Copyright © 2001, American Society for Microbiology. All Rights Reserved.
Simple Method for Determining Biovar and Serovar Types of
Ureaplasma urealyticum
Clinical Isolates Using
PCR–Single-Strand Conformation Polymorphism Analysis
DAVID PITCHER,1* MARGARET SILLIS,2ANDJANET A. ROBERTSON3Respiratory and Systemic Infection Laboratory, Central Public Health Laboratory, London NW9 5HT,1and Public
Health Laboratory, Norwich NR2 3TX,2United Kingdom, and Department of Medical Microbiology and Immunology,
University of Alberta, Edmonton, Alberta T6G 2H7, Canada3
Received 29 September 2000/Returned for modification 9 January 2001/Accepted 23 February 2001
Ureaplasma urealyticum has been associated with urethritis in men, obstetric problems in women, and
respiratory distress syndrome in preterm infants.U. urealyticumcan be divided into two biovars comprising 14
serovars. Partial sequences of genes encoding the multiple-banded antigens of the cell surface are known. Using a commercially available precast DNA mutation detection gel system, we have developed a simple and reproducible PCR–single-strand conformation polymorphism analysis method for differentiating the biovars of this species that reveals five patterns among the 14 serovars and enables clinical isolates to be typed directly from broth cultures.
Ureaplasma urealyticumis a common commensal of the
uro-genital tract in both men and women. It has been implicated in male nongonococcal urethritis (27) and associated with a num-ber of obstetric conditions, including chorioamnionitis and chronic respiratory distress in neonates, often with poor prog-nosis (1, 3). Use of polyclonal antisera raised against whole ureaplasmal cells has identifed 14 serovars (22). As shown earlier by DNA hybridization studies on serovars 1 to 8, these can be divided into two clusters or biovars. DNA homology between the biovars was less than 60% and sufficiently different to have separate species suggested for each of the biovars (5). More recently other phenotypic and genotypic traits, including results of restriction fragment analysis (7) and 16S rRNA se-quencing (20), and genome size (21) have confirmed this find-ing. Currently, separate species status is being sought for each of the biovars (J. A. Robertson, personal communication).
Biovar 1 (parvo) consists of serovars 1, 3, 6, and 14, and biovar 2 (T960) consists of serovars 2, 4, 5, 7, 8, 9, 10, 11, 12, and 13 (22). For many years there has been speculation about the association of particular serovars with disease. For exam-ple, in one study in which the role ofU. urealyticumin urinary tract disease was investigated, serotyping identified serovar 6 as the predominant type in urine samples (9). In another study, a statistical association was found between the occurrence of serovar 4 in women who had a history of recurrent abortions compared with healthy pregnant women (16). Despite these findings there has been no conclusive evidence linking a par-ticular serovar with a disease. In part, this could reflect diffi-culties in interpreting serological typing results. Multiple cross-reactions are common, and clinical samples frequently contain two or more serovars that cannot casually be separated (26).
Robertson et al. (19) were unable to ascribe any correlation between serovars isolated from tissues of subjects experiencing spontaneous and therapeutic abortion.
The difficulty in interpreting the results obtained in these studies was partly due to polyclonal antisera directed at the dominant antigens of the organism. A full set of set of mono-clonal antibodies (MAbs) against serovar-specific antigens has recently been developed and established that polyreactivity, a disadvantage of polyclonal serotyping, was not encountered when using MAbs (6). MAbs have been used to identify mul-tiple-banded antigens responsible for serovar specificity on the cell surface (34). Sequence variation in the genes encoding these antigens (mba genes) has been exploited to partially differentiate serovars using combinations of restriction en-zymes or DNA amplification with panels of primers (12, 15, 29). However, these methods all require multistep procedures. Single-strand conformation polymorphism (SSCP) analysis was originally developed to detect allelic variation in human genetic disorders (18). Double-stranded DNA is denatured, and the single-stranded products are separated by gel electro-phoresis under accurately controlled, nondenaturing condi-tions. During its migration through the gel, each strand as-sumes a folded conformation dependent on its internal base pairing and therefore on its sequence.
The migration distances of the conformers visualized as bands on the gel are reproducible and are determined by the structure. Base substitutions or deletions can alter the confor-mations, causing mobility shifts in the migration patterns and revealing sequence divergence in the specific genomic region amplified. In a small (100- to 200-bp) PCR product, as little as one base change can be detected by this method (2, 4)
Although SSCP analysis has not yet found wide acceptance in bacteriology, it has been used for the detection of rifampin-resistant mutants (rpoB gene) of Mycobacterium tuberculosis
(28) and to subtype Borrelia burgdorferi isolates (16S rRNA gene) (31). Bacterial 16S ribosomal DNA (rDNA) from clini-cal isolates has also been identified using fluorescent primers in
* Corresponding author. Mailing address: Respiratory and Systemic Infection Laboratory, Central Public Health Laboratory, 61 Colindale Ave., London NW9 5HT, United Kingdom. Phone: 44 (0) 208 200 4400. Fax: 44 (0) 208 205 6528. E-mail: dpitcher@phls.nhs.uk.
1840
on May 15, 2020 by guest
http://jcm.asm.org/
a PCR followed by SSCP analysis in an automated DNA se-quencer (30, 32).
Using commercially available precast gels in a horizontal electrophoresis system, we investigated the possibility of geno-typingU. urealyticumclinical isolates from the gel patterns of single-stranded conformers of PCR-amplified products of their
mbagenes.
MATERIALS AND METHODS
Strains, specimens, and culture conditions. Stock reference cultures were obtained from the American Type Culture Collection under accession numbers ATCC 27813 (serovar 1), 27814 (serovar 2), 27815 (serovar 3), 27816 (serovar 4), 27817 (serovar 5), 27818 (serovar 6), 27819 (serovar 7), 27618 (serovar 8), 33175 (serovar 9), 33699 (serovar 10), 33695 (serovar 11), 33696 (serovar 12), 33698 (serovar 13), and 33697 (serovar 14). These were maintained in the freeze-dried state and cultured in 4 ml of U4 broth (10).
Fluid clinical samples (endotracheal and nasopharyngeal aspirates) were in-oculated into 2 ml of arginine-urea broth (bioMe´rieux, Basingstoke, United Kingdom) (24). Swabs were dipped into the broth and swirled briefly. All cultures were incubated at 37°C and inspected daily for growth.
Eighteen isolates from tissues of subjects who had experienced abortion, whose serovars had been previously established using polyclonal antiserum and a colony epifluorescence method (19, 25), were cultured in broth, and the cells were centrifuged and resuspended in 70% ethanol until the DNA was extracted. The preparations used for PCR-SSCP analyses were later passages than those used for serotyping.
DNA extraction.Cells were harvested by centrifugation of broth cultures at 10,000⫻gfor 10 min. Pellets were washed twice in sterile phosphate-buffered saline (pH 7.4; Oxoid, Basingstoke, United Kingdom), resuspended by vigorously vortexing in 500l of sterilized Chelex 100 suspension (10%, wt/vol, in PCR quality water) (Bio-Rad, Hemel Hempstead, United Kingdom), and incubated in a 56°C water bath for 30 min. Suspensions were vortexed for 20 s, heated at 100°C for 8 min, and rapidly cooled on ice. Finally, samples were vortexed again, centrifuged at 10,000⫻gfor 5 min (17). The supernatants were transferred to clean tubes and stored at⫺20°C until required.
Amplification ofmbagene fragment.In PCR mixtures for SSCP analysis, the forward primer was UMS⫺120 (5⬘-TGCAATCTTTATATGTTTTCGTT-3⬘), located 120 bases upstream from the start codon of thembasequence of serovar 3 (29). Two similar reverse primers downstream of this position were UMA⫹46 (5⬘-CCTAGTGTAATTGCTCAAAATTT-3⬘) and UMA⫹46ⴱ(5⬘-CCTAATG TCATAGCTMAGAATTT-3⬘), which were used together to account for degen-eracies in bases in this region. Reaction mixtures (50l) contained 50 mM KCl, 2.5 mM MgCl2, 15 mM Tris-HCl (pH 8.0), a 200M concentration of each
deoxyribonucleotide triphosphate, 20 pmol of each primer and 1.5 U ofTaq DNA polymerase (Perkin-Elmer, Warrington, United Kingdom). Chelex extract (10l) was added. Samples were overlaid with oil, and 45 cycles of 95°C for 1 min, 54°C for 1 min, and 72°C for 1 min were carried out, followed by a 72°C,
10-min extension period. Reactions were performed in an Omnigene thermocy-cler (Hybaid, Ashford, United Kingdom).
SSCP gel electrophoresis.To 10l of PCR product, 10l denaturing buffer (1 ml of formamide containing 0.25% bromophenol blue, mixed immediately before use with 10l of 1 M NaOH) was added; the mixture was vortexed briefly, heated on a PCR heating block at 95°C for 5 min, and immediately cooled on ice; and 5l of 50% glycerol added.
The electrophoresis system was set up 1 h before the run. An SEA 2000 tank (Elchrom Scientific, Cham, Switzerland), which possesses a buffer-circulating pump, was filled with 1.5 liters of TAE buffer (30 mM Tris-acetate, 0.75 mM EDTA buffer [pH 8.2]). The external water jacket was connected to a temper-ature-controlled circulating water bath.
The temperature of the circulating buffer was adjusted to 9°C before the MDA 26-lane gels (Elchrom Scientific) were placed in the tank. Before loading the samples, the buffer-circulating pump was turned off. Wells were loaded with 20
l of ice-cold samples. Electrophoresis was carried out for 30 min at 48 V, the buffer pump was turned on, and electrophoresis continued for a further 17.5 h at 48 V. Gels were stained with SYBR gold (Molecular Probes, Leiden, The Netherlands) (1/10,000 in 10 mM TAE buffer) for 40 min, examined on a transilluminator at 254 nm, and photographed using a SYBR green filter and 667 Polaroid film.
The electrophoresis unit employed in this study enabled 26 samples, including controls to be run simultaneously. The approximate times required for process-ing 26 cultures were 1 h for DNA extraction, 15 min for preparation of the samples, 18 h for electrophoresis overnight, and 40 min for SYBR gold staining.
RESULTS
SSCP grouping of reference strains.The extraction of DNA
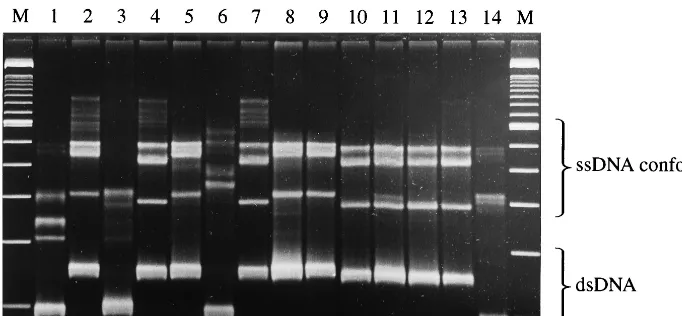
[image:2.612.132.473.74.232.2]from broth culture sediments using Chelex to suppress inhib-itors ofTaqDNA polymerase, when applied to both standard strain cultures and clinical isolates, yielded successful PCRs. Careful control of the electrophoresis conditions allowed iden-tification of gel banding patterns which unambiguously differ-entiated biovars 1 and 2 of U. urealyticum directly from the broth cultures of standard strains representing the 14 estab-lished serovars of this species and enabled placement of the serovars into fivembagene groups. A 100-bp ladder was in-cluded to estimate gel-to-gel reproducibility and does not re-late to the size of the conformers, whose migration distances are based on the shape of their folded structures rather than their sequence length (Fig. 1). Biovar 1 gave a PCR product of 173 bp from which three conformer band patterns were ob-tained; serovars 1 and 6 gave distinct patterns, and those of serovars 3 and 14 were identical. There was less discrimination
FIG. 1. PCR-SSCP patterns of the reference strains ofU. urealyticumused to define serovars. Lanes 1 to 14, serovars 1 to 14; lanes M, 100-bp ladder. ssDNA and dsDNA, single-stranded and double-stranded DNA, respectively.
on May 15, 2020 by guest
http://jcm.asm.org/
among the serovars of biovar 2, where the 217-bp product gave rise to two patterns. Pattern 2A contained serovars 2, 5, 8, and 9, and pattern 2B contained serovars 4, 7, 10, 11, 12, and 13. From these results we predicted that all isolates could be typed to one of the followingmbagroups: biovar 1 (1, 3/14, and 6) and biovar 2 (2A and 2B).
Correlation of SSCP analysis with serotyping.First we
[image:3.612.134.475.71.217.2]ex-amined the DNA from 18 coded, previously serotyped isolates of the abortion study (19). Their biovars had been predicted based on experience with serotyping, and more recently the isolates had been typed to the biovar level by 16S rRNA gene PCRs (23). PCR-SSCP band patterns for these are shown in Fig. 2. Correlation of the biovar designation between the two types of PCR (16S rRNA andmbagene sequences) and be-tween PCRs and the serotyping results were exact except for strain RH1139 (Table 1). On the basis of serology, RH1139, indicated as carrying serovars 11 and 13 of biovar 2, was placed in biovar 1 by both types of PCR.
For 12 of the other 17 strains, the SSCP analysis and sero-typing results were in agreement. The five remaining strains did not show such clear correlation. Isolate RH872 reacted with antiserum to serovar 3 but was identified as mbagene group 1 by SSCP analysis; isolate RH799, which reacted with antisera to serovars 12 and 13 (both in biovar 2), was identified correctly as group 2B by SSCP analysis, but it also reacted with serovar 9 antiserum, which is associated with biovar 2A. Sim-ilarly, isolate RH191 reacted with antisera to serovars 3 and 14 as well as 6, but onlymbagene group 6 was amplified.
From the SSCP patterns two cultures of mixed biovars (RH297 and RH541) and one (RH479) of mixed mbagene groups within a single biovar were detected. In the case of isolate RH297, serotyping resulted in reactions with antisera to serovars 6 and 13, which belong to biovars 1 and 2, respectively, and which should correlate with mbagene groups 6 and 2B. However, the SSCP analysis showed genes from each biovar but biovar 2 was associated withmbagene group 2A not 2B. In culture RH541, both serotyping and SSCP analysis iden-tified serovar 3, but SSCP analysis also indicated the presence of biovar 2A genes, and in RH479, for which serotyping re-vealed only the presence of serovar 4, equivalent to the mba
gene group 2B, SSCP analysis also detected the presence of group 2A.
Application of PCR-SSCP analysis to clinical isolates.
[image:3.612.310.548.448.680.2]Sec-ondly, we subjected the deposits from 44 Ureaplasma broth culture isolates from clinical specimens to PCR-SSCP analysis in order to confirm that the five SSCP patterns would cover most isolates. These specimens were randomly selected and consisted of 15 cultures from the tracheal and nasopharyngeal aspirates of neonates, both full term and preterm, and 29 cultures from high vaginal swabs taken from pregnant and
FIG. 2. PCR-SSCP patterns of serologically confirmed strains ofU. urealyticumlisted in Table 1. Lane 1, control serovar 1; lanes 2 to 5, RH303, RH313, RH1087, and RH872; lane 6, control serovar 3; lanes 7 to 10, RH297, RH541, RH666, and RH1139; lane 11, control serovar 6; lanes 12 to 14, RH555, RH677, and RH191; lane 15, control serovar 2; lanes 16 to 18, NIH5, T960, and RH479; lane 19, control serovar 4; lanes 20 to 23, RH122, RH507, RH539, and RH799; lane M, 100-bp ladder.
TABLE 1. Correlation of blind-tested PCR-SSCP analysis-determined groups with biovar as analysis-determined by PCR of 16S rDNA
and serovar and biovar as determined by serotyping
Strain no.
Serovar(s) or pattern(s) as determined by:
Serotypinga PCR
Serovar Predictedbiovar 16S rDNAbiovar b PCR-SSCP c
Biovar mbagene group
RH303 1 1 1 1 1
RH313 —d — 1 1 1
RH1087 1 1 1 1 1
RH872 3e 1 1 1 1
RH297 6, 13 1 1 1⫹2 6⫹A
RH541 3 1 1 1⫹2 3/14⫹A
RH666 3, 14 1 1 1 3/14
RH1139 11, 13 2 1 1 6
RH555 6 1 1 1 6
RH677 6 1 1 1 6
RH191 3, 6, 14 1 1 1 6
NIH5 5 2 2 2 A
T960 8 2 2 2 A
RH479 4 2 2 2 A⫹B
RH122 4 2 2 2 B
RH507 13 2 2 2 B
RH539 4, 13 2 2 2 B
RH799 9, 12, 13 2 2 2 B
aAs reported by Robertson and Stemke (22).
bAs reported by Jacobs et al. (11) and Robertson et al. (23).
cPCR-SSCP gel pattern groups: 1, serovar 1; 3/14, serovars 3 and 14; 6, serovar 6; 2A, serovars 2, 5, 8, and 9; 2B, serovars 4, 7, 10, 11, 12, and 13.
dDuplicate of RH303.
eBoldface type indicates discrepant results.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:3.612.310.550.449.679.2]from nonpregnant woman (Table 2). All gave easily recogniz-able patterns that could be assigned to one of the above groups; no mixed groups were detected. All five groups were represented among the isolates.
DISCUSSION
Genes encoding the multiple banded antigens (mbagenes)
ofU. urealyticumhave been fully sequenced for biovar 1
(se-rovar 3; GenBank accession no. L20329) and biovar 2 (se(se-rovar 10; GenBank accession no. U50459). The 3⬘ region of these genes shows marked differences in long stretches of tandem repeat units where serovar specificity is determined (34; X. Zheng, L. J. Teng, H. L. Watson, J. L. Glass, A. Blanchard, and G. H. Cassell, GenBank accession no. L20329 and U50459). Close to the 5⬘end of the gene, a 45-bp deletion in the biovar 1 gene accounts for the difference in PCR product length between biovars 1 and 2 (29, 33, 34). PCRs amplifying parts of the first 400 to 500 bases of the 5⬘end of thembagene region including 125 bases upstream of the start codon followed by restriction endonuclease typing have enabled the partial dif-ferentiation of serovars into groups (29). More detailed se-quence data were determined for this region for all 14 serovars by Kong et al. (15), who devised a stepwise system of biotyping and partial serovar identification based on PCR using two primer pairs and followed by restriction enzyme analysis of the products. Knox and colleagues (12, 13) have sequenced the
mba5⬘regions (nucleotide positions⫺104 to 207) of 33 clinical isolates ofU. urealyticumand compared them to the standard serovar sequences. Using a nested PCR with two outer and six inner primers, they defined nine subtypes within the biovar 1 isolates, including two subtypes of serovar 1, five of serovar 3, and two of serovar 6. Biovar 2 could be divided into two subtypes. In certain instances, the differences between subtypes of the same serovar was a single base difference. They also applied random amplified polymorphic DNA analysis (RAPD) to these isolates and, using seven primers, differentiated 13 RAPD subtypes. They concluded that RAPD provided the greatest discrimination as a typing method forU. urealyticum. To date, significant structural and functional differences have not been identified beyond the biovar level, and neither biovars or subdivisions of them have been convincingly corre-lated with disease states. Unlike RAPD and restriction digest patterns of nested PCR products, PCR-SSCP analysis requires a single set of primers and a single amplification step, making it a more-efficient approach for subdividing large numbers of ureaplasmas. Groups were designated on the basis of the SSCP patterns of serovar reference strains (Fig. 1), and most of the serovars of isolates from the abortion study were broadly in
agreement with their SSCP groups. The discrepant results are indicated in Table 1. One isolate (RH1139), when serotyped, belonged to biovar 2, but both 16S rDNA PCR and SSCP analysis placed it in biovar 1. The reason for this anomaly could not be explained. In two other instances where serotyping indicated mixed serovars, only one SSCP pattern was detected. These findings could represent greater precision of the SSCP analysis or changes in relative populations of subtypes in mixed cultures on later passages or the sensitivity of the methodolo-gies.
In three instances, mixed patterns were observed on SSCP gels, but each component could clearly be identified as belong-ing to one of the designated groups and probably indicatbelong-ing mixed cultures. Although the results of gene-based methods are expected to be more easily reproducible than those of phenotyping, experience has taught us that typing cloned, lab-oratory-adapted strains is much easier than working with wild-type isolates (26). The serowild-typed isolates that weremba geno-typed in this study emphasize this lesson. They had not been cloned; serotyping, biotyping, and PCR-SSCP analysis were not performed on identical cultures. All strains tested in this study provided unambiguous SSCP results, and the method could be used to group clinical isolates as shown in Table 2. However, because these were uncloned, randomly acquired isolates, we did not attempt to correlate the group designation with the pathogenic status of the patient. PCR-SSCP analysis did not detect any mixed groups among these specimens, al-though mixtures of serovars are commonly encountered on serotyping. In a mixed population of serovars, culture in broth could favor the growth of more-vigorous strains or those present at the highest initial density, and serotyping may reflect this. With PCR, there could be preferential amplification of the most abundant DNA template, resulting in a single group being detected.
Ideally, sequencing thembagene would be the most accu-rate way of typing isolates, but it is a time-consuming, expen-sive, and skilled procedure and not a practical proposition for analyzing large numbers of isolates. However, PCR-SSCP analysis could be a convenient way of rapid screening prior to selecting strains for sequencing. For most diagnostic laborato-ries that are unable to employ sequencing, the procedure could be used routinely to analyze clinical isolates.
The two biovars ofU. urealyticumcan now be readily differ-entiated by PCRs based upon differences in the 16S rRNA (11, 23), the 16S-23S rRNA intergenic region (8), and the mba
genes (29).
[image:4.612.52.295.92.169.2]Genotypic methods based on themba gene are effectively replacing the 14-member-serotyping scheme established with polyvalent antisera (12–15). Unlike MAbs, the hyperimmune sera used for serotyping may contain more than one antibody to each of the multiple antigens present in the whole-cell preparations used as immunogens. It has been shown, for in-stance, that many antisera contain antibodies to the biovar-specific urease enzyme (25). The infinitely greater complexity of antigen-antibody reactions compared to the variations in nucleotide sequences of a single gene means that complete correlation cannot be expected. In this study, we exploited sequence differences within thembagene both to separate the two biovars and also to indicate subgroups consisting of one or more serovars. A single PCR will thus allow differentiation of
TABLE 2. Distribution of PCR-SSCP groups among clinical samples
Clinical sample or group samplesNo. of (n⫽44)
No. of strains belonging to indicated serovars
Biovar 1 Biovar 2
1 3/14 6 2A 2B Babies (nasopharyngeal aspirates) 15 1 5 4 2 3
High vaginal swabs 29 10 8 4 1 6
on May 15, 2020 by guest
http://jcm.asm.org/
U. urealyticuminto five groups. Its application to controlled clinical studies may help to further an understanding of ure-aplasmal pathogenicity. We propose that the typing ofU.
urea-lyticumstrains from a wide variety of sources could be achieved
more rapidly, more cheaply, and in greater numbers by this technique than by previously described methods. Direct iden-tification of these genotypes in clinical specimens is our next goal.
ACKNOWLEDGMENT
We thank Robert C. George for constructive appraisal of the manu-script.
REFERENCES
1.Abele-Horn, M., C. Wolff, P. Dressel, F. Pfaff, and A. Zimmerman.1997. Association ofUreaplasma urealyticumbiovars with clinical outcome for neonates, obstetric patients, and gynecological patients with pelvic inflam-matory disease. J. Clin. Microbiol.35:1199–1202.
2.Ainsworth, P. J., L. C. Surh, and M. B. Coulter-Mackie.1991. Diagnostic single strand conformational polymorphism (SSCP): a simplified non-radio-active method as applied to a Tay-Sachs B1 variant. Nucleic Acids Res. 19:405–406.
3.Cassell, G. H., K. B. Waites, H. L. Watson, D. T. Crouse, and R. Harasawa. 1993.Ureaplasma urealyticumintrauterine infection: role in prematurity and disease in newborns. Clin. Microbiol. Rev.6:69–87.
4.Chaubert, P., D. Bautista, and J. Benhattar.1993. An improved method for rapid screening of DNA mutations by non-radioactive single-strand confor-mation polymorphism procedure. BioTechniques15:586.
5.Christiansen, C., F. T. Black, and E. A. Freundt.1981. Hybridization exper-iments with deoxyribonucleic acid fromUreaplasma urealyticumserovars I to VIII. Int. J. Syst. Bacteriol.31:259–262.
6.Echahidi, F., G. Muyldermans, S. Lauwers, and A. Naessens.2000. Devel-opment of monoclonal antibodies againstUreaplasma urealyticumserotypes and their use for serotyping clinical isolates. Clin. J. Diagn. Lab. Immunol. 7:563–567.
7.Harasawa, R., K. Dybvig, H. L. Watson, and G. H. Cassell.1991. Two genomic clusters among the 14 serovars ofUreaplasma urealyticum. Syst. Appl. Microbiol.14:393–396.
8.Harasawa, R., and Y. Kanamoto.1999. Differentiation of two biovars of Ureaplasma urealyticumbased on the 16S–23S rRNA intergenic spacer re-gion. J. Clin. Microbiol.37:4135–4138.
9.Hewish, M. J., D. F. Birch, and K. F. Fairley.1986.Ureaplasma urealyticum serotypes in urinary tract disease. J. Clin. Microbiol.23:149–154. 10. Howard, C. J., R. N. Gourlay, and J. Collins.1978. Serological studies with
bovine ureaplasma (T-mycoplasma) Int. J. Syst. Bacteriol.28:473–477. 11. Jacobs, E., M. Vonski, G. W. Stemke, and J. A. Robertson.1994.
Identifica-tion ofUreaplasmabiotypes. Med. Microbiol. Lett.3:31–35.
12. Knox, C. L., P. Giffard, and P. Timms.1998. The phylogeny ofUreaplasma urealyticumbased on thembagene fragment. Int. J. Syst. Bacteriol.48:1323– 1331.
13. Knox, C. L., and P. Timms.1998. Comparison of PCR, and random ampli-fied polymorphic DNA PCR for detection and typing ofUreaplasma urea-lyticumin specimens from pregnant women. J. Clin. Microbiol.36:3032– 3039.
14. Kong, F., G. James, M. Zhenfang, S. Gordon, B. Wang, and G. L. Gilbert. 1999. Phylogenetic analysis ofUreaplasma urealyticum: support for the es-tablishment of a new species,Ureaplasma parvum. Int. J. Syst. Bacteriol. 49:1879–1889.
15. Kong, F., X. Zhu, W. Wang, X. Zhou, S. Gordon, and G. L. Gilbert.1999.
Comparative analysis and serovar-specific identification of multiple-banded antigen genes ofUreaplasma urealyticumbiovar 1. J. Clin. Microbiol.37:538– 543.
16. Naessens, A., W. Foulon, J. Breynaert, and S. Lauwers.1988. Serotypes of Ureaplasma urealyticumisolated from normal pregnant women and patients with pregnancy complications. J. Clin. Microbiol.26:319–322.
17. Ochert, A. S., A. Boulter, W. Birnbaum, N. W. Johnson, and C. G. Teo.1994. Inhibitory effects of salivary fluids on PCR: potency and removal. PCR Methods Appl.3:365–368.
18. Orita, M., Y. Suzuki, T. Sekiya, and K. Hayashi.1989. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics5:874–879.
19. Robertson, J. A., L. H. Honore, and G. W. Stemke.1986. Serotypes of Ureaplasma urealyticum in spontaneous abortion. Pediatr. Infect. Dis. 5:S270–S272.
20. Robertson, J. A., L. A. Howard, C. L. Zinner, and G. W. Stemke.1994. Comparison of the 16S rRNA genes within the T960 and parvo biovars of ureaplasmas isolated from humans. Int. J. Syst. Bacteriol.44:836–838. 21. Robertson, J. A., L. Pyle, G. W. Stemke, and L. R. Finch.1990.Ureaplasma
urealyticumshows diverse genome size by pulse-field electrophoresis. Nucleic Acids Res.18:1451–1455.
22. Robertson, J. A., and G. W. Stemke.1982. Expanded serotyping system scheme forUreaplasma urealyticumstrains isolated from humans. J. Clin. Microbiol.15:873–878.
23. Robertson, J. A., A. Vekris, C. Be´be´ar, and G. W. Stemke.1993. Polymerase chain reaction using 16S rRNA sequences distinguishes the two biovars of Ureaplasma urealyticum. J. Clin. Microbiol.31:824–830.
24. Sillis, M.1993. Genital mycoplasmas revisited—an evaluation of a new culture medium. Br. J. Biomed. Sci.50:89–91.
25. Stemke, G. W., and J. A. Robertson.1981. Modified colony indirect epiflu-orescence test for serotypingUreaplasma urealyticumand an adaptation to detect common antigenic specificity. J. Clin. Microbiol.14:582–584. 26. Stemke, G. W., and J. A. Robertson.1985. Problems associated with
sero-typing strains ofUreaplasma urealyticum. Diagn. Microbiol. Infect. Dis. 3:311–320.
27. Taylor-Robinson, D., and P. M. Furr.1997. Genital mycoplasma infections. Wien. Klin. Wochenschr.109:578–583.
28. Telenti, A., P. Imboden, F. Marchesi, T. Schmidheini, and T. Bodmer.1993. Direct, automated detection of rifampicin-resistantMycobacterium tubercu-losisby polymerase chain reaction and single-strand conformation polymor-phism analysis. Antimicrob. Agents Chemother.37:2054–2058.
29. Teng, L.-J., X. Zheng, J. L. Glass, H. L. Watson, J. Tsai, and G. H. Cassell. 1994.Ureaplasma urealyticumbiovar specificity and diversity are encoded in multiple-banded antigen gene. J. Clin. Microbiol.32:1464–1469.
30. Turenne, C. Y., E. Witwicki, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 2000. Rapid identification of bacteria from positive blood cultures by fluo-rescence-based PCR-single stranded conformation polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol.38:513–520.
31. Weinecke, R., O. M. Koch, U. Neubert, U. Go¨bel, and M. Volkenandt.1993. Detection of subtype-specific nucleotide sequence differences in aBorrelia burgdorferispecific gene segment by analysis of conformational polymor-phisms of cRNA molecules. Med. Microbiol. Lett.2:239–246.
32. Widjojoatmodjo, M. N., A. D. C. Fluit, and J. Verhoef.1995. Molecular identification of bacteria by fluorescence-based PCR-single-strand confor-mation polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 33:2601–2606.
33. Zheng, X., K. Lau, M. Frazier, G. H. Cassells, and H. L. Watson.1996. Epitope mapping of the variable repetitive region within the MB antigen of Ureaplasma urealyticum. Clin. Diagn. Lab. Immunol.3:774–778.
34. Zheng, X., L.-J. Teng, H. L. Watson, J. L. Glass, A. Blanchard, and G. H. Cassell.1995. Small repeating units within theUreaplasma urealyticumMB antigen gene encode serovar specificity and are associated with antigen size variation. Infect. Immun.63:891–898.