5-(4-Methylpiperazin-1-yl)-2-nitroaniline
Chang-jun Luan, Cheng Guo,* Wei Wang, Jian-qiang Wang and Ren-jun Du
Department of Applied Chemistry, College of Science, Nanjing University of Technology, Nanjing 210009, People’s Republic of China
Correspondence e-mail: guocheng@njut.edu.cn
Received 7 April 2010; accepted 30 April 2010
Key indicators: single-crystal X-ray study;T= 293 K; mean(C–C) = 0.004 A˚;
Rfactor = 0.064;wRfactor = 0.192; data-to-parameter ratio = 13.5.
In the title compound, C11H16N4O2, the dihedral angle between the benzene ring and the plane of the four carbon atoms in the piperazine ring is 12.17 (3); the latter ring adopts a chair conformation. An intramolecular N—H O hydrogen bond generates an S(6) ring. In the crystal, the molecules are linked by N—H N hydrogen bonds, forming chains.
Related literature
For bond-length data, see: Allenet al.(1987). For the synthetic procedure and use of the title compound as an intermediate in the synthesis of tyrosine kinase inhibitors, see: Renhoweet al. (2009).
Experimental
Crystal data
C11H16N4O2
Mr= 236.28 Monoclinic,P21=c
a= 11.027 (2) A˚
b= 6.121 (1) A˚
c= 17.524 (4) A˚ = 103.79 (3) V= 1148.7 (4) A˚3
Z= 4
MoKradiation
= 0.10 mm1
T= 293 K
0.300.200.05 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Absorption correction: scan (Northet al., 1968)
Tmin= 0.971,Tmax= 0.995
2205 measured reflections
2090 independent reflections 1358 reflections withI> 2(I)
Rint= 0.042
3 standard reflections every 200 reflections
intensity decay: 1%
Refinement
R[F2> 2(F2)] = 0.064
wR(F2) = 0.192
S= 1.01 2090 reflections
155 parameters
H-atom parameters constrained
max= 0.25 e A˚3 min=0.18 e A˚
3
Table 1
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
N3—H3C N1i
0.86 2.39 3.156 (4) 148
N3—H3D O1 0.86 2.06 2.669 (4) 127
Symmetry code: (i)xþ2;yþ1 2;zþ
1 2.
Data collection: CAD-4 Software (Enraf–Nonius, 1985); cell refinement: CAD-4 Software; data reduction: XCAD4 (Harms & Wocadlo, 1995); program(s) used to solve structure: SHELXS97
(Sheldrick, 2008); program(s) used to refine structure:SHELXL97
(Sheldrick, 2008); molecular graphics:SHELXTL(Sheldrick, 2008); software used to prepare material for publication:SHELXTL.
The authors thank the Center of Test and Analysis, Nanjing University, for support.
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IM2192).
References
Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987).J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
Enraf–Nonius (1985).CAD-4 Software. Enraf–Nonius, Delft, The Nether-lands.
Harms, K. & Wocadlo, S. (1995).XCAD4. University of Marburg, Germany. North, A. C. T., Phillips, D. C. & Mathews, F. S. (1968).Acta Cryst.A24, 351–
359.
Renhowe, P. A., Pecchi, S., Shafer, C. M., Machajewski, T. D., Jazan, E. M., Taylor, C., Antonius-McCrea, W., McBride, C. M., Frazier, K., Wiesmann, M., Lapointe, G. R., Feucht, P. H., Warne, R. L., Heise, C. C., Menezes, D., Aardalen, K., Ye, H., He, M., Le, V., Vora, J., Jansen, J. M., Wernette-Hammond, M. E. & Harris, A. L. (2009).J. Med. Chem.52, 278–292. Sheldrick, G. M. (2008).Acta Cryst. A64, 112–122.
Acta Crystallographica Section E
Structure Reports Online
supporting information
Acta Cryst. (2010). E66, o1268 [https://doi.org/10.1107/S1600536810015953]
5-(4-Methylpiperazin-1-yl)-2-nitroaniline
Chang-jun Luan, Cheng Guo, Wei Wang, Jian-qiang Wang and Ren-jun Du
S1. Comment
The title compound, (I), has been reported as an intermediate for the synthesis of novel tyrosine kinase inhibitors
(Renhowe, P. A. et al., 2009). We herein report its crystal structure.
In the molecular structure of (I), (Fig.1), bond lengths (Allen et al., 1987) and angles are within normal ranges. N2, N3
and N4 atoms are almost coplanar with the benzene ring to which they are bonded [deviations of 0.078 (1), 0.052 (1) and
0.078 (1) Å]. The plane of C2—C3—C4—C5 is nearly parallel with the benzene ring plane (the torsion angle is 12.17 (3)
°). By contrast, due to the piperazine moiety adopting a chair conformation N1—C2—C5 and N2—C3—C4 form two
separate planes with torsion angle of 45.87 (2) ° and 25.92 (3) °, respectively, with respect to the benzene ring. The
crystal structure of the title compound exhibits N—H···O, C—H···O, and N—H···N intra- and intermolecular hydrogen
bonds to form a three dimensional network.
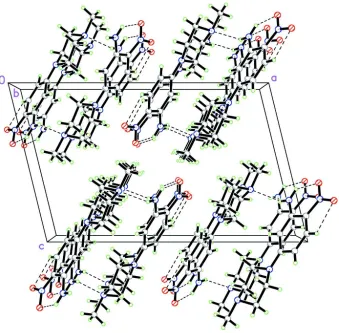
As can be seen from the packing diagram, (Fig. 2), the molecules are stacked along the b axis.
S2. Experimental
The title compound, (I) was prepared by a literature method (Renhowe, P. A. et al., 2009). Crystals suitable for X-ray
analysis were obtained by dissolving (I) (0.5 g) in methanol (20 ml) and evaporating the solvent slowly at room
temperature for about 7 d.
S3. Refinement
H atoms were positioned geometrically, with N—H = 0.86 Å, C—H = 0.93 Å for aromatic H, 0.97 Å for methylene and
0.96 Å for methyl groups. Refinement was performed using a riding model with Uiso(H) = xUeq(C), where x = 1.5 for
Figure 1
The molecular structure of (I), with the atom-numbering scheme. Displacement ellipsoids are drawn at the 50%
probability level.
Figure 2
[image:3.610.137.476.286.619.2]5-(4-Methylpiperazin-1-yl)-2-nitroaniline
Crystal data
C11H16N4O2
Mr = 236.28 Monoclinic, P21/c
a = 11.027 (2) Å b = 6.121 (1) Å c = 17.524 (4) Å β = 103.79 (3)° V = 1148.7 (4) Å3
Z = 4 F(000) = 504
Dx = 1.366 Mg m−3
Melting point: 428 K
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 25 reflections θ = 9–13°
µ = 0.10 mm−1
T = 293 K Block, yellow
0.30 × 0.20 × 0.05 mm
Data collection
Enraf–Nonius CAD-4 diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
ω/2θ scans
Absorption correction: ψ scan (North et al., 1968)
Tmin = 0.971, Tmax = 0.995
2205 measured reflections
2090 independent reflections 1358 reflections with I > 2σ(I) Rint = 0.042
θmax = 25.3°, θmin = 1.9°
h = 0→13 k = 0→7 l = −21→20
3 standard reflections every 200 reflections intensity decay: 1%
Refinement
Refinement on F2
Least-squares matrix: full R[F2 > 2σ(F2)] = 0.064
wR(F2) = 0.192
S = 1.01 2090 reflections 155 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained w = 1/[σ2(F
o2) + (0.1P)2 + 0.3P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.25 e Å−3
Δρmin = −0.18 e Å−3
Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4
Extinction coefficient: 0.038 (6)
Special details
Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
N1 0.6711 (2) 0.0465 (4) 0.16875 (13) 0.0438 (6)
C1 0.5525 (3) 0.1090 (6) 0.11543 (19) 0.0603 (9)
H1A 0.5683 0.2064 0.0760 0.090*
H1B 0.5007 0.1809 0.1447 0.090*
H1C 0.5107 −0.0193 0.0907 0.090*
N2 0.8586 (2) 0.0019 (4) 0.31264 (13) 0.0397 (6)
O2 1.2787 (2) −0.3532 (4) 0.58981 (13) 0.0662 (7)
C2 0.7330 (3) 0.2397 (5) 0.20898 (17) 0.0484 (8)
H2A 0.6826 0.3007 0.2422 0.058*
H2B 0.7409 0.3493 0.1705 0.058*
C3 0.8606 (3) 0.1827 (5) 0.25841 (16) 0.0466 (8)
H3A 0.9143 0.1440 0.2240 0.056*
H3B 0.8960 0.3103 0.2883 0.056*
N3 1.2683 (2) 0.2156 (4) 0.45524 (16) 0.0613 (8)
H3C 1.2595 0.3315 0.4267 0.074*
H3D 1.3359 0.1954 0.4909 0.074*
C4 0.7741 (3) −0.1801 (5) 0.28094 (18) 0.0478 (8)
H4A 0.7577 −0.2645 0.3242 0.057*
H4B 0.8149 −0.2755 0.2506 0.057*
N4 1.2841 (2) −0.1869 (5) 0.55073 (15) 0.0527 (7)
C5 0.6513 (3) −0.1026 (5) 0.22932 (17) 0.0506 (8)
H5A 0.6035 −0.2276 0.2048 0.061*
H5B 0.6033 −0.0292 0.2615 0.061*
C6 0.9667 (2) −0.0433 (4) 0.36840 (15) 0.0367 (7)
C7 1.0669 (2) 0.1023 (5) 0.38534 (15) 0.0398 (7)
H7A 1.0610 0.2307 0.3563 0.048*
C8 1.1758 (2) 0.0647 (5) 0.44396 (16) 0.0415 (7)
C9 1.1820 (2) −0.1317 (5) 0.48727 (15) 0.0422 (7)
C10 1.0839 (3) −0.2817 (5) 0.46870 (17) 0.0475 (8)
H10A 1.0901 −0.4120 0.4966 0.057*
C11 0.9799 (3) −0.2428 (5) 0.41112 (17) 0.0444 (7)
H11A 0.9169 −0.3471 0.3995 0.053*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
C8 0.0391 (15) 0.0386 (16) 0.0455 (16) −0.0015 (13) 0.0075 (13) −0.0057 (13) C9 0.0398 (15) 0.0469 (17) 0.0377 (15) 0.0063 (13) 0.0051 (12) 0.0029 (13) C10 0.0470 (17) 0.0444 (18) 0.0510 (17) 0.0014 (14) 0.0116 (14) 0.0123 (14) C11 0.0391 (15) 0.0395 (16) 0.0527 (17) −0.0031 (13) 0.0071 (13) 0.0101 (14)
Geometric parameters (Å, º)
N1—C5 1.455 (3) N3—H3C 0.8600
N1—C2 1.460 (4) N3—H3D 0.8600
N1—C1 1.466 (3) C4—C5 1.514 (4)
O1—N4 1.246 (3) C4—H4A 0.9700
C1—H1A 0.9600 C4—H4B 0.9700
C1—H1B 0.9600 N4—C9 1.422 (4)
C1—H1C 0.9600 C5—H5A 0.9700
N2—C6 1.377 (3) C5—H5B 0.9700
N2—C3 1.462 (3) C6—C7 1.395 (4)
N2—C4 1.473 (3) C6—C11 1.421 (4)
O2—N4 1.236 (3) C7—C8 1.401 (4)
C2—C3 1.507 (4) C7—H7A 0.9300
C2—H2A 0.9700 C8—C9 1.415 (4)
C2—H2B 0.9700 C9—C10 1.396 (4)
C3—H3A 0.9700 C10—C11 1.356 (4)
C3—H3B 0.9700 C10—H10A 0.9300
N3—C8 1.355 (3) C11—H11A 0.9300
C5—N1—C2 106.8 (2) N2—C4—H4B 109.1
C5—N1—C1 111.2 (2) C5—C4—H4B 109.1
C2—N1—C1 109.8 (2) H4A—C4—H4B 107.8
N1—C1—H1A 109.5 O2—N4—O1 120.6 (3)
N1—C1—H1B 109.5 O2—N4—C9 119.7 (3)
H1A—C1—H1B 109.5 O1—N4—C9 119.7 (3)
N1—C1—H1C 109.5 N1—C5—C4 111.3 (2)
H1A—C1—H1C 109.5 N1—C5—H5A 109.4
H1B—C1—H1C 109.5 C4—C5—H5A 109.4
C6—N2—C3 118.0 (2) N1—C5—H5B 109.4
C6—N2—C4 118.6 (2) C4—C5—H5B 109.4
C3—N2—C4 115.7 (2) H5A—C5—H5B 108.0
N1—C2—C3 110.8 (2) N2—C6—C7 122.0 (2)
N1—C2—H2A 109.5 N2—C6—C11 120.6 (2)
C3—C2—H2A 109.5 C7—C6—C11 117.4 (2)
N1—C2—H2B 109.5 C6—C7—C8 123.2 (3)
C3—C2—H2B 109.5 C6—C7—H7A 118.4
H2A—C2—H2B 108.1 C8—C7—H7A 118.4
N2—C3—C2 113.1 (2) N3—C8—C7 118.6 (3)
N2—C3—H3A 109.0 N3—C8—C9 124.2 (2)
C2—C3—H3A 109.0 C7—C8—C9 117.2 (2)
N2—C3—H3B 109.0 C10—C9—C8 119.9 (2)
H3A—C3—H3B 107.8 C8—C9—N4 123.3 (3)
C8—N3—H3C 120.0 C11—C10—C9 121.9 (3)
C8—N3—H3D 120.0 C11—C10—H10A 119.0
H3C—N3—H3D 120.0 C9—C10—H10A 119.0
N2—C4—C5 112.5 (2) C10—C11—C6 120.3 (3)
N2—C4—H4A 109.1 C10—C11—H11A 119.8
C5—C4—H4A 109.1 C6—C11—H11A 119.8
C5—N1—C2—C3 64.5 (3) C6—C7—C8—N3 179.3 (3)
C1—N1—C2—C3 −174.7 (2) C6—C7—C8—C9 −0.1 (4)
C6—N2—C3—C2 −170.8 (2) N3—C8—C9—C10 −177.0 (3)
C4—N2—C3—C2 40.4 (3) C7—C8—C9—C10 2.4 (4)
N1—C2—C3—N2 −53.1 (3) N3—C8—C9—N4 3.6 (4)
C6—N2—C4—C5 171.8 (2) C7—C8—C9—N4 −177.1 (2)
C3—N2—C4—C5 −39.5 (3) O2—N4—C9—C10 −5.1 (4)
C2—N1—C5—C4 −64.2 (3) O1—N4—C9—C10 175.8 (3)
C1—N1—C5—C4 175.9 (3) O2—N4—C9—C8 174.4 (3)
N2—C4—C5—N1 52.0 (3) O1—N4—C9—C8 −4.8 (4)
C3—N2—C6—C7 13.8 (4) C8—C9—C10—C11 −1.8 (4)
C4—N2—C6—C7 161.8 (2) N4—C9—C10—C11 177.7 (3)
C3—N2—C6—C11 −166.2 (2) C9—C10—C11—C6 −1.2 (5)
C4—N2—C6—C11 −18.2 (4) N2—C6—C11—C10 −176.7 (3)
N2—C6—C7—C8 177.3 (2) C7—C6—C11—C10 3.3 (4)
C11—C6—C7—C8 −2.7 (4)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
N3—H3C···N1i 0.86 2.39 3.156 (4) 148
N3—H3D···O1 0.86 2.06 2.669 (4) 127
C10—H10A···O2 0.93 2.35 2.671 (4) 100