3 likely is partial methylation of a BumHI site(s) in the pro- moter region of these two genes (Figure 5). Digestions with methylation insensitive restriction enzymes did not release two bands from these genes. The additional weakly hybridizing large fragment is of unknown origin but likely arises from partial methylation of BumHI or EcoRV sites in Sc. The locations of exchanges associated with reversion of R-st to R-sc are diagrammed for the representative alleles in Figure 8, and the structures of all 80 R-sc alleles tested are listed in Table 4.
As
found among the Lc-loss derivatives, the number of Nc2/Nc3R-St
sc Ncl Nc2 NC3
CENTROMERE
-
-
7e
TELOMEREFIGURE 8,"Diagrammatic representa-
tion of R-st's structure and origin of the four R-sc allele classes shown in Figure 7.
Designations as in Figure 5 .
relative intensity were noted, indicating that two copies of Nc2/Nc3like genes are present in R-st.
All R-sc derivatives of R-st were accompanied by loss of the same 3.3kb insertion that was lost in the seven R-SCA IC alleles described above. No Le-loss or R-sc deriva- tive was found to have this insert integrated at a new site in an rgene. Cloning and sequence analysis showed this insert to have structural features of known mobile elements and to be present in varying numbers in more than a dozen maize inbreds and accessions tested (W.
ECCLESTON and J. KERMICLE, unpublished observa- tions). These results confirm that this insertion is the genetically characterized mobile element I-R However, only
22
out of 80, or about one-fourth of all reversions of R-st to R-sc resulted from loss of the I-R from the complex without a coordinate loss in molecular com- plexity, a frequency of 5.12 X (22/80 X 18.61 XDue to the duplicate nature of the R-st complex, it has not been possible to determine whether such losses resulted from I-R excision, gene conversion be- tween r genes or a combination of both mechanisms. However, these results do show that I-R excision occurs infrequently if at all and, at most, can account for only about one-fourth of single kernel revertants.
The remaining 58 R-sc derivatives were associated with loss of molecular complexity and therefore re- sulted from unequal recombination between &::I-R and one of the three Ne genes. As shown in Figure 8, rever- sions necessitate the loss of I-R and retention of the Sc promoter region. Thus, at a minimum, identifiable exchanges between the r genes were constrained to oc- cur within the -5.8-kb region between the presumptive transcription start site in Sc and the insertion site of I-R in exon
7
of Sc. The distribution of unequal exchangesTABLE 4
Component structures of 80 R-sc alleles derived from R-st homozygotes
Class/allele" Molecular structureb
Strongly paramutagenic
57, 60, 64, 66, 67, 68, 72, 74, 77, 78, 82, 100, 104, 108, 109, 112, 121, 126,
56, 70, 76, 79, 88, 91, 95, 102, 105, 111, 128, 131
107
62, 69, 75, 81, 84, 86, 89, 103, 117 53, 80, 129, 130, 134
Very weakly paramutagenic 115
54, 55, 59, 63, 71, 73", 90, 92, 96, 99, 110, 114, 118, 119, 120, 127
58, 61, 65, 83, 85, 93, 94, 101, 106, 113, 122, 123, 125, 124, 135 133, 136
Weakly paramutagenic
Nonparamutagenic
132, Sc Ncl Nc2 Nc3
Scllncl Nc2 Nc3
Sc Ncl Nc2 Nc3 Scllncl Nc2 Nc3
Sc
11
nc2 Nc3 Scllncl Nc2 Nc3Scl
I
nc2 Nc3a Classes are from MCWHIRTER (1961) and are based on relative paramutagenic strengths.
Hybrid genes denoted as in Table 2.
356 W. B. Eggleston, M. Alleman and J. L. Kermicle
m
.:::::
sc 4B Ncl Nc2 Nc3
I- R 2 kbp
-
FIGURE 9.-Distribution of unequal exchanges among R-sc
derivatives of R-st homozygotes. Brackets indicate localization
of exchange positions based on loss of I-Rand restriction site
polymorphisms in Y genes. Restriction sites as in Figure 5. L C
transcription patterns are superimposed on all Y genes for
reference as in Figure 6.
among the R-sc alleles is shown in Figure 9. The region between the promoter and I-Rinsertion site in Screcom- bined with each of the NC'S from 15 to 22 times, at frequencies between 3.49 and 5.12 X A chi-square analysis detected no significant differences in the fre- quency of exchanges between Sc and the three Nc genes (d.f. =
2,
P = 0.48).As observed among the LC-loss derivatives, a strong tendency for exchanges to occur at the 3' ends of r
genes was evident. Where they could be localized, nearly all exchanges (34 out of 36) occurred in a 1.5- kb region bounded at the 5' end by the BumHI site just proximal to exon 3 in Nc2 and Nc3 and at the 3' end by the I-R element in &::I-R (Figures
5
and 9). This region is encompassed within the portion of the rgenes in which all exchanges occurred among the LC-loss de- rivatives. Exchanges between &::I-R and Ncl are de- limited to a broader region due to a lack of correspond- ing restriction site polymorphisms.Methylation of cytosine residues and r genes in R-st
LC
and R-st: To test whether and where cytosine residues inr genes in R-st were methylated, R-st and R-st LC DNA were double digested with HindIII, which is insensitive to cytosine methylation, plus either BstNI or EcoRII. The latter two enzymes recognize the sequence CCA/TGG. BstNI is insensitive to methylation of either C and EcoRII is sensitive to methylation of the second C. Blots were hybridized sequentially with pR-nj:l, pSc323:114 and pSc323320. Results are shown in Figure 10.
The right panel shows that when R-st and R-st LC are digested with HindIII plus BstNI or EcoRII and probed with pSc323g20, an -0.95-kb fragment is released from both alleles. This size fragment evidently is released from all r genes in R-st and R-st LC. The intensities of the fragment are identical in BstNI and EcoRII digests of each allele and no additional higher molecular weight fragments are observed in the EcoRII digests. Similar results were observed in hybridizations with pSc323:114 (not shown). These data indicate that methylation of EcoRII sites in the 1.9-kb segment of the coding region spanned by pSc323:114 and pSc323g20 is at low levels in all r genes in R-st and R-st LC.
S c > Sc::l-R
Ncl LC a Ne2 Nc3
Nc 1 Nc2 > Ne3
SC ScA- R
Ncl
,
Ne2 Nc3
LC
b P
7.1
6.1
5.1
4.1
3.0
2.0
1.6
,
1.0pR-nj: 1
pSc323:JZO
FIGURE 10.-Sequential hybridization of pR-nj:l and
pSc323j20 to DNA from R-st and R-st LC. All samples were
digested with HindIII. Some DNA's were additionally digested
with EcoRII (E) or RstNI (N). Assignment of fragment origins
is based on direct comparisons between R-st and R-st L C and
parallel analyses of R-sc and LC-loss alleles with one to four Y
genes. The faint additional higher molecular weight species
observed in Hind111 plus EcoRII digests in the right panel also
were present in the Y deletion stock r-A902, indicating that this fragment arises outside Y.
In the left panel, HindIII plus BstNI digests release fragments of 2.0, 1.6 and 0.7 kb with homology to pR- nj:1 from LC, Sc and the Nc genes, respectively. These fragments are absent in digestions of HindIII plus E m
RII, being replaced by higher molecular weight smears or by fragments equal or nearly equal in size to those released from L C and the Nc genes in digests with Hin-
dIII alone. These results indicate that the EcoRII sites in the HindIII fragments spanning the transcription start sites and well into the large second intron of LC
and the Nc genes (Figure 5) are heavily methylated. In contrast, the 1.6-kb fragment released from Sc by Hin-
dIII plus BstNI digests of both alleles is not replaced by the much larger fragment released from Sc by HindIII digestion alone. It currently is not known whether this fragment is replaced by the smear of fragment.. of slightly higher molecular weight or by the fragmentjust below the HindIII fragment released from the Nc genes in EcoRII digests or by both. Regardless, the results indi- cate that cytosine residues in the promoter region of
Organization
the Nc genes. Since a single filter was hybridized with all three probes, the results using pR-nj:l cannot be accounted for by incomplete EcoRII digestion.
DISCUSSION
Organization of pigmenting components in R-stip
pled
Molecular characterization of the deletion prod- ucts resulting from unequal recombination identified four r-hybridizing regions in R-st. Based on hybridiza- tion patterns and a comparison of the restriction maps of these regions with the restriction map of LC, each contains a minimum of one r transcription unit/gene oriented in the same direction as LC. Because the 5’ end of Sc is centromeric to its 3’ end (ALLEMAN andKERMICLE 1993), all four r genes identified in R-st also lie in this orientation. The possibility of additional r
sequences present in reverse orientation is not ex- cluded. Thus, one or more r-hybridizing regions/seg- ments could be comprised of tandem-inverted repeats of two identical r genes, as has been found for the S l - S2 subcomplex of R-rxtandard (WALKER et al. 1995).
Intensely pigmented R-st spots are encoded by Sc,
which is repressed by a 3.3-kb insertion in exon
7
of the identified r gene. The correlation between absence of this insert and reversion to full color identifies this insertion as the mobile element I-R Nearcolorless mot- tling is associated with three r genes arbitrarily desig- nated Ncl, Nc2 and Nc3 proceeding distally from Sc.These observations are consistent with previous reports on the genetic complexity of R-st and the basis for its somatic and germinal instability (ASHMAN 1960, 1965,
1970; KERMICLE 1970; SATYANARAYANA 1970; GAVAZZI and CALATI 1972; WILLIAMS et al. 1984).
Since unequal recombination products between Sc
and each of the Nc genes are fully colored, the 3’ end of the r gene identified in each Nc must be functional. Recombinants that contain only Nc3 express the near- colorless phenotype (KERMICLE et al. 1995), indicating
that it is a functional r gene. It has not been possible to isolate intact an Ncl or Nc2 separate from other Nc’s.
Hybrid genes containing the 5‘ end of Ncl express near- colorless, e.g, the Ncllllcgene in R-stAlc:g305. That both its ends are functional in such hybrid genes suggests that Ncl also is a functional r gene. None of the deriva- tives separately tests the 5’ end of Nc2. These results indicate that at least two and perhaps all three Nc genes contribute to the nearcolorless phenotype.
Molecular bases and patterns of R-slippled germinal instability: The evidence presented here indicates that meiotic instability of R-st results from a combination of at least two mechanisms. The primary mechanism is out-of-register synapsis leading to unequal recombina- tion between the r genes in the same orientation as
sc
and LC. All 41 LC-losses and 58 of 80 R-sc revertants occurred in conjunction with a loss in molecular corn-
plexity in R-st. With the exception of R-stAkg309, all
99 derivatives associated with reduction in complexity could be explained by a single recombination event. Even for this allele, the possibility that one of the events, a change from four to three r genes, occurred in the preceding generation could not be excluded.
Since recombination between genetic markers flank- ing r was not monitored among the derivatives charac- terized molecularly, it was not possible to determine from these populations the proportions of inter- VS.
intrachromosomal unequal exchange events. In a previ- ous study, KERMICLE (1970) found that 45% of R-SC’S
recovered from R-st homozygotes as female parent were associated with recombination between genetic markers flanking r. Considering the possibility of coincidental reversion and crossing over within the marked region, the percentage attributable to interchromosomal cross- ing over was estimated to be one-third. In the current report, 72.5% (58/80) of R-SC’S were associated with reduction in molecular complexity. Combining the two results indicates that inter- and intrachromosomal ex- changes account for -33 and 39.5% (72.5-33%) of R- sc’s, respectively. This proportion appears to differ from unequal crossovers involving R-r:standard, where most were interchromosomal (DOONER and KERMICLE 1971).
The frequency of unequal exchanges between com- parable sites in rgenes in the two experiments reported here do not differ appreciably. Among the unequal recombinationderived R-sc’s, the average frequency of unequal recombination per Nc gene was 4.49
x
loc4
((58/80 X 18.61 X 10-4)/3). These exchanges neces- sarily involve the promoter to I-R interval in Sc. The same is true for the R-scAlc derivatives from R-st LC. In this instance, the frequency (2.85 X was only slightly lower. This indicates that the larger physical distance separating LC from the r genes in R-st did not significantly reduce its ability to recombine with them, relative to the frequency of exchanges between r genes within the complex.
Among neither set of derivatives was there strong evidence for preferential exchanges between specific r
genes. There were no significant differences between the frequencies that Sc recombined with each Nc or that
LC recombined with each r gene in R-st. These results also appear to rule against physical distance as having a large impact on recombination frequencies.
358 W. B. Eggleston, M. Alleman and J. L. Kermicle
reciprocal exchanges occurring preferentially in pro- moter regions, have been observed in yeast (NICOLAS et al. 1989; STAPLETON and PETES 1991; R. MALONE, personal communication cited in STAPLETON and PETES 1991; Wu and LICHTEN 1994). This polarity has been related to more open or accessible chromatin structure
(Wu and LICHTEN 1994).
What is the basis for the concentration of unequal exchange sites in r? It appears not to arise from the I- R element in Sc inducing recombination since the same polarity was observed in exchanges between LC and the Nc genes, which lack I - R Alternatively, the polarity could be associated with differential methylation levels in the 5' and 3' end of the r genes. High levels of methylation were detected in the 5' ends of the rgenes in R-st, including LC when present. Conversely, little or no methylation was detected in the region encom- passing exons 3-9, where nearly all exchanges oc- curred. Thus, the pattern of unequal exchange is in- versely correlated with cytosine methylation levels, perhaps indicating that some aspect of cytosine methyl- ation or the chromatin structure of methylated regions, interferes with unequal recombination. Extensive tests have detected little cytosine methylation at b (PAT- TERSON et al. 1993).
The possibility that cytosine methylation interferes with unequal exchange at r also is suggested by results from the R-rstandard complex. Molecular analyses of unequal exchanges found that most occur at the 3' ends of the rgenes (ROBBINS et al. 1991) as found here for R-st. M. ALLEMAN and J. KERMICLE (unpublished observations) have found that cytosine methylation lev- els are high in promoter regions and low in the region of exons 3-9 and that paramutation results in an in- crease in promoter but not coding region methylation in the r genes in R-rxtandard. In an early study of r allele instability, BRAY and BRINK (1966) found the frequency of meiotic instability within R-r:standard and its deriva- tives to be reduced following paramutation. Such re- gional effects of methylation are consistent with results from mammals in which V(D)J recombination is sig- nificantly reduced by chromatin structure associated with methylated regions but not by cytosine methylation
per
se (HSIEH and LIEBER 1992).Among LC-loss derivatives, the detection of unequal exchange was not confined to sites within r genes as defined by the transcription units, as it was for R-sc derivatives. LC is known to be a part of a larger duplica- tion that extends distally to r and encompasses the nearby isr locus (DOONER and KERMICLE 1976). The physical size of this duplication has not been deter- mined. Here, unequal exchanges distal to the r genes were recovered as LC-losses that did not produce hybrid r genes containing the 3' end of LC (g346 and the four R-stAlc (III) alleles). These results confirm the presence of homologous sequences distal to R-st and to LC that can participate in unequal exchanges. These recombi-
nants were recovered at a frequency, 2.26 X within the range observed for exchanges between specific r genes. Whether these exchanges occurred in or be- tween genes distal to r and to LC could not be deter- mined. In all, nearly 90% (36 out of 41) of exchanges occurred between sites within r genes. This result is consistent with previous observations that meiotic re- combination in maize occurs much more frequently in genes than in the genome as a whole (DOONER et al. 1985; DOONER 1986; CIVARDI et al. 1994).
The nearly one-fourth of R-sc derivatives not associ- ated with recombination could have arisen by excision of I-R from Sc or by gene conversion events that re- moved I-R. Thus, the maximal rate for I-R excision or for gene conversion events that remove I-R is 5.12 X (22/80 X 18.61 X The two possibilities may not be independent. Breaks made by the I-Rtransposase in or adjacent to the I-R element could increase the frequency of gene conversion events in that region, as has been observed for the P transposable element in Drosophila (ENGELS et al. 1990; GLOOR et al. 1991). To date, it has not possible to estimate separately the rate of I-R excision from R-st and the frequency of gene conversion events that remove I - R
Origins
and
evolution of complex r alleles: Numer- ous alleles of the r locus behave as if comprised of multiple r cistrons with a variety of expression patterns and abilities to recombine (STADLER 1948a,b; ASHMAN 1965). The molecular organization of two now has been determined. The organization and tissue-specific ex- pression of R-st differs significantly from that of R- r:standard. R-rxtandard is comprised of one complete and three truncated r genes (ROBBINS et al. 1991; WALKER et al. 1995). The P gene regulates expression in plant parts. In the same orientation, but 190 kb distal, liesq,
a P-homologous 5' sequence lacking an r coding region. Twenty kilobase pairs more distal to q is an inverted duplication of two partially truncated but func- tional r genes, designated S1 and S2, regulated by a single promoter region located between the two tran- scription units. The S genes regulate expression in the aleurone. WALKER et al. (1995) have postulated that R- r:standard arose from duplication of an r gene express- ing plant color. This event was followed by one or more additional rearrangements induced by a mobile ele- ment inserted into the more distal copy, producing the truncatedq
gene and the inverted S genes. The initial duplication should have encompassed the progenitor r gene as well as the nearby isr locus to account for the presence of isr loci between the P and q genes as wellas distal to S1 and S2.