Copyright 8 1988 by the Genetics Society of America
Naturally Occurring Variation in the Restriction Map of the Amy Region of
Drosophila melanogaster
Charles H. Langley,* Antony E. Shrimpton,*.’ Tsuneyuki Yamazaki; Naohiko Miyashita,*
Yoshinori MatsuotP2 and Charles
F.
Aquadro*9s
*
Laboratory o Molecular Genetics, National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina 27709, andf
Laboratory of Population Genetics, Department of Biology, Faculty of Sciences, Kywhu University, Fukuoka, JapanManuscript received February 1, 1988 Accepted March 1 1, 1988
ABSTRACT
The restriction maps of 85 alleles of the Amy region of Drosophila melanogaster from natural populations were surveyed. A subset of these were also scored for allozyme phenotype and adult enzyme activity of a-amylase. Large insertions were found in 12% of the alleles in a 15-kb region surrounding the two transcriptional units of the duplicated Amy locus. The low frequencies at which each of these large insertions were found are consistent with earlier reports of variation in other loci. Four small deletions were found in the region 5’ to the Amy genes. Each was also rare in the population. Restriction site variation provided an estimate of per nucleotide heterozygosity of 0.006. Several statistically significant linkage disequilibria were observed between four polymorphic restriction sites and the allozymes. Adult a-amylase activity was correlated with the allozymes and with the polymorphism at one restriction site close to the transcriptional units.
S
IZE variation in the DNA surrounding transcrip- tional units appears to comprise a significant portion of the variability to be found among naturally occurring alleles in Drosophila melanogaster (LANGLEY,MONTGOMERY and QUATTLEBAUM 1982; LEIGH
BROWN 1983; AQUADRO et al. 1986; CROSS and BIRLEY 1986; LANGLEY and AQUADRO 1987; SCHAEFFER, LANG- LEY and AQUADRO 1988). Most of these size variants
can be attributed to insertions of middle repetitive dispersed sequences, suspected or known to be trans- posable elements. From restriction map surveys (LEIGH BROWN 1983; AQUADRO et al. 1986; KREITMAN and AGUADI? 1986; LANGLEY and AQUADRO 1987) and DNA sequence surveys (KREITMAN 1983) information is emerging concerning the levels of diversity at the nucleotide level and the amount of linkage disequi- librium to be expected in this species. &Acting elements affecting levels of gene expression are ex- pected in the regions flanking each gene. Thus the study of molecular variation in the gene and its surrounding region may shed light on various theo- retical models proposed to explain existing molecular genetic variation in natural populations (KIMURA
1983; GILLESPIE 1987). In the interest of extending these studies we have investigated the restriction map variation in and around the structural genes for a-amylase (Amy, 2-80; 54A-B1; GEMMILL, LEVY and
Buildings, West Mains Road, Edinburgh EH9 3JN, Scotland. University of Illinois, Champaign, Illinois 61820.
versity, Emerson Hall, Ithaca, New York 14853.
Genetics 119: 619-629 (July, 1988).
’
Present address: Department of Genetics, Edinburgh University, King’s Present address: Department of Ecology, Ethology and Evolution, Present address: Section of Genetics and Development, Cornell Uni-DOANE 1985; LEVY, GEMMILL and DOANE 1985; GEM- MILL, SCHWARTZ and DOANE 1986; BOER and HICKEY 1986; BENKEL and HICKEY 1986). Together with information on allozyme type and adult a-amylase activity these results provide corroborations of many of the results from Adh (AQUADRO et ai. 1986) and raise several interesting questions for further study.
T h e Amy locus has been cloned and partially char- acterized by GEMMILL, LEVY and DOANE (1985). They reported that (as suspected from earlier biochemical genetics studies) it is a duplicated locus (see Figure 1). T h e two transcriptional units are some 5 kb apart and transcribed divergently (from different strands) (BENKEL et al. 1988). Using a heterologous expression system LEVY, GEMMILL and DOANE (1985) have been able to assign the protein electrophoretic phenotypes, AMY’ and AMY3, to the right and left genes, respec- tively (see Figure 1) in the case of the cloned Amy’,’ haplotype from the Canton-S stock. GEMMILL, SCHWARTZ and DOANE (1986) analyzed seven labo- ratory strains by Southern blot analysis. They report several restriction site differences and two large insertions. They also identified one apparent inver- sion of the Amy region (GEMMILL, SCHWARTZ and DOANE 1986) and concluded that all strains had the same duplicated structure and indicated the apparent chromosome-proximal and -distal orientation of the restriction map.
620 C. H. Langley et al.
that most chromosomes in nature bear amy alleles expressing the allozyme phenotype Amy’ (except in equatorial Africa). Some 12 rarer allozyme haplotypes have been recognized involving more than eight different electrophoretic variants. There is consid- erable genetic variation in a-amylase activity. trans- Acting loci have been found that modify the expres- sion of a-amylase in various tissues (ABRAHAM and DOANE 1978; KLARENBERG and SCHARLOO 1986). a- Amylase expression responds strongly to dietary car- bohydrates, especially glucose during the larval stages (ABE 1958; DOANE 1969b; HICKEY 1977; HOORN and SCHARLOO 1978; YAMAZAKI and MATSUO 1984; BEN-
KEL and HICKEY 1986). T h e variant allozyme haplo-
types are often associated with high adult levels of enzyme activity (DOANE et al. 1987; BENKEL and HICKEY 1986), although substantial evidence indicates that this variation is largely attributable to levels of expression rather than specific activity differences in the proteins (HICKEY 198 1; HICKEY and BENKEL 1982). These variant Amy haplotypes have also been found to be in linkage disequilibrium with the poly- morphic inversion Zn(2R)NS (LANGLEY, TOBARI and KOJIMA 1974; LANGLEY, ITO and VOELKER 1977, MUKAI, WATANABE and YAMAGUCHI 1974, YAMAZAKI et al. 1984).
In this report we describe the restriction map variation in and around the Amy region (15 kb) for 85 haplotypes from natural populations of
D.
melan- oguster. This variation is combined with data on allozymes, adult a-amylase activity and polymorphic inversions. T h e relationship of these various types of variation in the Amy region and their comparison to the results of previous studies of other regions of the Drosophila genome contribute to the emerging un- derstanding of population genetic variation at the DNA level.MATERIALS AND METHODS
Strains: Two sources of D. melanogaster were used. The first are 49 isogenic second chromosome stocks in a com- mon homozygous genetic background (LAURIE-AHLBERG et al. 1980). These second chromosome lines were established from collections from Rhode Island (RI), Wisconsin (WI), North Carolina (NC) and Kansas (KA). Each of these lines was characterized as to Amy region restriction map (see below), as well as Amy allozyme phenotype (using the technique in MCCUNE 1969). a-Amylase activity and total soluble protein of adult females (reared on “normal media,” 260 hr posteclosion; YAMAZAKI and MATSUO 1984) were assayed for two replicate blocks, consisting of two samples (ten flies) of each line. Amylase activity was measured as the AOD at 550 nm using a starch-iodine assay as described in YAMAZAKI and MATSUO (1984). The total soluble protein was assayed from the same homogenates (LOWRY et al. 195 1). LAURIE-AHLBERG et al. (1980) have scored these lines for second chromosome gene arrangement. The second set of 36 lines are isofemale lines collected from many different locales worldwide within the last 4-10 yr. A few of these lines were still segregating more than one restric-
tion map haplotype. In each of these cases there was a preponderant and distinguishable haplotype that is re- ported in Table 1.
Restriction mapping and analysis: The 49 isogenic sec- ond chromosome lines were scored by single enzyme digests with five six-cutter restriction enzymes ( B a m H I , BglII, EcoRI, HindIII and SalI) as described in AQUADRO et al. (1986). All variation within the region probed by hDm65 (GIMMELL, LEVY and DOANE 1985) could be ascribed to either a polymorphism of a restriction site or to presence of an insertion or deletion (NC25 is the exception, see below). The localization of these polymorphisms could only be made to within the indicated fragments in Figure 1, since no additional restriction mapping or cloning has been carried out. The restriction map of NC25 was most simply understood as an inversion between the two Amy genes similar to the Amy null described in GEMMILL, SCHWARTZ and DOANE (1986), SCHWARTZ (1986) and DOANE et al. (1987). The 36 isofemale lines from around the world were scored by four single-enzyme digests ( B a m H I , HindIII, EcoRI and SalI). Here too all the variation could be attributed to either insertions or deletions or to polymorph- isms in restriction sites.
Statistical analysis: Linkage disequilibria between the polymorphisms were analyzed pair-wise utilizing Fisher’s exact test to determine significance and the correlation coefficient as a measure of the dependence. The rarer multiple Amy allozyme haplotypes were pooled in some tests. The levels of nucleotide variation were estimated
using the methods of EWENS, SPIELMAN and HARRIS (1981), HUDSON (1982), NEI and LI (1979) and NEI and TAJIMA (1981). Each polymorphism in Table 1 was examined by analysis of variance for association with activity and activity adjusted for total protein (see below). The model of analysis of variance (ANOVA) for the line effect is as follows:
Y y k = u
+
Bi+
Ll+
(B*L)zj+
eijk,where u is overall mean, Bi is the ith block effect ( i = 1, 2), Lj is thejth line effectj = (1 - 49), ( B * L ) , is the interaction effect and eqk is the residual term ( k = 1, 2). The effects were assumed to be random. F-tests were conducted acccordingly.
Since significant correlation between the activity and the total amount of protein over line means ( r = 0.39, P <
0.01) was detected, the raw activity was adjusted by the total amount of protein (analysis of covariance). The line means or the raw activities ( Y + ) were regressed on the line mean of the amount of protein (Pq.) for each block. The sums of squares and products were pooled over blocks to obtain a single slope ( b ) . The adjusted activity was then obtained as follows:
?ijb = Y i j k - b
*
(Pi,k - P...).-
RESULTS
Am,: Region
I I I I I 1 1 1 1 1 1 1 I I
- 9 - 7 - 8 - 6 - 5 - 4 - 3 - 2 - 1 0 +1 + 2 + 3 +4
62 1
FI(:I.RE I.-Organimtion o f the D N A it1 the A t t r , ~ region [Irasetl on the pul)lid~etl restriction m ; l p a w l S C Y I I I C I K C ~ 01 (;I \ I \ I I I . I . . IL\.Y and
DOASF. (1985), LEVY, GF.VMII.I. and D ~ A S F . (198.5). ( ~ : M M I I . I . . S ( : I I W A K I Z and DOASI.. ( I W i ) , I%OEK and 1 I ~ x t . 1 ( I W i ) . I%I.SKII. m t l ~ ~ I K : K I . S
(I9Xtl)l antl a sununary of the restriction map variation olnervetl ill alleles samples from mtur;d populations (see text). Along the t o p is a
coordinate in kilobases centered between the t w o trmscriptional units. the ;Ipproxim;tte 1)ountlaries and direction of transcription are
indicated I I V the arrows. n e tloul)lr-heatletI arrow li~l~eletl inversion a indicates the apparent inversion of the region 1)etween the t w o
transcriptional units that was IOuntl in N(X5. T h e restriction sites ( B = H r r e r H I . Bgl = ilrplll, E = E w K I . H = IlitrtlllI and S = . S n / l )
above the bar are monomorphic in the sample. Those restriction sites marked l)elow the bar are poly~norphic in the sample. T h e approximate positions (within designated fragments) antl sizes of large insertions are indicated. T h e sizes o f ' the small insertion and
deletions are as follows: ! t u ( / ) ) 0 . 1 kl). I h / ( r ) 0 . 1 kh, D r / ( g ) 0 . 1 kl). f ) d ( r r ) 0 . 1 kl) and ! ) r / ( / ) 0 . 3 kl).
HLDSOS (1982) provides an estimate of 8 = 4Nep, where Ne is the effective population size and p is the mutatio? rate per nucleotide site to selectively neutral alleles; 8 = 0.006 (Var(8) = 4.9 x 1 O " j or 1.4 x
lo-', assuming either high levels of recombination or no recombination respectively; see HL'DSON 1982). An alternative measure of within population variation at the DNA level is the average pairwise genetic distance among alleles in the sample, r (NEI and LI 1979; NEI and TAJIMA 1981). T h e data in Table 1 yield an estimate of .ir = 0.008. This latter estimate may be more appropriate in situations such as this where some observations are missing (LANGLEY and AQUADRO 1987). When their values are small (as in this case)
6
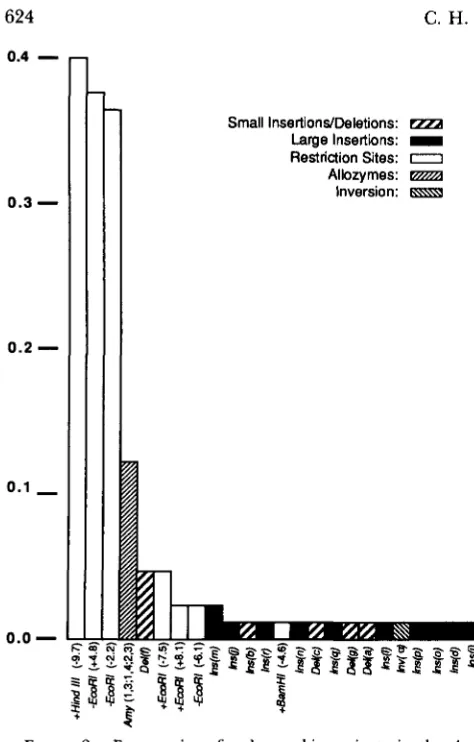
and .ir can be interpreted as the probability that an average nucleotide position in the Amy region would be different in two randomly chosen alleles from natural populations.Insertions and deletions: Figures 1 and 2 show that nine large insertions (>300 bp) were found among the 85 alleles examined. Eight of these were found only once, while the insertion Zns(m) was found
twice among the three alleles collected in Argentina. T h e frequency of large insertion bearing haplotypes was 0.12. T h e large insertions are distributed across the probed region. Although several were found to be inserted into fragments that are partially com- posed of transcriptional unit sequences, they are shown in Figure 1 in the more probable locations outside the two transcritional units. Four small dele- tions were found in the region between the two Amy transcriptional units
(Zns(c)
could lie within the left transcriptional unit). One of these small deletions, Del(/) was found four times (Table 1 and Figure 2), while the remainder were unique. One small inser- tion,Zns(b)
was found near - 7.8 on the map in Figure 1. T h e pattern of restriction fragments from the digests of KC25 was most easily interpreted as an inversion of the map between the two transcrip- tional units (Figure 1).622 C . H. Langley et al.
.CI
8
t
M
f
- 4
3
.B9 e
CI
-3
-2
1
3
s
""""""""""""""""""""""~
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
m m m m m m m m m m m ~ m m m m m ~ m m m m m ~ m ~ m m ~ m m m ~ m m m m m m m m ~ m ~ m ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
u """""""""""""W"~W"""""""
g
G G G t G G G G G G G t G I G G P P G P G G G G G G G G t G G G G G G I G G G G G G G G G
2
iiiiiiiiiiiiiiiiiiiiiiviiiiiiiiiviiiiiiiiiiii
4 4 4 d 4 4 4 4 4 4 4 4 4 4 d 4 4 d 4 4 4 d 4 4 4 d ~ ~ 4 d 4 4 4 4 d ~ 4 ~ 4 4 4 4 4 4
u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u u E
e c c e e e e e e e e e e e e e c e e e ~ e e e ~ e e e e e e e e e e e e ~ ~ ~ e e e e e
~ ~ ~ ~ Y ~ Y Y Y ~ Y Y Y Y Y Y ~ Y Y ~ Y Y Y Y Y Y Y Y Y ~ Y ~ ~ ~ ~ ~ Y ~ ~ Y ~ ~ Y Y Ya a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a m ~ a a a a a a a a a a a a 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 5 1 1 3 3 3 1 3 1 1 1 3 3 1 1 5 3 3 3 3 3 3 3 3 3 3
~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ J J ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~
d
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~a a a a a a a a a a a a a a ~ a a a a m a a a a m a a a a a a a a a a a a a a a a m a m
2 2 2 2 2 2 2 2 2 2 1 2 2 2 2 1 2 2 1 2 2 2 1 1 2 2 2 2 2 2 1 ~ 1 2 2 2 2 2 2 2 2 2 2 ~ 2
< < < < < < < < < < ~ ~ < < < < < < < < < < < 4 < < 4 < < < 4 4 4 < e < < < <
'5 & E
.s
,"a
-
6"
Q J W
3 2
. T . m
C E :
2 2
a
Y
.s
3 .;7 %
$ 2
a 4
.-. i ? E
6'
-
*
2
22
I
2 f d
2:
2 7
e
I h
Z 6
Z I
QJ
.-
ei
~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ W W W ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ W W
, 5 , 5 . 5 C . 5 3 3 3 3 9 3 3 9 9 9 9 9 9 9 9 9 9 2 2 9 9 9 9 9 9 9
m n n ' ~ m u u u o u u u ~ w w w w w w w w w ~ w w w w w w w w
e
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
s
s
s
s
s
s
s
s
s
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
3
3
3
3
3
2
z
z
z
z
z
z
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
~
m m m m m m m
~ ~ ~ ~ ~ ~ ~ C C c C C C C c C c c C C c c C c c C
n r A n n n a n n n n w l w l * C ~ c e C
a n . n m m n n m n n n n n m n m n n m
u u n n n n n n n u ~ 3 u l - l n n n n
a m m m m m m m m d m m m .'""Y.~ 0 0 0 0 0 0 0 C C C C
h
-
v4
-
2 %" h
9,
a s
+ F!
5
.=B
- F ! ,a,
4
-
v>
z
%
-
v
F!
4
* * ~ * ~ o w o ~ ~ ~ ~ o r n ~ - - ~ e 4 - m ~ - r n m o ~ r n ~ r n ~ r n - r n e 4 w ~ ~ ~ w w r n ~ ~ r n
o w w w e 4 m o r n w o ~ ~ ~ e 4 e 4 e 4 ~ w ~ ~ ~ ~ ~ o r n e 4 ~ r n e 4 ~ w e 4 - - - e 4 w * r n ~ ~ o o ~ w
oe4-e4-e4-we4**"e4*e4-e4e4-**o-~e4e4e4*~~*~e4o~e4e4"o-e4~~
0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
1 1 1 1 1 1 1 + 1 1 + 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 I I I I I I I
+ + I + I + I + I I + + + I + I + I I + + + l + l l + + l + l + l I + + + + I + I + + I +
I I I I l l ~ l l l l l l l l l l l l I I I I I I I I I I I I I I I I I I I I I I I I I I
I I I I I I I I I I l l + l l l l l l I I l l + l l l l l l l l l l l l l l I I I I I I I
~~"--"3~*""-3"""-""--"-*~---~"-*~"*"
-
e e 4 e4I I + + + I + I + I I I + + l + + + l + + + + + l + l l l l I + I + + I I I + + I + I I I
. . . .
I I I I I I I I I I I I I I I I I I I I I I I I I I I I I I l l + l l l l l 1 1 + 1 1 1 1
I I I I I I I I I I I I I I I I I I I I I I I I I I I I I l l + l l l l l l I I I I I I I
l l l l l l l l l l l l l + l l + + l l + + + l l + l + + l I ~ I 1 ~ + + + + ~ 1 ~ 1 1 1
~ - * w O e 4 * ~ ~ w - ~ ~ o * ~ ~ m r n - e 4 ~ w * ~ ~ o o o ~ - 3 ~ " ~ e 4 e 4 o ~ ~ ~ o ~ ~ ~ - ~ e 4 e 4 * - e 4 * ~ w ~ m r n ~ w o ~ ~ - ~ ~ ~ ~ -J < < <
Amy Region 623
1
0 0 0 0
w m m w
""
2 2 2 z
""
4 4 4 4 u u u u
z z z z
g ; z z
m m m mz z z x - 1 - 1 - 1 - 1
+:Be-?
d l Z d d
2 3 2 3
d i d <
d 4 d d
Y e 2 2
4 4 4 4
""
2=21
B B % %
s 2 2 2
w o o 0
0 0 0 0
*T-sgl
4 1 ? ?I l l 1
+
I+ +
I I I I
I 1 + 1
-Gee-
-
I
+ 1 + 1
+ I
+ +
I l l 1
I
1 + 1 I
I I I ?
W $ 1
& M d d
s
; $ 8 + 2 . E"""h
$ c 3 , s L A m
g z g g $ j g g
mzz.g-pz
" m s s c z c s s ~ c z $ j ~ z s s
zcnznzzz
"""I.s.g.s
" 3 v1 u c ? h , g # ; c i ? u LL=39 9 g 9 9 9 9 9 9 9 9 9 c ~ " E n n n a n a n
.E
2:
z w my
$ $ b $ $ $ 2 $ $ $ $ < b b + h % $ 5 5 5 r 5 5 5 m o w -
a a
q a a a a a a a a a3 3 3
q a a < 6 4 < < 6 1.p.%*LSX
9 3 3 9 5 9 % 9 9 9 9 5 ' s ' s 3 3 5 5
c E c c e e E$ 3 2
G"3m m m m m m m
m m L C I p i &
6
~
~
6
~
6
~
~
~
6
~
m = o E 5 f z~
~
~
~
~
~
6
6
6
~
6
6
~
~
~
~
~
~
~
6
~
~
6
~
~
z z
I l I l a Y y Y P S P Y ~ P m m ~ u y B ~ ~ ~ ~ ~ ~ m m m~ z z z z z z z z e r
9
5
9
s z z
4 d 6 d.,==9
2 z z z z x z z
r u y y y u g y . % : S , = S Z Z - 0 0 , r d U Oo o o o o o o o o o o o o o o o o o ~ ~ ~2" ~i X5 5 ~s" 2" 2" ~ ~ ~ ~ ~ o o o o o o o
~ ~ ~ z z z z ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ 3 ~ V ~ ~ ~ W W W ~ ~ ~ ~ ~ ~ ~ ~ ~
s
e
0 Z " . x pE z , s
E ..-u2
L.5rGL-Z.9 .E .E .B
.e
4:
E 2
s-
m l ;g 2
& p z
z z z E z z z
hl- 0
E % +
m m m
i
ii
5;
c E . S . , e U Um $
Scq
P
E;;;."S 5
2 2 2
5 5 5 82
g
g
8 8
8
z
z s s s s s
0 0 0 0 3 3 u u u u u u u C ' 9 W e - 98 8
E
3 3 3.2.3 0 a J . Z . 2 m "L. L. ~L sa, e, a 0 0 0 0 L L " 0.4.2..9.9,, ~ ~ U U ~m m m u ~ m ~m Ed m E 3 , . S $ . ' O S j m L - - S E C c ~ ec 4 c 4 u u d ~ ~ ~ ~ ~ ~ ~ z 4 ~ ~ ~ m
v,;--f%2
z a z z z ~ 3 3 3 o o ~ ~ ~ ~ ~ ~ ~$
3 8 : 5;2s#E 5 E 0'3 2 L
m E 0 - 0
m L m L'"2
E
as
z
G Z ' C $ % J X5
- 22,
m5 %
al a c)"$9- f
2
-.i:?
e, 2.M-2qLL v 1 h 9
E m - c ,065
y ) y ) y ) y ) y ) m m y ) y ) y ) m y ) y ) y ) y ) y ) m y ) m y ) y ) y ) m m y ) y ) y ) y ) m ~ * m m y ) y ) m T 2 b M g Y C z E z z z z z z z z z z z z z z z z z z z z z z z z z z z z z z z z z z & u ~ . z 4 ' - 0 G L E ' 3
3s
o = %.- 2 5.9
z z z z z z z z z z z z z z z z z z z z z z ~ z z z z z z.$ E 3 a J w ( O m ~ ~
".-
~ ~aJ L ~x
~ ~ ~+ + l + + + + + + + + l + + + + + l + l + + I I + + + + + + + l l l l l
pKJc4~xo
bC;m
E
Sd m. _ - - * s n
I I I I I I I I I I I I l l + + l l l I I I I I I I I I I I I I I I I I
ss2Eamo,
m z + ; ? z i
2
. k S s : ?
I I I I I I I I r r l l l l l r r l l l l l l l l I l + l ~ l l r l .&";"c;8 ~ l
20 Lm L0
G 2 5 C
o?,c o.s=:,
+ + + + + + + I + I I + I
+ + + + + + + + + + + + + + +
I+
I I+ + + +
~ ~ 1 1 P ~ ~-.- u::.
-
0 f x y) o 2 e 5 z c .- %.E:
"E:
9
" """"h
c X U 4 CUGVNCUG4CUCJNCU
w w m m w m m m m m m G G
~ ~ ~ m m m m m m m m m - - - - m m m m
m m m m
U m I~cI-o~TS5. U'Z Z -113a rTl l~Q a a a c 3 n a F I m m m T Q * %
2(a:gz4g
X X U X X X X X X X X X U U U U X X ~ ~ ~ ~ ~ m w m w O O X~X EX Z~ XC ~ X
i 5 3 2 " s " 2 " ~ 5 2 " x - 5 2 " x x x 5 . 5 ~ d d d d d y y y y u u
z z z z z z z z z z z z z z z z z z
m m m m m m
.- ""A
Lk
5 %
d m m m
- a s = E E E E E:
E : z . s . ~ . ~
0 O U U U U U ' ~ ' ~ VI ~1m m m m m m m m e c E e ~ ~ o w o 0 o oE
i s k
o M - 2I
v e
v
h
-
h2 %
h i?
-
v
t
z
2
p g d 2 - g
+
y ) y ) m y ) y ) y ) m y ) y ) y ) y ) y ) y ) y ) y ) y ) m y ) y ) y ) y ) m ~ m y ) y ) m m y ) ~ m y ) m y ) y ) ~ "'sIYM,d
y ) y ) y ) y ) m m y ) y ) y ) y ) m m y ) y ) m m m y ) y ) y ) y ) y ) m m m m y ) y ) y ) m y ) m m y ) * m
~ Z Z Z Z z Z z Z 2 z Z 2 2 z Z ~ z ~ Z Z Z Z L Z Z ~ ~ Z Z % % L ~ Z z ~ ~ ~ ~ ~ g ; ~
. :
; i-
&?a
g uY m 4 c i
. . .
$ O ~ % z r - m1 1 1 1 1 1 1 + 1 + 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 I I I
.s
c P m W -;
11 x o - m ."9 p ..z
E
Iz"
3%
LI I I I I I I I I I I I I I I I I I I I I I I I I I I I I I I I I I I I z - c s < c h r n a 2 - 0
2 " 8 r & a g z -
I l l + l + l l l f l + l + + + l + l I I + + + + + I + I I + + + + + + 3 -
a&""
U . ~ ~ b " ~ B O
$ 8
f p > % . z
~ = ~ O J w z ~ & 5 + 4 o I .
m t . 0 m O J " m f?-
-
E $ + S g b E . g
e E ~ C I C U " C S O J 5 m ~ m a ~ e u - I " ~ ~ m * C U ~ , ~ ~ ~ - o
&fiT$&
3 ~ ~ ~-2 - m
0 o b
3:
3 . 2 2 . 2 . 2 . z . z m P Y F T z ~ ~ E E E 5 ? c . ? c . ? ~ > ~ " - m > > b m - - w Z - 0 cn c "
sx
5
ga ~ u d 4 4 ~ ~ ~ ~ ~ ~ x 4 4 4 m $ 2 . g s Q a < m - x ~ 2 ~0) ~ ~ ~ ~ t . ~ O J 4 ~ ~ > ~ ~ ~ ~ ~
.% E d 0
C. H. Langley et al. 624
0.4
-
0 . 3
-
0 . 2
-
0.1
-
0.0
-
I
Small InsertiondDeletions: mLarge Insertions:
-
Restriction Sites: 0
Allozymes:
Inversion: tsm
FIGURE 2.-Frequencies of polymorphic variants in the Amy
region (see text). The frequencies are calculated based on numbers of alleles actually scored for each polymorphism. The rarer class for each polymorphism is plotted in descending order of frequency and from left to right along the map in Figure 1 .
(0.88) and the frequency of the next most common allozyme type, Amy’,’ (0.08) are expected from pre- vious studies of variation at the Amy locus in natural populations from North America (SINGH, HICKEY and DAVID 1982).
Nonrandom associations: T h e individual popula- tion samples in this study are inappropriately small for the investigation of linkage disequilibrium. In the two largest samples, RI and KA, there were only two statistically significant cases, both in the larger RI sample. Table 2 shows the three comparisons that were statistically significant when RI and KA were pooled. T h e frequencies of the four polymorphisms [EcoRI ( - 7.8), EcoRI ( - 2.2), Amy and EcoRI (+4.8)] involved in these comparisons did not differ between RI and KA. Therefore pooling of the two population samples may be justified. The direction of the linkage disequilibria were also consistent between these to samples. Amy’ and EcoRI ( - 7.8) were found to be significantly correlated ( P
<
0.02). Amy’ and EcoRI (+4.8) also appeared dependent ( P<
0.01). Amy’ and EcoRI ( - 2.2) were marginally significantly cor-related. Thus the allozyme variation was in linkage disequilibrium with three polymorphic restriction sites closely linked to it. But EcoRI ( - 2.2) and EcoRI
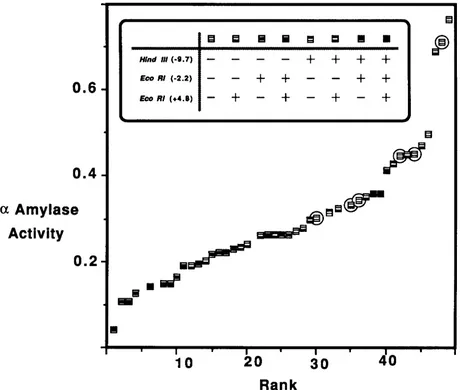
( + 4.8) showed no linkage disequilibrium with one another. However, inspection of Figure 3 shows that the nine haplotypes lacking these two restriction sites are consistently among the highest activities and five of the six alleles bearing electrophoretic variants (Le., nondmy’) are among these nine. Two of these, Amy’P bearing haplotypes, also uniquely lack the EcoRI
( - 7.5). No association was detected between any of the insertionldeletion variation and AMY activity. Since only two of the chromosomes bore inversion Zn(2R)NS there is little statistical power to discern any association between this polymorphism and any of the other variants in the Amy region. Nevertheless, the adjusted activities of these two chromosomes are significantly higher than the average (see below) and they both have the same haplotype (identical for all restriction sites and allozyme phenotype and neither has any insertions or deletions relative to the most common and standard haplotype).
Activity variation: AMY activity varies greatly not only with Amy genotype and the genotypes at other loci but also with developmental stage, tissue and environmental conditioning such as carbohydrate in the medium (DOANE et al. 1987; BENKEL and HICKEY 1986). In this study only the activity of adult females reared on “standard media” was measured and anal- ysis of variance of activity adjusted for soluble protein showed a large line effect ( P
<
0.001). Previous studies indicated that the variant AMY allozyme types (in this case Amy’,’, Amy’,4, Amy2.’) generally exhibit higher adult levels of amylase expression than the average Amy’ (DOANE et al. 1987; BENKEL and HICKEY1986). Figure 3 and Table 1 show that this is true of the six variants found in this study. Analysis of variance of the adjusted adult activities of the 49 lines showed a significant difference
(P
<
0.02) between the average adjusted activities of Amy’ and non-Amy’ lines, 0.277 ? 0.022 ( N = 43) and 0.432 k 0.061 ( N = 6), respectively. Among the restriction map polymorphisms scored for the 49 second chromo- some lines only one showed any association with activity (+EcoRI (+2.2), 0.216 ? 0.020; -EcoRI (+2.2), 0.367 k 0.031; P<
0.005). As mentioned,above the two lines bearing the polymorphic inversion Zn(2R)NS have significantly higher activity (0.522 2
0.168 us. 0.286 k 0.021; P
<
0.04).DISCUSSION
Amy Region 625
TABLE 2
Nonrandom associations between polymorphic sites in the Amy region in the KA and XI population samples
Sites Gametic types
1 2 Population
+ + + -
- +
" D' R PCEcoRI ( - 7.5) Amy' KA" 0 0 1 2 1
RI 0 2 1 8 2
+
1.oo
+
0.67 1 0.03Pooled 0 2 3 0 3
+
1.00+
0.603 0.02EcoRI ( - 2.2) Amy' KA 7 05 1 - 1.00 -0.312 0.46
RI 8 0 1 0 4 - 1.00 - 0.356 0.25
Pooled 15 0 15 5 - 1
.oo
- 0.354 0.06Amy' EcoRI ( + 4.8) KA 7 5 0 1 - 1
.oo
-0.312 0.46RI 1 4 4 0 4 - 1.00 - 0.624 0.01
Pooled 2 1 9 0 5 - 1.00 - 0.624 0.01
D' is a normalized measure of nonrandom association proposed by LEWONTIN (1964). R is the correlation coefficient. P indicates the " No estimation of linkage disequilibrium (D' or R ) was made because of monomorphisms at EcoRI ( - 7.5).
probability of the observed or greater level of nonrandom association as determined from Fisher's exact test.
0.6
0.4
a
Amylase
Activity
0.2
- + + - -
+ - + - + - +
Rank
FIGURE 3.-Distribution of adjusted amylase activities (see text). The average adult female activity ( AOD at 550 nm adjusted for total protein) is plotted against rank of adjusted activity. The restriction site haplotype at Hind111 ( - 9.7), EcoRI ( - 2.2) and EcoRI ( + 4.8) is indicated by the pattern of fill in the individual boxes. The circles (0) surround those alleles with a non-Amy' allozyme phenotype, i e . ,
626 C. H. Langley et al.
that of the Hsp70 region in that all chromosomes examined have the same basic structure of two di- vergent transcription units (sharing great sequence similarity) separated by several kilobases of unique sequence (GEMMILL, LEVY and DOANE 1985; LEVY, GEMMILL and DOANE 1985; GEMMILL, SCHWARTZ and DOANE 1986; BOER and HICKEY 1986). The apparent inversion of the interlocus region in one of the chromosomes we have analyzed (NC25, see Figure l ) , is analogous to the process suggested by LEIGH BROWN and ISH-HORWITZ (1981) to explain the con- certed evolution of Hsp7U and Amy (PAYANT et al.
1988) transcriptional units over longer evolutionary periods. This inverted and divergently transcribed gene duplication might represent an evolved econ- omy of regulatory mechanism, e.g., shared 5' se- quences. O r it may be a more genetically stable gene duplication, e.g., less likely than tandemly duplicated genes to asymmetrically pair leading to unequal ex- change.
Large insertions: The Adh, Hsp7U and white regions of D. melanogaster from natural populations often contain a large insertion, greater than one kilobase in size (LANGLEY, MONTGOMERY and QUATTLEBAUM
1982; LEIGH BROWN 1983; AQUADRO etal. 1986; CROSS and BIRLEY 1986; LANGLEY and AQUADRO 1987). These large insertions from Adh and Hsp70 were found to be members of middle repetitive dispersed families (presumed transposable elements) previously cloned and characterized from laboratory stocks (LEIGH BROWN 1983; AQUADRO et al. 1986). Labora- tory stocks are known to carry many such elements and many spontaneous mutations in laboratory stocks are due to the insertion of these elements (FINNEGAN 1986). It is also known that most insertions of these transposable elements throughout the genome occur at low frequencies in natural populations of D . me- lanogaster (MONTGOMERY and LANGLEY 1983; MONT- GOMERY, CHARLESWORTH and LANGLEY 1987; LEIGH
BROWN and Moss 1987).
T h e Amy region is typical in that large insertions were found in many alleles, 10 of 85, and that these were all unique except for one, Zns(m), found in two alleles from the same population in Argentina (see below). T h e distribution of these large insertions within the Amy region is not remarkable (see Figure
1). Although none could be unambiguously localized within either of the transcriptional units, several are definitely within one kilobase of the transcriptional units. No relationship between presence of large insertions and low adjusted a-amylase activity is ap- parent. If such large insertions drastically interrupted a gene's expression, such a severe mutation will certainly be detected by natural selection and be removed quickly (HALDANE 1927). Presumably this is why such disruptive insertions (and the clearly mutant alleles they cause) are rarely seen in wild-type chro-
mosomes (VOELKER et al. 1980; LANGLEY et al. 1981), but are the staple of laboratory Drosophila genetics. T h e insertions in the flanking regions are clearly milder mutations, if mutant at all, with respect to their effect on adult a-amylase activity. Their frequencies are consistently low indicating that some force is preventing alleles bearing such insertions from rising to higher frequencies typical of other kinds of varia- tion in the region, e.g., restriction site polymorphisms, allozymes and some small insertion/deletion variants. GOLDING, AQUADRO and LANGLEY (1986) showed that the large insertions in the Adh region could not be interpreted as a simple equilibrium between insertion and copy-number independent excision. Two hypoth- esis (not necessarily mutually exclusive) might be put forward to account for this situation. The first hy- pothesis is that these transposable element insertions are indeed deleterious mutants and natural selection does detect them. Transposable elements inserted
into the regions flanking genes may be less deleterious than those that inactivate the gene and thus they are found in small samples such as ours. Insertions having a small effect on level, timing or distribution of gene expression at a nearby locus might have selection coefficients as small as l o p 4 or lo-' and still be held at low frequencies by mutation-selection balance in populations as large as that of D . melanogaster (LANG-
LEY et al. 1981). But recent results from the survey of the numbers of transposable elements on the X
and autosomes call into question the hypothesis of natural selection against a very subtle phenotype. MONTGOMERY, CHARLESWORTH and LANGLEY (1987) found that for two of the three transposable element families surveyed there was no apparent reduction of the numbers on the X chromosome as expected for sex linked, partially recessive deleterious mutants (HALDANE 1927). Based on these results and other relevant data, these authors suggested a second hy- pothesis: that transposable elements might be elimi- nated by a process associated with meiotic recombi- nation that occurs only in females in D. melanogaster and thus is the same for x's and autosomes. Meiotic asymmetric pairing and unequal exchange between transposable element insertions is known to occur in Drosophila as well as yeast (GREEN 1959; JUDD 1959;
GOLDBERG et al. 1983; ROEDER 1983; DAVIS, SHEN and
J U D D 1987; JINKS-ROBERTSON and PETES 1986; LICH- TEN, BORTS and HABER 1987). The products of such
exchanges are either duplications, deficiencies or translocations; the great majority of which are likely to suffer relatively severe natural selection. Further studies at specific loci and on the cytogenetic level can address this issue directly and quantitatively.
Amy Region 627
found more than once in different populations. Al- though such small deletions may arise recurrently in different populations there is no evidence to suggest that D e l 0 is not a geographically widespread poly- morphism of unique origin. This situation is similar to that found at the other loci studied, where small deletion/insertion polymorphisms are found at inter- mediate frequencies and 5‘ to transcriptional units (LEIGH BROWN 1983; KREITMAN and AGUADJ? 1986; AQUADRO et al. 1986; LANGLEY and AQUADRO 1987). Two types of evidence suggest that these small length variants may not be completely immune to culling by some form of genetic or phenotypic selection. Al- though occasionally a small insertion or deletion may reach intermediate frequencies such as D e l 0 in the Adh region studied by AQUADRO et al. (1986), most are rare if not unique. As noted by BARRIE, JEFFREYS
and SCOTT (1981) for the hemoglobin region of primates and LANGLEY, MONTGOMERY and QUATTLE- BAUM (1982) for the Adh region of Drosophila, the
sequences flanking these genes (and others) are con- served in terms of spacing and homology. This is inconsistent with rapid and random sequence turn- over interstitial to transcriptional units. T h e pheno- typic and evolutionary significance of sequence varia- tion in the interstitial regions between many eukar- yotic genes is somewhat of a paradox in that most of these regions are rarely required for “normal” expression yet often have mild quantitative effects (HAZELRIGG, LEVIS, and RUBIN 1984; LAURIE-AHL-
BERG and STAM 1987). While the sequences in flank-
ing regions generally evolve more rapidly than coding sequences, evidence of conservation can be found.
a-Amylase-dlozymes
and
activity: Although the number of polymorphic sites in this study is too few to make a clear analogy, the comparison with Adh seems striking in that the allozyme variation appears to be strongly associated with a change in enzyme activity and with polymorphisms at the DNA level (linkage disequilibrium). It has been known that the Amy haplotypes bearing the rarer, nondmy’ allo- zymes were generally associated with higher adult enzyme activity. From the data presented above and depicted in Figure 3 it can be suggested that this common property has a single origin, since all six non-Amy’ haplotypes were associated with the absence of the EcoRI (-2.2) and Hind111 (-9.4) sites. And five of the six were also found to lack EcoRI (+4.8) site. As KREITMAN (1983) and many others before have argued, amino acid polymorphisms, especially those that involve changes in net charge, may rep- resent selectively favored variants in a regime of natural selection that discerns significant differences among all amino acid changes. Another curious ob- servation is that allozyme variants at Adh and Amy are strongly associated with variation in levels of expression and with flanking restriction site poly-morphisms. These charged amino acid polymorph- isms are more likely to affect the function of the molecule. Note, however, that the evidence indicates that the associated differences in enzyme activities are largely attributable to variation in the amounts of protein expression rather than specific activity of the allozymes (LAURIE-AHLBERG 1985; DOANE et al.
1987; BENKEL and HICKEY 1986). T h e results of the studies from these two loci and other recent reports argue for continuing skepticism toward the view that polymorphic allozymes are free from the forces of natural selection (KIMURA 1983; GILLESPIE 1987).
Restriction site polymorphisms: T h e estimated level of nucleotide variation (either 8 = 4Nep. or expected heterozygosity) based on restriction enzyme site polymorphism in the Amy region (-0.006) is similar to that observed at other loci, such as Adh (0.006), Hsp70 (0.002), or white (0.014). Thus an average pair of randomly chosen alleles will differ at 90 nucleotide positions over the 15-kb Amy region we have examined (Figure 1). Until more detailed data are available little can be concluded about the Amy region from these crude measures. It does
appear that the estimates of variation in D . melano- gaster are converging toward a value near 0 = 0.008. It remains to be seen if this value applies to all regions of the D . melanogaster genome and to other Drosophila species.
We thank W. W. DOANE, D. A. HICKEY, R. M. GEMMILL and J.
N. LEVY for kindly sharing their clones, stocks and constructive criticisms of this paper.
LITERATURE CITED
ABE, K., 1958 Genetical and biochemical studies on amylase in
Drosophila melanogarter. Jpn. J. Genet. 33: 138-145.
ABRAHAM, I., and W. W. DOANE, 1978 Genetic regulation of tissue-specific expression of amylase structural gene in Droso- phila melanogaster. Proc. Natl. Acad. Sci. USA 75: 4446-4450. AQUADRO, C. F., S. F. DEESE, M. M. BLAND, C. H. LANGLEY and
C. C. LAURIE-AHLBERG, 1986 Molecular population genetics of the alcohol dehydrogenase gene region of Drosophila melan-
ogaster. Genetics 114: 1 165- 1 190.
BARRIE, P. A., A. J. JEFFREYS and A. F. SCOTT, 1981 Evolution of the beta-globin gene cluster in man and primates. J. Mol. Biol. 149: 319-336.
BENKEL, B., S . ABUKASHAWA, P. H. BOER and D. A. HICKEY, 1988 Molecular cloning of DNA complementary to Drosophila melanogaster alpha-amylase mRNA. Genome 29: 5 10-5 15. BENKEL, B. F., and D. A. HICKEY, 1986 The interaction of genetic
and environmental factors in the control of amylase gene expression in Drosophila melanogaster. Genetics 114: 943-954. BOER, P. H., and D. A. HICKEY, 1986 The alpha-amylase gene in
Drosophila melanogaster. Nucleotide sequence, gene structure and expression motifs. Nucleic Acids Res. 14: 8399-841 1. CROSS, S. R. H., and A. J. BIRLEY, 1986 Restriction endonuclease
map variation in the Adh region in populations of Drosophila melanogarter. Biochem. Genet. 2 4 415-433.
628 C. H. Langley et al.
DEJONG, G. A., A. J. W. HOORN, G. E. W. THORIG and W. SCHARLOO, 1972 Frequencies of amylase variants in Droso- phila melanogaster. Nature 238: 452-453.
DOANE, W. W., 1969a Drosophila amylases and problems in cellular differentiation. In RNA in Development, Edited by E. W. HAWLEY. University of Utah Press, Salt Lake City, Utah. DOANE, W. W., 1969b Amylase variants in Drosophila melanogaster:
linkage studies and characterization of enzyme extracts. J. Exp. Zool. 171: 321-342.
DOANE, W. W., R. M. GEMMILL, P. E. SCHWARTZ, S. A. HAWLEY and R. A. NORMAN, 1987 Structural organization of the alpha-amylase gene locus in Drosophila melanogaster and Dro- sophila miranda. Isozymes Curr. Top. Biol. Med. Res. 14: 229- 266.
EWENS, W. J., R. S. SPIELMAN and H. HARRIS, 1981 Estimation of genetic variation at the DNA level from restriction endo- nuclease data. Proc. Natl. Acad. Sci. USA 78: 3748-3750.
FINNEGAN, D. J., 1986 Transposable elements in eukaryotes. Int. Rev. Cytol. 93: 281-326.
GEMMILL, R. M., J. N. LEVY and W. W. DOANE, 1985 Molecular cloning of alpha-amylase genes from Drosophila melanogarter.
I. Clone isolation by use of mouse probe. Genetics 110: 299- 312.
GEMMILL, R. M., P. E. SCHWARTZ and W. W. DOANE, 1986 Struc- tural organization of the Amy locus in seven strains of Drosophila melanogaster. Nucleic Acids Res. 14: 5337-5352.
GILLESPIE, J. H., 1987 Molecular evolution and the neutral allele theory. Oxf. Surv. Evol. Biol. 4: 10-37.
GOLDBERG, M. L., J.-Y. SHEEN, W. J. GEHRING and M. M. GREEN,
1983 Unequal crossing-over associated with asymmetrical
synapsis between nomadic elements in the Drosophila melano- gaster genome. Proc. Natl. Acad. Sci. USA 80: 5017-5021.
GOLDING, G. B., C. F. AQUADRO and C. H. LANGLEY, 1986 Se- quence evolution within populations under multiple types of mutation. Proc. Natl. Acad. Sci. USA 83: 427-431.
GREEN, M. M., 1959 Non-homologous pairing and crossing over in Drosophila melanogaster. Genetics 44: 1243-1256.
HALDANE, J. B. S., 1927 A mathematical theory of natural and artificial selection. Part V. Selection and mutation. Proc. Camb. Philos. SOC. 23: 838-844.
HAZELRIGG, T., R. LEVIS and G. M. RUBIN, 1984 Transformation of white DNA in Drosophila: dosage compensation, reste inter- action, and position effects. Cell 36: 469-481.
HICKEY, D. A., 1977 Selection for amylase allozymes in Drosophila melanogaster. Evolution 31: 800-809.
HICKEY, D. A., 1979 The geographical pattern of enzyme poly- morphism in D . melanogaster. Genetics 51: 1-4.
HICKEY, D. A , , 1981 Regulation of amylase activity in Drosophila melanogaster: variation in the number of enzyme molecules produced by different amylase genotypes. Biochem. Genet.
HICKEY, D. A,, and B. BENKEL, 1982 Regulation of amylase expression in Drosophila melanogaster: effects of dietary carbo- hydrate. Biochem. Genet. 20: 1 1 17-1 129.
HOORN, A. J. W., and W. SCHARLOO, 1978 The functional significance of amylase polymorphism in Drosophila melanogas-
ter. V. The effect of food components on amylase and a-glucosidase activity. Genetica 49: 181-187.
HUDSON, R., 1982 Estimating genetic variability with restriction endonucleases. Genetics 100: 71 1-719.
JINKS-ROBERTSON, S., and T. D. PETES, 1986 Chromosomal trans- locations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics 114: 731-752.
JUDD, B. H., 1959 Studies on some position pseudoalleles at the
white locus region in Drosophila melanogaster. Genetics 44: 34- 42.
19: 783-796.
KIKKAWA, H., and K. ABE, 1960 Genetic control of amylase in
Drosophila melanogaster. Annot. Zool. Jpn. 33: 14-23.
KIMURA, M., 1983 The Neutral Theoly of Molecular Evolution. Cam- bridge University Press, Cambridge.
KLARENBERG, A. J., and W. SCHARLOO, 1986 Nonrandom asso- ciation between structural Amy and regulatory map variants in
Drosophila melanogaster. Genetics 1 1 4 875-884.
KREITMAN, M., 1983 Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304:
KREITMAN, M., and M. A G U A D ~ , 1986 Excessive polymorphism at the Adh locus in Drosophila melanogaster. Genetics 114: 93- 110.
LANGLEY, C. H., and C. F. AQUADRO, 1987 Restriction map variation in natural populations of Drosophila melanogaster: white
locus region. Mol. Biol. Evol. 4: 651-663.
LANGLEY, C. H., K. ITO and R. A. VOELKER, 1977 Linkage disequilibrium in natural populations of Drosophila melanogas- ter: seasonal variation. Genetics 86: 447-454.
LANGLEY, C. H., E. A. MONTGOMERY and W. F. QUATTLEBAUM,
1982 Restriction map variation in the Adh region of Droso-
phila. Proc. Natl. Acad. Sci. USA 79: 5631-5635.
LANGLEY, C. H., Y. N. TOBARI and K. KOJIMA, 1974 Linkage disequilibrium in natural populations of Drosophila melanogas- ter. Genetics 78: 92 1-936.
LANGLEY, C. H., R. A. VOELKER, A. J. LEIGH BROWN, S . OHNISHI, B. DICKSON and E. MONTGOMERY, 1981 Null allele frequen- cies at allozyme loci in natural populations of Drosophila melanogaster. Genetic 99: 151-156.
LAURIE-AHLBERG, C. C., 1985 Genetic variation affecting the expression of enzyme-coding genes in Drosophila: an evolu- tionary perspective. Isozymes Curr. Top. Biol. Med. Res. 12:
LAURIE-AHLBERG, C. C., and L. F. STAM, 1987 Use of P-element transformation to identify the molecular basis of naturally occurring variants affecting Adh expression in Drosophila me- lanogaster. Genetics 115: 121-140.
LAURIE-AHLBERG, C. C., G. MARONI, G. C. BEWLEY, J. C. LUCCHESI and B. S. WEIR, 1980 Quantitative genetic variation of en- zyme activities in natural populations of Drosophila melanogaster.
Proc. Natl. Acad. Sci. USA 77: 1073-1077.
LEIGH BROWN, A. J., 1983 Variation at the 87A heat-shock locus in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 80: 5350- 5354.
LEIGH BROWN, A. J., and D. ISH-HOROWICZ, 1981 Evolution of the 87A and 87C heat-shock loci in Drosophila. Nature 290:
LEIGH BROWN, A. J., and J. E. Moss, 1987 Transposition of the I element and copia in a natural population of Drosophila
melanogaster. Genet. Res. 49: 121-128.
LEVY, J. N., R. M. GEMMILL and W. W. DOANE, 1985 Molecular cloning of alpha-amylase genes from Drosophila melanogaster.
11. Clone organization and verification. Genetics 110: 313- 324.
LEWONTIN, R. C., 1964 The interaction of selection and linkage. I. General considerations: heterotic models. Genetics 49: 3 1- 41.
LICHTEN, M., R. H. BORTS and J. E. HABER, 1987 Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics
LOWRY, 0. H., N. J. ROSEBROUGH, A. L. FARR and R. J. RANDALL,
1951 Protein measurement with the Fohn phenol reagent.
J. Biol. Chem. 193: 265-275.
MCCUNE, T. B., 1969 A genetic study of the amylase isozyme polymorphism in Drosophila melanogaster. Ph.D. dissertation, University of Hawaii.
412-417.
33-88.
677-682.
Amy Region 629
MONTGOMERY, E. A., and C. H. LANGLEY, 1983 Transposable elements in Mendelian populations. 11. Distribution of three copia-like elements in a natural population of Drosophila melun- ogaster. Genetics 104: 473-483.
MONTGOMERY, E., B. CHARLESWORTH and C. H. LANGLEY, 1987 A test for the role of natural selection in the stabilization of transposable element copy number in a population of Droso- phila melanogaster. Genet. Res. 49: 3 1-4 1 .
MUKAI, T., T. K. WATANABE and 0. YAMAGUCHI, 1974 The genetic structure of natural populations of Drosophila melano- gaster. XII. Linkage disequilibrium in a large local population. Genetics 77: 771-793.
NEI, M., and W.-H. LI, 1979 Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269-5273.
NEI, M., and F. TAJIMA, 1981 DNA polymorphism detectable by restriction endonucleases. Genetics 97: 145-163.
PAYANT, V., S. ABUKASHAWA, M. SASSERVILLE, B. F. BENKEL, D. A.
HICKEY and J. DAVID, 1988 Evolutionary conservation of the structure and regulation of amylase genes among eight species of Drosophila melanogaster species subgroup. Mol. Biol. Evol. In press.
ROEDER, G. S., 1983 Unequal crossing over between yeast trans- posable elements. Mol. Gen. Genet. 190 117-121.
SCHAEFFER, S. W., C. F. AQUADRO and C. H. LANGLEY,
1988 Restriction map variation in the Notch region of Dro-
sophila melanogaster. Mol. Biol. Evol. 5: 30-40.
SCHWARTZ, P. E., 1986 Molecular analysis of the Amy locus from strains of Drosophila melanogaster with reduced amylase activity. Master's thesis, Arizona State University.
SINGH, R. S., and M . B. COULTHART, 1982 Genic variation in abundant soluble proteins of Drosophila melanogaster and Dru-
sophila pseudoobscura. Genetics 102: 437-453.
SINCH, R. S., D. A. HICKEY and J. DAVID, 1982 Genetic differ- entiation between geographically distant populations of' Dro- sophila melanogaster. Genetics 101: 235-256.
VOELKER, R. A., C. H. LANGLEY, A. J . LEIGH BROWN, S. OHNISHI, B. DICKSON, E. MONTGOMERY and S. C. SMITH, 1980 Enzyme null alleles in natural populations of Drosophila melanugaster. Frequencies in a North Carolina population. Proc. Natl. Acad. Sci. USA 77: 1091-1095.
YAMAZAKI, T., and Y. MATSUO, 1984 Genetic analysis of natural populations of Drosophila melanugaster in Japan. 111. Genetic variability of inducing factors of amylase and fitness. Genetics
108: 223-235.
YAMAZAKI, T., Y. MATSUO, Y. INOUE and Y. MATSUO,
1984 Genetic analysis of natural populations of Dro.wphiln
melanogaster in Japan. I. Protein polymorphism, lethal gene, sterility gene, inversion polymorphism, and linkage disequilib- rium. Jpn. J. Genet. 59: 33-49.