Search for the decay D0→π+π−μ+μ−
Full text
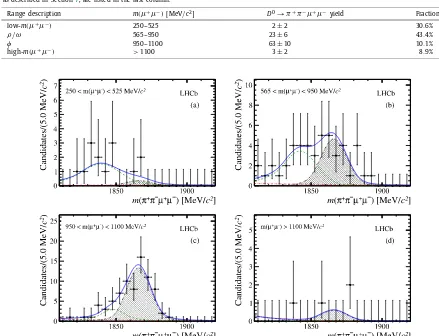
Figure



Related documents
This demographic and clinical pattern of presentations of patients in HARVARD PEPFAR (Presidential Emergency Plan For AIDS Relief), supported ARV clinic at UMTH
RAPD analysis for genetic variations among four mutants of black gram (high seed protein, tall, bushy, and dwarf mutants) induced by physical gamma rays and chemical
This paper is based on a descriptive survey of the relationship between policy framework governing ECDE and actual practices in Early Childhood Development Centres (ECDCs)
Vol. In the germinative zone, the oogonia having prominent nuclei and clear cytoplasm, divide mitotically and are arranged around a central anucleate tube like rachis which
A combination of multiple input multiple output (MIMO) and orthogonal frequency division multiplexing (OFDM) (MIMO- OFDM) is an emerging technology for high speed
A comparison with the perform- ance (ISI) of unprimed burnout patients (M = 1716, SD = 888) and healthy controls (M = 1089, SD = 351) from an earlier study (Van Dam et al., 2011)
(2006) Metal based antibacterial and antifungal agents: Synthesis, characterization and in-vitro biological evalua- tion of Co(II), Cu(II), Ni(II), and Zn(II) complexes with
M: protein marker; Lane 1: the purified enzyme via Ni-affinity chromatography; Lane 2: the crude extract from the bacterial cells transformed with the recombinant pET23a plasmid;
