Distribution and drug resistance of pathogens isolated from 4142 blood
cultures
Xing-Wang Ning, Si-Yu Jiang, Hui-Bing Zhu, Xiao-Bing Xie, Yu Cheng
The Medical Laboratory Center, The First Hospital of Hunan University of Chinese Medicine, Changsha, Hunan, 410007, China
Journal of Hainan Medical University
http://www.hnykdxxb.com
ARTICLE INFO ABSTRACT Article history:
Received 3 Feb 2018
Received in revised form 18 Feb 2018 Accepted 26 Feb 2018
Available online 14 Mar 2018
Keywords:
Blood culture Distribution Drug resistant Antibacterial
Corresponding author: The Medical Laboratory Center, The First Hospital of
Hunan University of Chinese Medicine, Changsha, Hunan, 410007, China. E-mail: chengyu2005166@163.com
Fund Project: Scientific Research Project of Hunan Provincial Education Department (17B195).
1. Introduction
With the widespread use of broad-spectrum antibiotics and immunosuppressants, as well as various invasive diagnosis and treatment technologies, such as interventional therapy, venous catheterization and so on; the prevalence of microbial bloodstream infection is increasing, which seriously threatens the safety of
patients.
Blood culture is an important method to diagnose blood infection and monitor the condition of critically ill patients. It is also an important evidence to diagnose and treat bacteremia and septicemia. Therefore, analysising the distribution and drug sensitivity of pathogens isolated from blood culture is particularly important. In this paper, we analyses the distribution and the antibiotics susceptibility of the main pathogens isolated from positive blood culture specimens, from January 2015 to December 2016, which provides an important basis for the clinical to use antibiotics correctly. And we expect clinicians could pay more attention to blood culture.
Objective:To investigate the distribution and drug resistance of pathogens isolated from 4 142 blood cultures, and provide a scientific foundation for guiding clinical rational use of antimicrobial agents for bloodstream infections. Methods: Blood cultures obtained from 4 142 inpatients and outpatients who were hospitalized from Jan 2015 to Dec 2016. The culture was detected by automatic BACT/ALERT 3D blood culture system of biomerieux. Bacteria isolated from positive blood cultures were further identified, and the drug susceptibility tests were conducted by VITEK-2 Compact automatic microbial analysis system and ATB Expression microbial analysis system. The drug susceptibility results were evaluated according to CLSI 2014 standard. Statistical analysis was performed by using WHONET 5.6 software. Results:A total of 396 unique strains were isolated from 4 142 blood cultures, and the positive rate is 9.6%. Among the positive blood cultures, 194 (49.0%) strains were identified as Gram-positive, 185 (46.7%) strains were identified as Gram-negative, and 17(4.3%) strains were identified as fungi. The coagulase-negative Staphylococcus were the most frequently detectable (29.2%), followed by Escherichia coli (18.4%), Klebsiella pneumonia (7.3%), Staphylococcus aureus (5.1%), Acinetobacter baumannii(5.1%), and Enterococcus genera (5.1%). The incidences of methicillin resistant coagulase negative staphylococcus (MRCNS) and methicillin resistant staphylococcus aureus (MRSA) were 81.9% and 50.0% respectively. However, vancomycin resistant staphylococcus and enterococcus were not detected. The prevalence of extended spectrum β lactamases (ESBLs) in E. coli and K. pneumoniae were 56.2% and 37.9%, respectively. All the E. coli strains were sensitive to amikacin, piperacillin/ tazobactam, imipenem and meropenem, and the sensitive rate of K. pneumoniae strains to
imipenem and meropenem were 93.1% and 89.7%, respectively. Conclusions MRCNS
2. Data and methods
2.1 Specimen source
A total of 4,142 blood samples from outpatients and inpatients were collected, from January 2015 to December 2016, in the First Hospital of Hunan University of Chinese Medicine. Duplicate samples from the same region of the same patient were removed, and 396 strains pathogenic bacteria were isolated.
2.2 Instruments and reagents
The blood culture was detected by automatic BACT/ALERT 3D blood culture system and its matching culture bottles from BioMerieux. Bacteria identification and drug susceptibility tests were conducted by VITEK-2 Compact automatic microbial analysis system, ATB Expression microbial analysis system and their matching bacteria identification and drug susceptibility card. M-H plate, blood plate and chocolate plate were purchased from the BioMerieux. Antimicrobial susceptive disk purchased
from Oxid company. The control strains--Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Enterobacter cloacae (ATCC 700323), Staphylococcus aureus (ATCC 25923), Enterococcus faecalis (ATCC 29212), Yeast (ATCC 34449) were purchased from the National Center of Clinical Laboratory.
2.3 Method
(1) Sterile collect 3-5 mL newborn or children's blood into children's blood culture bottles; sterile collect 8-10 mL adult blood into the adult aerobic and anaerobic blood culture bottles. After mixing, it should be immediately sent to examine.
(2) Put the blood culture bottles into automatic BACT/ALERT 3D blood culture system. As soon as it is warned--in other words, positive reaction were detected in some blood culture bottles, we take the blood culture bottles out immediately, smear, make gram stain, and use microscope to examine them. At the same time, inoculate them onto Columbia blood agar plate and chocolate plate. Incubate at 35 ℃, 5% CO2 incubator for 18-24 h to observe the growth of colony. Then, according to the requirements of systems,
Table 2.
Sensibility of the main staphylococcus isolated from blood culture to common antibiotics (%).
Antibacterial drugs Coagulase negative staphylococcus aureus Staphylococcus aureus
Sensitivity rate Resistance rate Sensitivity rate Resistance rate
Penicillin 3.4 96.6 0.0 100.0
Phenazole 18.1 81.9 50.0 50.0
Erythromycin 19.8 80.2 25.0 75.0
Clindamycin 49.2 50.8 65.0 35.0
Gentamicin 70.0 30.0 70.0 30.0
Compound Sulfamethoxazole 46.6 53.4 80.0 20.0
Rifampicin 84.5 15.5 90.0 10.0
Vancomycin 100.0 0.0 100.0 0.0
Linezolid 100.0 0.0 100.0 0.0
Table 1.
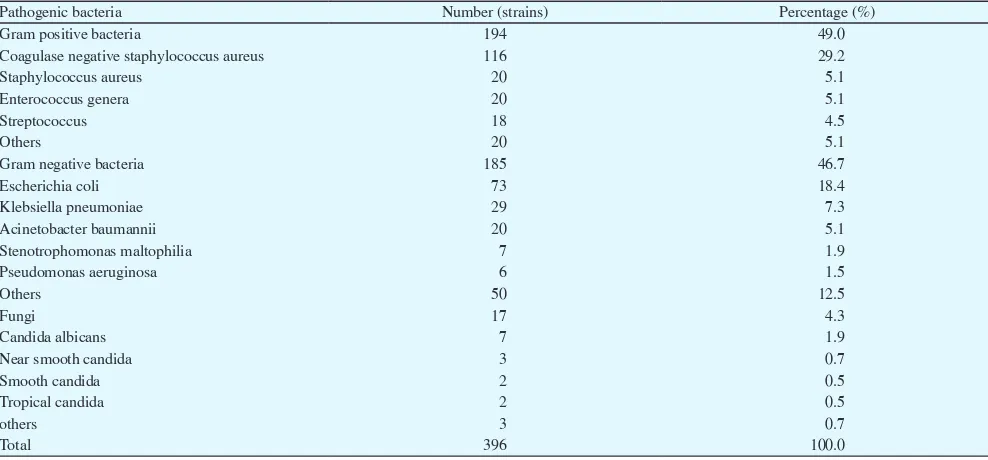
Pathogens isolated from blood culture and their constituent ratio.
Pathogenic bacteria Number (strains) Percentage (%)
Gram positive bacteria 194 49.0
Coagulase negative staphylococcus aureus 116 29.2
Staphylococcus aureus 20 5.1
Enterococcus genera 20 5.1
Streptococcus 18 4.5
Others 20 5.1
Gram negative bacteria 185 46.7
Escherichia coli 73 18.4
Klebsiella pneumoniae 29 7.3
Acinetobacter baumannii 20 5.1
Stenotrophomonas maltophilia 7 1.9
Pseudomonas aeruginosa 6 1.5
Others 50 12.5
Fungi 17 4.3
Candida albicans 7 1.9
Near smooth candida 3 0.7
Smooth candida 2 0.5
Tropical candida 2 0.5
others 3 0.7
conduct bacteria identification and make drug susceptibility tests. If system had not detected positive-reaction in blood culture bottles in 5th day, they would be considered as negative blood culture bottles.
(3) Bacterial identification and drug sensitivity test were carried out by VITEK-2 Compact automatic microbial analysis system and ATB Expression microbial analysis system. Based on the 2015 CLSI standard, the results of drug sensitivity were read out.
2.4 Statistical analysis
The results were statistically analyzed by software WHONET 5.6.
3. Results
3.1. Distribution of pathogens isolated from blood culture
In this study, 4 142 samples of blood culture specimens were detected and 396 strains of pathogens were isolated, in which the positive rate was 9.6%. Among them, there were 194 Gram-positive
bacteria (49%), 185 Gram-negative bacteria (46.7%), and 17 fungi (4.3%).
The main pathogens we isolated were 116 strains of coagulase negative staphylococcus (29.2%), 73 strains of Escherichia coli (18.4%), 29 strains of Klebsiella pneumoniae (7.3%), 20 strains of Staphylococcus aureus (5.1%), 20 strains of Bauman Acinetobacter (5.1%), 20 strains of Enterococcus (5.1%; among them, there were 11 strains of Enterococcus faecium, 6 strains of Enterococcus faecalis, 3 strains of Enterococcus gallinarum), 17 strains of fungi (4.3%). The major pathogens isolated from blood culture and their compositions are shown in Table 1.
3.2 Sensibility of the main staphylococcus isolated from blood
culture to common antibiotics
Coagulase-negative staphylococci and Staphylococcus aureus were more resistant to penicillin and erythromycin, and the detection rate of MRCNS and MRSA were 81.9% and 50% respectively. No strains were found resistant to linezolid and vancomycin. The susceptibility of the main staphylococcus isolated from blood culture to common antibiotics is shown in table 2.
Table 3.
Sensibility of the main enterococcus isolated from blood culture to common antibiotics (%).
Antibacterial drugs Enterococcus faecium Enterococcus faecalis
Sensitivity rate Resistance rate Sensitivity rate Resistance rate
Clindamycin 0.0 100.0 0.0 100.0
Levofloxacin 0.0 100.0 0.0 100.0
Penicillin 11.1 88.9 100.0 0.0
Ampicillin 11.1 88.9 100.0 0.0
Erythromycin 11.1 88.9 33.3 66.7
Ciprofloxacin 11.1 88.9 66.7 33.3
Tetracycline 33.3 66.7 0.0 100.0
Gentamicin (high concentration) 66.6 33.4 66.7 33.3 Linezolid 88.9 0.0 (11.1 intermediate) 100.0 0.0
Vancomycin 100.0 0.0 100.0 0.0
Teicoplanin 100.0 0.0 100.0 0.0
Table 4.
Sensibility of the main enterobacteriaceae isolated from blood culture to common antibiotics (%).
Antibacterial drugs Escherichia coli Klebsiella pneumoniae
Sensitivity rate Resistance rate Sensitivity rate Resistance rate
Aztreonam 68.5 31.5 79.3 20.7
Ampicillin 23.3 76.7 0.0 100.0
Ampicillin/ sulbactam 65.8 34.2 75.9 24.1
Piperacillin/tazobactam 100.0 0.0 89.7 10.3
Ceftriaxone 38.4 61.6 75.9 24.1
Ceftazidime 69.9 30.1 79.3 20.7
Cefepime 72.6 27.4 79.3 20.7
Meropenem 100.0 0.0 89.7 10.3
Imipenem 100.0 0.0 93.1 6.9
Cotrimoxazole 47.9 52.1 79.3 20.7
Ciprofloxacin 76.7 23.3 82.8 17.2
Gentamicin 67.1 32.9 82.8 17.2
Amikacin 100.0 0.0 100.0 0.0
Levofloxacin 80.0 19.2 86.2 13.8
3.3 Sensibility of the main enterococcus isolated from blood
culture to common antibiotics
20 strains of enterococcus have been isolated during the study. All the strains are susceptible to vancomycin and teicoplanin except enterococcus gallinarum which is naturally resistant to vancomycin. However, all the strains are not resistant to linezolid.And it is reported that all strains are resistant to clindamycin and levofloxacin. The sensitivity of different strains to ampicillin was quite difference, the sensitivity rate of Enterococcus faecalis was 100%, but the sensitivity of Enterococcus was only 11.1%. Sensibility of the main enterococcus isolated from blood culture to common antibiotics were show in Table 3.
2.4 Drug resistance rate of the main enterobacteriaceae
isolated from blood culture to common antibiotics
In this study, 73 strains of Escherichia coli were isolated, of which 41 strains were ESBLs-producing Escherichia coli, accounting for 56.2%. 29 strains of Klebsiella pneumoniae were isolated, of which 11 strains were ESBLs producing strains, accounting for 37.9%. Escherichia coli and Klebsiella pneumoniae had highest resistance rates to third-generation cephalosporins and had lower resistance rates to piperacillin / tazobactam. No strains resistant to amikacin were found. All Escherichia coli we isolated were sensitive to imipenem and meropenem, while Klebsiella pneumoniae were resistant to imipenem and meropenem, and the resistance rates of imipenem is 6.9% and of meropenem is 10.3%. Sensibility of the main enterobacteriaceae isolated from blood culture to common antibiotics were show in Table 4.
4. Discussion
In accordance with the research results, there are 4142 blood culture being sent to our hospital for laboratory test from January, 2015 to December, 2016. 396 distinct pathogenic bacteria in total have been isolated after removing the duplicate specimen of the same region of the same patient and the positive rate is 9.6% which is similar to the positive rate reported in the literature[1] while there exists significant difference compared with the positive rate of 16.12% reported by SHEN Cuihua and others[2]. The cause of the difference could be attributed to patients’s variation in different hospitals. Among the isolated pathogenic bacteria, gram-positive bacteria make up 49% which is higher than gram-negative bacteria. The most common type of gram-positive bacteria is coagulase negative staphylococcus with the percentage of 29.2%. Staphylococcus aureus and enterococcus
enterobacteriaceae[7]. Yet escherichia coli and klebsiella pneumoniae are susceptible to antimicrobial agents with enzyme inhibitor such as ampicillin/sulbactam and piperacillin/tazobactam which can be used as empirical medication for bacterial infection. The susceptibility rate of escherichia coli to meropenem and imipenem is 100% while klebsiella pneumoniae is resistant to meropenem and imipenem at a certain extent which should be emphasized in order to avoid clinical and hospital infection. In addition, relevant personnel should take strict precautions against invalid antibiosis and prevent the transmission of klebsiella pneumoniae which is resistant to carbapenem.
The detection rate of fungus (mainly Candida) is 4.3% which is close to 3.4% reported by GUO Pu and others[8], and is lower than 7.8% reported by LIANG Jun and others[9]. The fungus isolated from the blood culture is mainly distributed in the samples collected from Center ICU, Cardio-Thoracic Surgery ICU and department of hematopathology. The low immunity of ICU patients with severe underlying diseases, invasive procedures such as endotracheal intubation and peripherally inserted central catheterization and the extensive use of broad spectrum antibiotics,and hypoproteinemia will lead to dysbacteria in the body, which increases the infection risks of candida[10]. The weakness of patients with hematological disease, neutropenia or hypofunction of neutrophils, and chemotherapy for leukemia can greatly increase the risks of candidiasis. The number of fungus strains isolated during this study is very small, so no analysis of antimicrobial susceptibility test results has been carried out. However, relevant clinical departments should attach great importance to this aspect. The major reasons of fungal bloodstream infection are the patients’ low immunity, invasive procedures and the extensive use of broad spectrum antibiotics.
Currently, as a consequence of rapid spread of worldwide drug-resistant strain, it is much more difficult to select suitable antimicrobial agents during clinical practice. Clinical medicine should put more emphasis on bacterial cultures, especially the blood culture and antimicrobial susceptibility test results. Strictly follow corresponding procedures and regulations when collecting the blood culture and sending the sample for laboratory test, strengthen the surveillance of drug resistance of pathogenic bacteria, grasp drug-resistance rules of pathogenic bacteria of our hospital and local region, follow the indications of medication usage , use antibacterial
agents standardly and reasonably in accordance with antimicrobial susceptibility test results in order to decrease the development of drug-resistant strains and prevent the wide spread of infection within the hospital.
Reference
[1] Zhou Hongjing, Feng Ranran, Zhang Lixia, Zhang Li. Analysis on distribution and drug resistance of pathogenic bacteria in blood culture. J Huaihai Med 2016; 34(1): 29-31.
[2] Shen Cuihua, Zhang Jing, Jin Yan. Distribution and drug resistance of pathogens isolated from 4211 blood specimens. Chin J Nosocomiol 2012; 22(7): 1509-1511.
[3] Li Fang, Cao Bing, Liu Yingmei, Li Binbing, Wang Shanshan, Yang Chunxia, et al. Distribution and resistance analysis of isolates from blood culture samples. Chin J Nosocomiol 2010;
20(9): 1319-1322.
[4] Rao Rong, Liu Zhijun. 215 strains of blood culture pathogens and their resistance analysis. Prac Geriatrics 2012; 26(2): 174-176.
[5] Xie Liangyi, Yang Qiwen. Distribution and alarm time analysis of blood culture positive bacteria. Chin J Med Infect 2015; 25(2): 279-281.
[6] Yang Qing, Yu Yunsong, Lin Jie. Surveillance of drug resistance of Enterococcus CHINET in 2005-2014 years. Chin J Infect Chemother 2016; 16(2): 146-152.
[7] Wu Shimu, Shi Xiaoyan, Xia Min. Surveillance and drug resistance of extended spectrum beta lactamases produced by Enterobacteriaceae. Int J Lab Med 2009; 30(11): 1115-1116. [8] Guo Pu, Qiao Yan, Zhang Haitao. Distribution and drug
resistance of 76 strains of Candida blood stream infection. J Bengbu Med Coll 2016; 41(5): 661-663.
[9] Liang Jun, Zhang Zhou, Xu Yuanhong. Analysis of the distribution and drug resistance of bacterial fungal bloodstream infection. J Clin Transfusion Lab Med 2013; 25(10): 101. [10] Sunson, Wen Miaoyun, Zeng Hongke. Flora distribution and