0095-1137/07/$08.00
⫹
0
doi:10.1128/JCM.02289-06
Copyright © 2007, American Society for Microbiology. All Rights Reserved.
Evaluation of the Invader Assay with the BACTEC MGIT 960 System
for Prompt Isolation and Identification of Mycobacterial Species
from Clinical Specimens
䌤
Sadahiro Ichimura,
1,3* Makoto Nagano,
2Nobuko Ito,
2Masahiro Shimojima,
1Toru Egashira,
2Chikara Miyamoto,
2Kiyofumi Ohkusu,
3and Takayuki Ezaki
3Department of Microbiology, BML, Inc.,
1and Division of Advanced Technology, BML, Inc.,
21361-1 Matoba, Kawagoe,
Saitama 350-1101, Japan, and Department of Microbiology, Regeneration and Advanced Medical Science,
Gifu University Graduate School of Medicine, Gifu, Gifu 501-1194, Japan
3Received 10 November 2006/Returned for modification 29 January 2007/Accepted 27 July 2007
Rapid and accurate identification of mycobacterial species is essential for patient management. We describe
the use of the Invader assay in conjunction with the BACTEC MGIT 960 system that together provide an
efficient procedure for clinical use. This assay discriminates base differences (e.g., genotyping
single-nucleotide polymorphisms) under homogeneous and isothermal conditions and can measure directly on
genomic DNA without prior target DNA amplification. To identify a wide variety of mycobacterial species, 20
Invader probes were designed to target the 16S rRNA gene and the 16S-23S rRNA gene internal transcribed
spacer 1 (ITS-1) region. To validate the Invader probes, we used 78 ATCC strains, and 607 clinical
mycobac-terial strains, which were identified by DNA sequencing of the 16S rRNA gene and ITS-1. The Invader assay
could accurately identify and differentiate these strains according to target sequences. Moreover, it could detect
and identify 116 (95.1%) of 122 positive liquid cultures from the BACTEC MGIT 960 system and did not react
to 83 contaminated MGIT cultures. Species identification takes 6.5 h by the Invader assay: 2.0 h for DNA
extraction, 0.5 h for handling, and up to 4 h for the Invader reaction. The Invader assay has the speed, ease
of use, and accuracy to be an effective procedure for the bacteriological diagnosis of mycobacterial infections.
Rapid and accurate identification of mycobacteria is
essen-tial for determining appropriate therapies and for
epidemio-logical studies (7, 11, 14, 37). For example, the U.S. Centers for
Disease Control and Prevention (CDC) recommends
turn-around times of 2 to 3 weeks for processing of
Mycobacterium
tuberculosis
(4, 33, 36). A definitive diagnosis of mycobacterial
infection depends on growth and identification of the bacteria
(2, 3). To speed the bacterial culturing, time-consuming
cul-tures on egg-based solid media, such as Lo
¨wenstein-Jensen and
Ogawa slants, are being replaced by faster liquid culture
meth-ods, such as the BACTEC MGIT 960 system
(Becton-Dickin-son, Sparks, MD) and the MB/BacT system (Organon
Teknika, Boxtel, The Netherlands) (1, 13, 15, 21, 22, 33).
Now a method is urgently needed to rapidly identify a wide
variety of
Mycobacterium
species directly from liquid cultures.
Unfortunately, the available methods have several limitations.
For example, the widely used AccuProbe system (GenProbe,
San Diego, CA) (8, 21, 25, 29) identifies only a limited number
of the many mycobacterial species seen in a clinical laboratory.
Although much faster and more accurate, DNA sequencing
(16, 26, 39), PCR restriction fragment length polymorphism
assays (6, 28, 35) and InnoLiPA Mycobacteria (Innogenetics,
Ghent, Belgium) (23, 38) require expensive equipment and an
exclusive workspace for PCR. In addition, DNA sequencing
and PCR-restriction fragment length polymorphism assay
re-quire pure cultures, and if the sample was the mixed culture,
separate culture on a solid medium would further slow these
assays.
Many routine procedures in clinical laboratories have been
simplified by homogeneous fluorescent detection systems.
Here we report our study of one of these, the Invader assay
(Third Wave Technologies, Madison, WI) (17). The Invader
assay can accurately discriminate single-base differences, such
as single nucleotide polymorphisms, and can measure directly
on genomic DNA without prior target amplification. We
eval-uated the suitability of the Invader assay for directly identifying
mycobacteria from the MGIT 960 system.
MATERIALS AND METHODS
Bacterial strains.The validation of the Invader probes was performed by testing 78 ATCC strains (Table 2) and 607 clinical mycobacterial strains (Table 3), which were identified and confirmed the target sequences by DNA sequenc-ing of the 16S rRNA gene and the 16S-23S rRNA gene internal transcribed spacer 1 (ITS-1). These clinical strains cultured on Ogawa slants mainly for nontuberculousMycobacteriuminfections were tested in the BML general lab-oratory from December 2003 to June 2005 (excluding overlapping patients).
Clinical specimens for the BACTEC MGIT 960 system.Between 21 and 25 November 2004, 1,390 consecutive clinical specimens were received for routine mycobacterial detection in the BML general laboratory. These included 1,040 sputum and 155 other respiratory specimens, 28 digestive samples, 15 sterile body fluids, 14 urine samples, 6 wound samples, and 132 samples from unspec-ified sources.
Inoculation and cultivation of clinical specimens. Before inoculation, the MGIT 960 tubes were prepared as described by the manufacturer (Becton Dickinson). A 0.5-ml portion of the processed specimen was inoculated into the MGIT, and the tubes were introduced into the MGIT 960 instrument and incubated until they were found to be positive by the instrument or for 42 days. A 0.1-ml portion of the processed specimen was inoculated onto a solid slant of Ogawa egg medium (Kyokuto Pharmaceutical, Tokyo, Japan), and the slants
* Corresponding author. Mailing address: Department of
Microbi-ology, BML, Inc., 1361-1 Matoba, Kawagoe, Saitama 350-1101, Japan.
Phone: 81-049-232-0940. Fax: 81-049-232-0529. E-mail: ichi-s@bml
.co.jp.
䌤
Published ahead of print on 8 August 2007.
3316
on May 16, 2020 by guest
http://jcm.asm.org/
were incubated in 5 to 10% CO2. For 56 days, the growth on the slants was
examined for visible colonies. All positive media were examined by Ziehl-Neelsen and Gram staining to confirm the presence of only acid–fast bacteria, and the colonies were subcultured onto Trypticase soy agar II with 5% sheep blood (TSA II; Becton Dickinson) to check for contaminants.
DNA extraction.For Ogawa slants, a sample of DNA was extracted from a loopful (3-mm3
sphere) of bacterial colony. Bacterial cells were mechanically disrupted with glass beads. After phenol-chloroform treatment (DDH Mycobac-teria; Kyokuto Pharmaceuticals), DNA in the aqueous phase was extracted and purified on a robotic liquid handler AGE-96 (Biotec, Tokyo, Japan) with mag-netic silica particles (MagneSil blood genomic max yield system; Promega, Mad-ison, WI). For the MGIT 960 system, a 4.0-ml aliquot of culture broth was centrifuged for 10 min at 13,000⫻g. The pellet was extracted with the bacterial DNA/RNA extraction kit (MORA-EXTRACT; Kyokuto Pharmaceuticals). Bac-terial cells were mechanically disrupted with zirconia beads, and phenol-chloro-form treatment was perphenol-chloro-formed, as recommended by the manufacturer. A 100-l aliquot of TE buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA) was added to the extracted DNA pellet, and DNA concentrations were determined by using the PicoGreen system (Molecular Probes, Eugene, OR) as recommended by the manufacturer.
Amplicor PCR.For an MGIT-positive culture, an aliquot of each culture was tested in parallel by the Amplicor PCR (Roche Diagnostic Systems) forM. tuberculosiscomplex,M. avium, andM. intracellulareas described in the manu-facturer’s instructions.
DNA sequencing. The 25-l reaction mixture contained ExTaq HS buffer
(Takara Shuzo, Ohtsu, Japan) with 2.0 mM MgCl2, 200M concentrations of
each of the deoxynucleoside triphosphates, 1.0 U of ExTaq HS DNA polymer-ase, 10 ng of template, 10 pmol of each of the primers SSU-bact-27f (5⬘-AGA GTT TGA TCM TGG CTC AG-3⬘) and SSU-bact-907r (5⬘-CCG TCA ATT CMT TTR AGT TT-3⬘) for the 16S rRNA gene and 16S-1511f (5⬘-AAG TCG TAA CAA GGT ARC CG-3⬘) and 23S-23r (5⬘-TCG CCA AGG CAT CCA CC-3⬘) for the ITS-1 region (18). Amplification was performed with a GeneAmp PCR system 9700 thermocycler (Applied Biosystems, Foster City, CA) for 30 cycles (30 s at 94°C, 30 s at 53°C, and 90 s at 72°C), followed by an extension step at 72°C for 7 min. The PCR products were visualized with ethidium bromide staining and UV illumination. Purification of the amplicons was performed with the AMPure PCR purification system (Agencourt, Beverly, MA), according to the manufacturer’s instructions. The ABI Prism BigDye Terminator v1.1 cycle sequencing ready reaction kit (Applied Biosystems) was used for the sequencing of the PCR products. The sequencing reaction mixture contained 0.5l of BigDye premix, 1.75l of 5⫻sequencing buffer, 1.6 pmol of sequencing primer, and approximately 10 ng of PCR product template in a total volume of 10l. SSU-bact-27f, SSU-bact-907r, 16S-1511f, and 23S-23r were used for sequencing primers for both DNA strands. The sequencing reaction was performed with a GeneAmp PCR system 9700 thermocycler (Applied Biosystems). A denaturation step at 95°C for 2 min proceeded 25 cycles (10 s at 96°C, 15 s at 53°C, and 2.5 min at 60°C). Sequencing products were purified with a CleanSEQ Sequencing Re-action Clean-Up system (Agencourt) and analyzed with a 3130xl genetic analyzer (Applied Biosystems), according to the manufacturer’s instructions. Raw se-quencing data were edited to resolve discrepancies by evaluating the electro-phoretograms with sequencing analysis software (v3.3; Applied Biosystems). The edited sequence data from both strands were aligned with the DNASIS Pro (Hitachi Software Engineering, Yokohama, Japan). We analyzed the consensus sequence of approximately 500 bp of the 5⬘end of the 16S rRNA gene for comparison with the sequence databases stored in GenBank (BLAST; http: //www.ncbi.nlm.nih.gov/BLAST/) or the Ribosomal Differentiation of Medical Microorganisms (RIDOM; http://www.ridom-rdna.de/) (9, 10).
Probe design for Invader assay.The 16S rRNA gene and ITS-1 sequences were aligned with those from the GenBank database and quality-controlled RIDOM database. With Invader Creator software (Third Wave Technologies), 20 specific probes were designed to identify and differentiate mycobacteria in the conserved regions of ITS-1 and 16S rRNA gene hypervariable regions A and B, including species-specific sites. In addition, mixed genusMycobacteria(GNS) probes for the mycobacteria genus-specific region and broad-range bacterial (BRB) probe for the bacteria universal region were designed from conserved regions of 16S rRNA gene (Table 1). Signal probes and Invader oligonucleotide for the specific nucleotide sequences were designed to have theoretical annealing temperatures of 63 and 77°C, respectively, using a nearest-neighbor algorithm on the basis of final probe and target concentrations. The signal probes and Invader oligonucleotides used to detectMycobacteriumspecies by the Invader assay are shown in Table 1.
Biplex Invader assay.The Invader assay utilizes the thermostable flap endo-nuclease Cleavase XI, which cleaves invasive structures formed from single-base
overlap between the Invader oligonucleotide and the signal probe when hybrid-ized to a complementary target DNA. This method can accurately discriminate single-base differences, such as single-nucleotide polymorphisms, and can mea-sure genomic DNA (ⱖ104
copies/assay). The Invader assay combines structure-specific cleavage enzymes and a universal FRET (Fo¨rster resonance energy transfer) system, and the biplex format of the Invader assay enables simultaneous detection of two kinds of species in a single well. At first, 3l of genomic DNA (0.03 to 0.33 ng/l) was added to a 384-well plate, 6l of mineral oil (Sigma) was overlaid into all reaction wells, the mixtures were denatured by incubation at 95°C for 10 min, and 3l of the appropriate reaction mixture was added. The reaction mixture contained 32 ng of Cleavase XI enzyme, 0.817mol of each signal probe/liter, 0.163mol of each Invader oligonucleotide/liter, 0.65mol of each of FAM dye and Redmond Red dye FRET cassettes (Epoch Bioscience, Redmond, WA)/liter, 5.04% PEG 8000, 30.7 mmol of MgCl2/liter, and 24.5
mmol of morpholinepropanesulfonic acid/liter. After the reagent was dispensed, the plate was spun for 10 s at 400⫻gand then sequentially incubated isother-mally at 64°C up to 4 h in the thermal fluorescence microtiter plate reader (Fluodia; Otsuka Electronics, Osaka, Japan), whereas the fluorescence intensi-ties were measured at 15-min intervals for FAM (excitation, 486 nm; emission, 530 nm) and Redmond Red (RED) (excitation, 560 nm; emission, 620 nm).
Invader assay data analysis.Raw data were analyzed by using a Microsoft Excel-based spreadsheet (Microsoft, Redmond, WA). For each specific signal, fold-over-zero (FOZ) values were calculated as follows for the signal obtained with each dye: FOZ⫽raw counts from sample/raw counts from no target control.
DNA samples from 54 mycobacterial reference strains were used to determine the cutoff value of FOZ. The mean plus five standard deviations of the FOZ value of the nontarget sequence in each well was calculated to 1.22 to 1.95 (probe set 1 to 10) of FAM-FOZ and 1.23 to 1.70 (probe set 1 to 10) of RED-FOZ. Therefore, the cutoff level was set at 2.00.
RESULTS
Probe design and validation.
The most widely accepted gene
for bacterial identification is the 16S rRNA gene, but this gene
alone lacks sufficient resolution to identify all mycobacterial
species (5, 32, 37, 39). For example, the 16S rRNA gene could
not distinguish between
M. kansasii
and
M. gastri
, and also
between
M. chelonae
and
M. abscessus
. However, since the
ITS-1 region had greater diversity than the 16S rRNA gene
(27, 18), the Invader probes designed in the ITS-1 region could
distinguish these strains. Therefore, the Invader assay was set
up with a more effective combination of the 16S rRNA gene
and the ITS-1 region.
To validate the Invader probes, we evaluated 65 type and 13
reference strains and 607 mycobacterial clinical strains.
Spe-cific signals were obtained for the target sequences of the type
and reference strains, according to the criteria of the Invader
assay. The GNS and the BRB probes were designed for the
mycobacterial genus-specific region and the bacteria universal
regions, respectively. Using the Invader assay, strains that
re-acted only to both the BRB and GNS probes were
mycobac-teria other than target species, and strains that reacted to only
the BRB probe were bacteria other than mycobacteria (Table
2). Next, we confirmed the variation and conservation of target
sequences in 607 clinical mycobacterial strains, which were
iden-tified by DNA sequencing of the 16S rRNA gene and ITS-1. For
all 607 clinical strains, the identities determined by DNA
sequenc-ing corresponded exactly to those determined by the Invader
assay (Table 3).
Identification directly from the BACTEC MGIT samples.
By
examining a serial dilution of reference strains, the detection
limit of genomic DNAs for all Invader probes was found to be
0.03 ng/
l. DNA extracted from 122 positive samples of the
MGIT 960 system had a minimum concentration of
⬍
0.01
ng/
l (less than the lower limit of the PicoGreen system), a
on May 16, 2020 by guest
http://jcm.asm.org/
maximum value 24.38 ng/
l, and an average of 4.40 ng/
l.
Zirconia beads efficiently extracted DNA for
M. tuberculosis
and NTM. Six MGIT samples were negative in the Invader
assay, and the DNA concentrations of four MGIT samples
were below the detection limit (
⬍
0.01 ng/
l). Although the
remaining two MGIT samples had sufficient DNA for the
Invader assay (from
ⱖ
1 ng/
l to
⬍
10 ng/
l), smears and
cul-tures of these samples showed contamination with
S. aureus
(
ⱖ
10
7CFU/ml). Also, the Invader probes of these two samples
[image:3.585.44.548.79.594.2]reacted only with the BRB probe. Overall, when more than
TABLE 1. Invader probes designed for this study
Set no. Target Species (sequevars)a Invader probeb Sequencec(5⬘–3⬘)
Set1 16S rRNA gene M. tuberculosiscomplex MTC signal probe cgcgccgaggTAAAGCGCTTTCCACC MTC Invader oligo GCGGGCTCATCCCACACCGCc ITS-1 M. gordonae(A, B, C, and D) Mgo signal probe acggacgcggagCTCGGGTGCTGTCC
Mgo Invader oligo AGACACACTATTGGGTCCTGAGGCAACACCt Set2 16S rRNA gene M. avium(I and II) MAV signal probe cgcgccgaggAGGTCCTATCCGGTATTAG
MAV Invader oligo CACCGCAAAAGCTTTCCACCAGAAGACATGCGT CTTGc
16S rRNA gene M. kansasii/M. gastri(I, II, III, IV, MKN/MGS signal probe acggacgcggagCCGGGATAAGCCTGG V, and VI) MKN/MGS Invader oligo ACGTGGGCAATCTGCCCTGCACAa Set3 16S rRNA gene M. intracellulare(I and V) MIN signal probe 1 cgcgccgaggTAAAGACATGCGCCTAAA
M. intracellulare(II, III, and IV) MIN signal probe 2 cgcgccgaggAAAAGACATGCGTCTAAAG MIN Invader oligo CCATCCCACACCGCAAAAGCTTTCCACCc ITS-1 M. chelonae(A, B, and C) Mch signal probe acggacgcggagATGGATAGTGGTTGCG
Mch Invader oligo GCTTGGTGGTGGGGTGTGGTCTTTGACTTt Set4 16S rRNA gene M. chelonae/M. abscessus(I, II) MCH/MAB signal probe cgcgccgaggACTTCATGGTGAGTGGT
MCH/MAB Invader oligo GGGAAACTGGGTCTAATACCGGATAGGACCA CACt
16S rRNA gene Mycobacteriumspp. GNS signal probe 1 acggacgcggagACACCGCAAAAGCTTT GNS signal probe 2 acggacgcggagACACCGCTAAAGCGC GNS signal probe 3 acggacgcggagACACCGCTACCAAACA GNS signal probe 4 acggacgcggagACACCACTACCACACG GNS Invader oligo 1 CTGATAGGCCGCGGGCTCATCCCc GNS Invader oligo 2 GGCCGCGGGCCCATCCCc Set5 16S rRNA gene M. scrofulaceum MSC signal probe cgcgccgaggGCCTTCGTCGATGG
MSC Invader oligo TCTTCTCCACCTACCGTCAACCCACAAAGTGAt 16S rRNA gene M. gordonae(I) MGO signal probe 1 acggacgcggagTGTGTCCTGTGGTCC
M. gordonae(II, III, IV, and V) MGO signal probe 2 acggacgcggagTGTGTTCTGTGGTCCT MGO Invader oligo CACCGCAAAAGCTTTCCACCACAGGACAt Set6 16S rRNA gene M. fortuitum(I) MFO signal probe cgcgccgaggATATTCGGTATTAGACCCAG
MFO Invader oligo ACCACACACCATGAAGCGCGTGGTCt
ITS-1 M. gastri(A) Mgs signal probe acggacgcggagTGCTTGCTGTTGCG
Mgs Invader oligo GCCTCAGGACCCAATAGTGTGTCTGGCTa Set7 16S rRNA gene M. peregrinum/M. septicum MPE/MSE signal probe cgcgccgaggGCACTTCCTGGTGTG
MPE/MSE Invader oligo CCTGGGAAACTGGGTCTAATACCGAATATGAC CGCt
16S rRNA gene M. lentiflavum(I and II) MLE signal probe acggacgcggagAAAGGCATGCGCCA
MLE Invader oligo CCATCCCACACCGCAAAAGCTTTCCACCAc Set8 16S rRNA gene M. terrae(I and II) MTE signal probe cgcgccgaggTGGGATGCATGTTCTG
MTE Invader oligo CTGGGAAACTGGGTCTAATACCGGATAGGA CCAa
16S rRNA gene M. nonchromogenicumgroupd MNG signal probe 1 acggacgcggagTCGGCGAAGCTCC
MNG signal probe 2 acggacgcggagTCGGCGAAGCTTG
MNG Invader oligo TGACGGCCTTCGGGTTGTAAACCTCTTTCAGTAc Set9 16S rRNA gene M. xenopi(I, II, and III) MXE signal probe cgcgccgaggAGAATGGTCCTATCCGG
MXE Invader oligo ACACTTTCCACCACCCCACATGCGCt ITS-1 M. scrofulaceum/M. Msc/Mps signal probe acggacgcggagACCACTCAGAACGAGC
parascrofulaceum Msc/Mps Invader oligo CACCCCACCACCAAGATGGAGGGACc
Set10 16S rRNA gene Broad-range bacteria BRB signal probe cgcgccgaggCCGGGAACGTATTCACC BRB Invader oligo TGACGGGCGGTGTGTACAAGGCa 16S rRNA gene M. szulgai MSZ signal probe acggacgcggagGGGTCCTATCCGGTATT
MSZ Invader oligo CACCCCAAGGCATGCGCCTCGt
aSequevars (sequence variants) were referred to Ribosomal Differentiation of Medical Microorganisms (RIDOM). bOligo, oligonucleotide.
cThe flap sequences of “cgcgccgagg” and “acggacgcggag” were used for the detection of FAM and Redmond Red, respectively (see Materials and Methods). The
invasive nucleotides of Invader oligonucleotides are indicated in lowercase characters.
dM. nonchromogenicumgroup (M. nonchromogenicumgroup organisms):M. nonchromogenicum,M. terrae(sequevar I, II, and III),M. hiberniae,M. arupense, and M. kumamotonense.
on May 16, 2020 by guest
http://jcm.asm.org/
0.10 ng of mycobacterial genomic DNA/
l was obtained from
the MGIT, the Invader assay detected individual species in a
simultaneous assay (Table 4).
After MGIT cultivation of the 1,390 samples, 122 (8.8%)
samples were found to be positive for mycobacteria. The
breakdown of 122 MGIT-positive samples was 63 (51.6%)
M. tuberculosis
, 29 (23.8%)
M. avium
, 17 (13.9%)
M.
intra-cellulare
, 4 (3.3%)
M. kansasii
, 2 (1.6%)
M. abscessus
, 2
(1.6%)
M. chelonae
, 2 (1.6%)
M. fortuitum
, 1 (0.8%)
M.
gordonae
, and 6 (4.9%) negative isolates. Two (1.6%)
sam-ples had both
M. avium
and
M. abscessus
. Although six
samples were negative as determined by the Invader assay,
they were positive by the Amplicor
M. tuberculosis
complex
PCR. The four negative samples except for two negative samples
contaminated with
S. aureus
changed to positive for
M.
tubercu-losis
complex by the Invader assay after additional cultivation of
the remaining MGIT broth for 1 week (Table 4). In addition, 83
(6.0%) MGIT cultures showed contamination of bacteria other
than mycobacteria (Table 5). In the Invader reactions, these
sam-ples were positive only for the BRB probe.
DISCUSSION
A rapid and accurate method for diagnosing mycobacterial
infection is urgently needed in the clinical laboratory. This
study describes the combined use of the BACTEC MGIT 960
system and the Invader assay. The Invader assay correctly
identified and differentiated total 888 clinical and reference
strains according to a target sequence. In particular, it detected
and identified 116 (95.1%) of 122 positive liquid cultures soon
after automatic positive signal of the MGIT 960 system. Thus,
the Invader assay has the accuracy and sensitivity to be an
effective procedure for a definitive diagnosis of mycobacterial
infection in a clinical setting.
The key to any assay based on DNA sequences is the probe.
The target region for DNA probes must be sufficiently
con-served among clinical strains of the species, and the sequence
data should be reliable, definitive, and commonly used. To
identify mycobacteria, probes have traditionally been based on
sequences from the 16S rRNA gene that corresponds to
Esch-erichia coli
positions 130 to 210 and positions 430 to 500.
However, the 16S rRNA genes of some species have the same
or very similar sequences (5, 32, 37, 39). For example, using the
first 500 bases of the 5
⬘
end of the 16S rRNA gene to analyze
sequences, RIDOM or MicroSeq (Applied Biosystems) (24)
cannot differentiate
M. abscessus
and
M. chelonae
;
M. kansasii
and
M. gastri
;
M. marinum
and
M. ulcerans
;
M. senegalense
,
M.
houstonense
ATCC 49403, and
M. farcinogenes
;
M. porcinum
and
M. neworleansense
ATCC 49404; and
M. septicum
and
M.
peregrinum.
In the present study, probes were based on a
com-TABLE 2. ATCC strains identified by the Invader assay
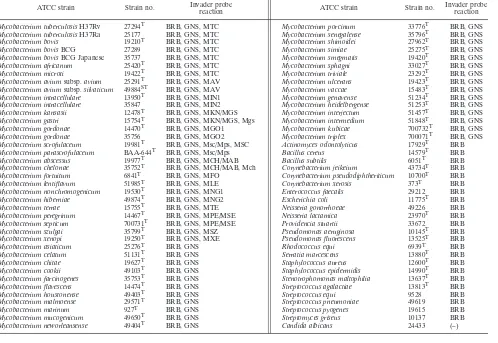
ATCC strain Strain no. Invader probe
reaction ATCC strain Strain no.
Invader probe reaction
Mycobacterium tuberculosisH37Rv 27294T BRB, GNS, MTC Mycobacterium porcinum 33776T BRB, GNS
Mycobacterium tuberculosisH37Ra 25177 BRB, GNS, MTC Mycobacterium senegalense 35796T BRB, GNS
Mycobacterium bovis 19210T BRB, GNS, MTC Mycobacterium shimoidei 27962T BRB, GNS
Mycobacterium bovisBCG 27289 BRB, GNS, MTC Mycobacterium simiae 25275T BRB, GNS
Mycobacterium bovisBCG Japanese 35737 BRB, GNS, MTC Mycobacterium smegmatis 19420T BRB, GNS
Mycobacterium africanum 25420T BRB, GNS, MTC Mycobacterium sphagni 33027T BRB, GNS
Mycobacterium microti 19422T BRB, GNS, MTC Mycobacterium triviale 23292T BRB, GNS
Mycobacterium aviumsubsp.avium 25291T BRB, GNS, MAV Mycobacterium ulcerans 19423T BRB, GNS
Mycobacterium aviumsubsp.silvaticum 49884ST BRB, GNS, MAV Mycobacterium vaccae 15483T BRB, GNS
Mycobacterium intracellulare 13950T BRB, GNS, MIN1 Mycobacterium genavense 51234T BRB, GNS
Mycobacterium intracellulare 35847 BRB, GNS, MIN2 Mycobacterium heidelbergense 51253T BRB, GNS
Mycobacterium kansasii 12478T BRB, GNS, MKN/MGS Mycobacterium interjectum 51457T BRB, GNS
Mycobacterium gastri 15754T BRB, GNS, MKN/MGS, Mgs Mycobacterium intermedium 51848T BRB, GNS
Mycobacterium gordonae 14470T BRB, GNS, MGO1 Mycobacterium kubicae 700732T BRB, GNS
Mycobacterium gordonae 35756 BRB, GNS, MGO2 Mycobacterium triplex 700071T BRB, GNS
Mycobacterium scrofulaceum 19981T BRB, GNS, Msc/Mps, MSC Actinomyces odontolyticus 17929T BRB
Mycobacterium parascrofulaceum BAA-644T BRB, GNS, Msc/Mps Bacillus cereus 14579T BRB
Mycobacterium abscessus 19977T BRB, GNS, MCH/MAB Bacillus subtilis 6051T BRB
Mycobacterium chelonae 35752T BRB, GNS, MCH/MAB, Mch Corynebacterium jeikeium 43734T BRB
Mycobacterium fortuitum 6841T BRB, GNS, MFO Corynebacterium pseudodiphtheriticum 10700T BRB
Mycobacterium lentiflavum 51985T BRB, GNS, MLE Corynebacterium xerosis 373T BRB
Mycobacterium nonchromogenicum 19530T BRB, GNS, MNG1 Enterococcus faecalis 29212 BRB
Mycobacterium hiberniae 49874T BRB, GNS, MNG2 Escherichia coli 11775T BRB
Mycobacterium terrae 15755T BRB, GNS, MTE Neisseria gonorrhoeae 49226 BRB
Mycobacterium peregrinum 14467T BRB, GNS, MPE/MSE Neisseria lactamica 23970T BRB
Mycobacterium septicum 700731T BRB, GNS, MPE/MSE Providencia stuartii 33672 BRB
Mycobacterium szulgai 35799T BRB, GNS, MSZ Pseudomonas aeruginosa 10145T BRB
Mycobacterium xenopi 19250T BRB, GNS, MXE Pseudomonas fluorescens 13525T BRB
Mycobacterium asiaticum 25276T BRB, GNS Rhodococcus equi 6939T BRB
Mycobacterium celatum 51131T BRB, GNS Serratia marcescens 13880T BRB
Mycobacterium chitae 19627T BRB, GNS Staphylococcus aureus 12600T BRB
Mycobacterium cookii 49103T BRB, GNS Staphylococcus epidermidis 14990T BRB
Mycobacterium farcinogenes 35753T BRB, GNS Stenotrophomonas maltophilia 13637T BRB
Mycobacterium flavescens 14474T BRB, GNS Streptococcus agalactiae 13813T BRB
Mycobacterium houstonense 49403T BRB, GNS Streptococcus equi 9528 BRB
Mycobacterium malmoense 29571T BRB, GNS Streptococcus pneumoniae 49619 BRB
Mycobacterium marinum 927T BRB, GNS Streptococcus pyogenes 19615 BRB
Mycobacterium mucogenicum 49650T BRB, GNS Streptomyces griseus 10137 BRB
Mycobacterium neworleansense 49404T BRB, GNS Candida albicans 24433 (–)
on May 16, 2020 by guest
http://jcm.asm.org/
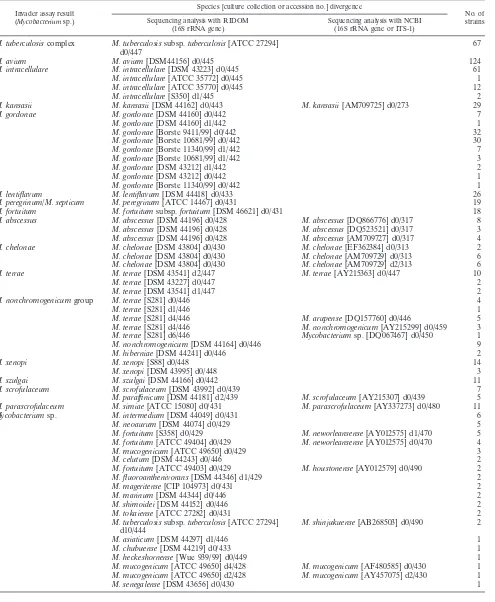
[image:4.585.45.543.82.425.2]TABLE 3. Comparison of the Invader assay and DNA sequencing
Invader assay result (Mycobacteriumsp.)
Species [culture collection or accession no.] divergence
No. of strains Sequencing analysis with RIDOM
(16S rRNA gene)
Sequencing analysis with NCBI (16S rRNA gene or ITS-1)
M. tuberculosis
complex
M. tuberculosis
subsp.
tuberculosis
[ATCC 27294]
d0/447
67
M. avium
M. avium
[DSM44156] d0/445
124
M. intracellulare
M. intracellulare
[DSM 43223] d0/445
61
M. intracellulare
[ATCC 35772] d0/445
1
M. intracellulare
[ATCC 35770] d0/445
12
M. intracellulare
[S350] d1/445
2
M. kansasii
M. kansasii
[DSM 44162] d0/443
M. kansasii
[AM709725] d0/273
29
M. gordonae
M. gordonae
[DSM 44160] d0/442
7
M. gordonae
[DSM 44160] d1/442
1
M. gordonae
[Borste 9411/99] d0/442
32
M. gordonae
[Borste 10681/99] d0/442
30
M. gordonae
[Borste 11340/99] d1/442
7
M. gordonae
[Borste 10681/99] d1/442
3
M. gordonae
[DSM 43212] d1/442
2
M. gordonae
[DSM 43212] d0/442
1
M. gordonae
[Borste 11340/99] d0/442
1
M. lentiflavum
M. lentiflavum
[DSM 44418] d0/433
26
M. peregrinum/M. septicum
M. peregrinum
[ATCC 14467] d0/431
19
M. fortuitum
M. fortuitum
subsp.
fortuitum
[DSM 46621] d0/431
18
M. abscessus
M. abscessus
[DSM 44196] d0/428
M. abscessus
[DQ866776] d0/317
8
M. abscessus
[DSM 44196] d0/428
M. abscessus
[DQ523521] d0/317
3
M. abscessus
[DSM 44196] d0/428
M. abscessus
[AM709727] d0/317
4
M. chelonae
M. chelonae
[DSM 43804] d0/430
M. chelonae
[EF362384] d0/313
2
M. chelonae
[DSM 43804] d0/430
M. chelonae
[AM709729] d0/313
6
M. chelonae
[DSM 43804] d0/430
M. chelonae
[AM709729] d2/313
6
M. terrae
M. terrae
[DSM 43541] d2/447
M. terrae
[AY215363] d0/447
10
M. terrae
[DSM 43227] d0/447
2
M. terrae
[DSM 43541] d1/447
2
M. nonchromogenicum
group
M. terrae
[S281] d0/446
4
M. terrae
[S281] d1/446
1
M. terrae
[S281] d4/446
M. arupense
[DQ157760] d0/446
5
M. terrae
[S281] d4/446
M. nonchromogenicum
[AY215299] d0/459
3
M. terrae
[S281] d6/446
Mycobacterium
sp. [DQ067467] d0/450
1
M. nonchromogenicum
[DSM 44164] d0/446
9
M. hiberniae
[DSM 44241] d0/446
2
M. xenopi
M. xenopi
[S88] d0/448
14
M. xenopi
[DSM 43995] d0/448
3
M. szulgai
M. szulgai
[DSM 44166] d0/442
11
M. scrofulaceum
M. scrofulaceum
[DSM 43992] d0/439
7
M. paraffinicum
[DSM 44181] d2/439
M. scrofulaceum
[AY215307] d0/439
5
M. parascrofulaceum
M. simiae
[ATCC 15080] d0/431
M. parascrofulaceum
[AY337273] d0/480
11
Mycobacterium
sp.
M. intermedium
[DSM 44049] d0/431
6
M. neoaurum
[DSM 44074] d0/429
5
M. fortuitum
[S358] d0/429
M. neworleansense
[AY012575] d1/470
5
M. fortuitum
[ATCC 49404] d0/429
M. neworleansense
[AY012575] d0/470
4
M. mucogenicum
[ATCC 49650] d0/429
3
M. celutum
[DSM 44243] d0/446
2
M. fortuitum
[ATCC 49403] d0/429
M. houstonense
[AY012579] d0/490
2
M. fluoroanthenivorans
[DSM 44346] d1/429
2
M. mageritense
[CIP 104973] d0/431
2
M. marinum
[DSM 44344] d0/446
2
M. shimoidei
[DSM 44152] d0/446
2
M. tokaiense
[ATCC 27282] d0/431
2
M. tuberculosis
subsp.
tuberculosis
[ATCC 27294]
d10/444
M. shinjukuense
[AB268503] d0/490
2
M. asiaticum
[DSM 44297] d1/446
1
M. chubuense
[DSM 44219] d0/433
1
M. heckeshornense
[Wue 939/99] d0/449
1
M. mucogenicum
[ATCC 49650] d4/428
M. mucogenicum
[AF480585] d0/430
1
M. mucogenicum
[ATCC 49650] d2/428
M. mucogenicum
[AY457075] d2/430
1
M. senegalense
[DSM 43656] d0/430
1
on May 16, 2020 by guest
http://jcm.asm.org/
bination of the 16S rRNA gene and the ITS-1 region. The
specificity of these probes was confirmed by extensive
compar-ison with well-studied databases.
Although the 16S rRNA gene has a low mutation rate, it
does display microheterogeneity within a species.
Microhet-erogeneity was examined by mixed probes within the
vari-able regions of
M. intracellulare
(sequevars [sequence
vari-ants] I to V), and
M. gordonae
(sequevar I to V) (Table 1).
For example,
M. intracellulare
(sequevar III and IV)
dis-played microheterogeneity for type strain and were
deter-mined to be negative by the Amplicor
M. intracellulare
PCR.
As an additional control for the accuracy of mycobacterial
identification, mixed GNS probes were designed to select
for the mycobacteria genus-specific region. These probes
allowed differentiation of families closely related to the
My-cobacteriaceae
, including the
Gordoniaceae
,
Nocardiaceae
,
and
Tsukamurellaceae
. A BRB probe based on the bacterial
conserved region was designed for confirmation of the
quan-tity and quality of sample DNA. In addition, mixed
myco-bacteria could be recognized by comparing the signal of a
species probe with the signals of the BRB and GNS probes.
In the clinical laboratory, the MicroSeq 500 assay is a
com-mercial sequencing assay which can be used for the routine
identification of clinical mycobacterial isolates. The
turn-around time for this assay is 2 days and requires approximately
4 h of a technologist’s time. On the other hand, the turnaround
time for an Invader assay of 20 samples was
⬍
6.5 h and
re-quired only about 0.5 h of a technologist’s time. DNA
sequenc-ing requires less judgment on the part of technologists for
interpretation and can identify a wide range mycobacterial
species. However, labor, the reagents, equipment, and software
necessary for this assay are significantly more expensive than
the ribosomal probe hybridization. These factors limit the
abil-ity of hospital-based laboratories to use this assay (24).
More-over, DNA sequencing requires more time and effort to target
mixed cultures and multiple copy regions, such as ITS-1, since
subcloning or separate cultures are needed. Setup of the
In-vader assay system requires no expensive measuring
equip-ment, such as an automated DNA sequencer, and no exclusive
workspace for PCR.
In the present study, the Invader probe setting
cost-effec-tively detected more than 90% of the mycobacteria isolated
in a routine clinical laboratory. On the other hand, each
species probe could also perform an independent assay as
needed.
In supplemental studies, although
M. tuberculosis
complexes
were difficult to identify at the species or strain level, these
complex species and BCG strains could be discriminated by
designing the Invader probes for
gyrB
and RD1 (12, 19, 34;
[image:6.585.44.544.81.232.2]data not shown). Since the Invader assay can measure genes
with different GC contents, future versions may measure drug
resistance genes simultaneously (41, 40, 20, 30, 31).
TABLE 4. One hundred twenty-two MGIT-positive samples identified by the Invader assay and concentrations of the extracted DNA
Identificationa
(no. of isolates) Smear of
cultureb Invader assay
Distribution (no. of isolates) of extracted genomic DNA at a concn (ng/l) of:
⬍0.01 ⱖ0.01 to⬍0.1 ⱖ0.1 to⬍1 ⱖ1 to⬍10 ⱖ10
M. tuberculosis
complex (57)
AFB
M. tuberculosis
complex
11
37
9
M. tuberculosis
complex (4)*
AFB
All negative
4
M. tuberculosis
complex (2)*
AFB, GPC
Bacteria (BRB)
2
M. avium
(29)
AFB
M. avium
28
1
M. intracellurare
(17)
AFB
M. intracellulare
16
1
M. kansasii
(4)
AFB
M. kansasii
3
1
M. abscessus
(2)
AFB
M. abscessus
2
M. chelonae
(2)
AFB
M. chelonae
2
M. fortuitum
(1)
AFB
M. fortuitum
1
M. fortuitum
(1)
AFB, GPC
M. fortuitum
1
M. gordonae
(1)
AFB
M. gordonae
1
M. avium
and
M. abscessus
(2)
AFB
M. avium
and
M. abscessus
2
a
Isolates were identified by DNA sequencing except as noted. *, Isolates identified by Amplicor PCR (M. tuberculosiscomplex,M. avium, andM. intracellulare).
b
[image:6.585.43.281.466.715.2]Acid fast bacteria (AFB) were confirmed by microscopy of the Ziehl-Neelsen staining. Contaminations of gram-strain positive coccus (GPC) wereStaphylococcus aureus.
TABLE 5. Bacteria other than mycobacteria detected from the
MGIT 960
Speciesa
No. of isolates
Staphylococcus aureus
...28
Staphylococcus
sp. (CNS) ...13
Providencia stuartii
... 6
Bacillus cereus
... 4
S
treptomyces
sp... 4
Actinomadura
sp. ... 3
Serratia marcescens
... 3
Capnocytophaga sputigena
... 2
Gordonia sputi
... 2
Pseudomonas aeruginosa
... 2
Streptococcus genomo
sp. ... 2
Arthrobacter
sp. ... 1
Bacillus aminovorans
... 1
Bacillus subtilis
... 1
Brevibacterium
sp. ... 1
Corynebacterium jeikeium
... 1
Corynebacterium urealyticum
... 1
Enterococcus faecium
... 1
Nocardia farcinica
... 1
Paenibacillus
sp. ... 1
Rhodococcus equi
... 1
Stenotrophomonas maltophilia
... 1
Streptococcus parasanguis
... 1
Streptococcus pneumoniae
... 1
Streptococcus salivaris
... 1
aThese isolates were identified by DNA sequencing.
on May 16, 2020 by guest
http://jcm.asm.org/
ACKNOWLEDGMENTS
We thank Yuko Kazumi and Isamu Sugawara (Mycobacterial
Ref-erence Center, The Research Institute of Tuberculosis, Tokyo, Japan)
for helpful discussions. We thank Fuminori Hoshino for help in
pre-paring the manuscript and Gary Howard for critical reading of the
manuscript.
REFERENCES
1.Alcaide, F., M. A. Benitez, J. M. Escriba, and R. Martin.2000. Evaluation of the BACTEC MGIT 960 and the MB/BacT systems for recovery of mycobacteria from clinical specimens and for species identification by DNA AccuProbe. J. Clin. Microbiol.38:398–401.
2.American Thoracic Society, Medical Section of the American Lung Associ-ation.1997. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Am. J. Respir. Crit. Care Med.156:S1–S25.
3.American Thoracic Society.1997. Rapid diagnostic tests for tuberculosis: what is the appropriate use? Am. J. Respir. Crit. Care Med.155:1804–1814. 4.Centers for Disease Control and Prevention.2002. TB elimination cooper-ative agreement. Centers for Disease Control and Prevention, Atlanta, GA. 5.Clarridge, J. E., III.2004. Impact of 16S rRNA gene sequence analysis of identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev.17:840–862.
6.da Silva Rocha, A., C. da Costa Leite, H. M. Torres, A. B. de Miranda, M. Q. Pires Lopes, W. M. Degrave, and P. N. Suffys.1999. Use of PCR-restriction fragment length polymorphism analysis of thehsp65gene for rapid identi-fication of mycobacteria in Brazil. J. Microbiol. Methods37:223–229. 7.Falkinham, J. O.1996. Epidemiology of infection by nontuberculous
myco-bacteria. Clin. Microbiol. Rev.9:177–215.
8.Goto, M., S. Oka, K. Okuzumi, S. Kimura, and K. Shimada.1991. Evaluation of acridinium–ester-labeled DNA probes for identification ofMycobacterium tuberculosisandMycobacterium avium-Mycobacterium intracellularecomplex in culture. J. Clin. Microbiol.29:2473–2476.
9.Harmsen, D., J. Rothganger, C. Singer, J. Albert, and M. Frosch.1999. Intuitive hypertext-based molecular identification of micro-organisms. Lan-cet353:291.
10.Harmsen, D., S. Dostal, A. Roth, S. Niemann, J. Rothga¨nger, M. Sammeth, J. Albert, M. Frosch, and E. Richter.2003. RIDOM: comprehensive and public sequence database for identification ofMycobacteriumspecies. BMC Infect. Dis.3:26.
11.Henry, M. T., L. Inamdar, D. O’Riordain, M. Schweiger, and J. P. Watson.
2004. Nontuberculous mycobacteria in non-HIV patients: epidemiology, treatment, and response. Eur. Respir. J.23:741–746.
12.Kasai, H., T. Ezaki, and S. Harayama.2000. Differentiation of phylogeneti-cally related slowly growing mycobacteria by theirgyrBsequences. J. Clin. Microbiol.38:301–308.
13.Katila, M.-L., P. Katila, and R. Erkinjuntti-Pekkanen.2000. Accelerated detection and identification of mycobacteria with MGIT 960 and COBAS AMPLICOR systems. J. Clin. Microbiol.38:960–964.
14.Katoch, V. M.2004. Infections due to non-tuberculous mycobacteria (NTM). Indian J. Med. Res. Rev.4:290–304.
15.Kent, P. T., and G. P. Kubica.1985. Public health mycobacteriology: a guide for a level III laboratory. Centers for Disease Control, Atlanta, GA. 16.Kirschner, P., and E. C. Bottger.1998. Species identification of mycobacteria
using rDNA sequencing. Methods Mol. Biol.101:349–361.
17.L. Lyamichev, V., A. Mast, J. Hall, J. Prudent, M. Kaiser, T. Takova, R. Kwiatkowski, T. Sander, M. de Arruda, D. Arco, B. Neri, and M. A. Brow.
1999. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat. Biotechnol.17:
292–296.
18.Lane, D. J.1991. 16S/23S rRNA sequencing, p. 115–175.InE. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
19.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover.
1996. Molecular analysis of genetic differences betweenMycobacterium bovis
BCG and virulentM. bovis. J. Bacteriol.178:1274–1282.
20.Mdluli, K., R. A. Slayden, Y. Zhu, S. Ramaswamy, X. Pan, D. Mead, D. D. Crane, J. M. Musser, and C. E. Barry III.1998. Inhibition of a Mycobacte-rium tuberculosis-ketoacyl ACP synthase by isoniazid. Science 280:1607– 1610.
21.Metchock, B. G., F. S. Nolte, and R. J. Wallace.1999.Mycobacterium, p.
399–437.InP. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, DC.
22.Nolte, F. S., and B. Metchcock.1995.Mycobacterium, p. 400–437.InP. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. ASM Press, Washington, DC. 23.Padilla, E., V. Gonzalez, J. M. Manterola, A. Perez, M. D. Quesada, S.
Gordillo, C. Vilaplana, M. A. Pallares, S. Molinos, M. D. Sanchez, and V. Ausina.2004. Comparative evaluation of the new version of the INNO-LiPA Mycobacteria and genotypeMycobacteriumassays for identification of My-cobacteriumspecies from MB/BacT liquid cultures artificially inoculated with mycobacterial strains. J. Clin. Microbiol.42:3083–3088.
24.Patel, J. B., D. G. B. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nackamkin.2000. Sequence–based identification ofMycobacteriumspecies using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol.38:246–251.
25.Reisner, B. S., A. M. Gaston, and G. L. Woods.1994. Use of Gen–Probe AccuProbes to identifyMycobacterium aviumcomplex,Mycobacterium tuber-culosiscomplex,Mycobacterium kansasii, andMycobacterium gordonae di-rectly from BACTEC TB broth cultures. J. Clin. Microbiol.32:2995–2998. 26.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche,
J. L. Gaillard, and C. Pierre-Audigier.1999.hsp65sequencing for identifi-cation of rapidly growing mycobacteria. J. Clin. Microbiol.37:852–857. 27.Roth, A., M. Fisher, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch.
1998. Differentiation of phylogenetically related slowly growing mycobacte-ria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol.36:139–147.
28.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch.2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S– 23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol.
38:1094–1104.
29.Salfinger, M., and G. E. Pfyffer.1994. The new diagnostic mycobacteriology laboratory. Eur. J. Clin. Microbiol. Infect. Dis.13:961–979.
30.Scorpio, A., and Y. Zhang.1996. Mutations in pncA, a gene encoding amidase/nicotinamidase, cause resistance to the antituberculosis drug pyrazin-amide in tubercle bacillus. Nat. Med.2:662–667.
31.Sreevatsan, S., K. E. Stockbauer, X. Pan, B. N. Kreiswirth, S. L. Moghazeh, W. R. Jacobs, Jr., A. Telenti, and J. M. Musser.1997. Ethambutol resistance inMycobacterium tuberculosis: critical role ofembBmutations. Antimicrob. Agents Chemother.41:1677–1681.
32.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kampfer, M. C. J. Maiden, X. Nesme, R. Rossella–Mora, J. Swings, H. G. Truper, A. Vauterin, A. C. Ward, and W. B. Whitman.2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteri-ology. Int. J. Syst. Evol. Microbiol.52:1043–1047.
33.Styrt, B. A., T. M. Shinnick, J. C. Ridderhof, J. T. Crawford, and F. C. Tenover.1997. Turnaround times for mycobacterial cultures. J. Clin. Micro-biol.35:1041–1042.
34.Talbot, E. A., D. L. Williams, and R. Frothingham.1997. PCR identification ofMycobacterium bovisBCG. J. Clin. Microbiol.35:566–569.
35.Taylor, T. B., C. Patterson, Y. Hale, and W. W. Safranek.1997. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J. Clin. Microbiol.35:79–85. 36.Tenover, F. C., J. T. Crawford, R. E. Huebner, L. J. Geiter, C. R. Horsburgh,
Jr., and R. C. Good.1993. The resurgence of tuberculosis: is your laboratory ready? J. Clin. Microbiol.31:767–770.
37.Tortoli, E.2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev.16:319–354. 38.Trueba, F., M. Fabre, and P. Saint-Blancard.2004. Rapid Identification of
Mycobacterium genavensewith a new commercially available molecular test, INNO-LiPA MYCOBACTERIA v2. J. Clin. Microbiol.42:4403–4404. 39.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani.2001. Necessity of
quality–controlled 16S rRNA gene sequence databases: identifying nontu-berculousMycobacteriumspecies. J. Clin. Microbiol.39:3637–3648. 40.Wilson, T. M., and D. M. Collins.1996.ahpC, a gene involved in isoniazid
resistance of theMycobacterium tuberculosiscomplex. Mol. Microbiol.19:
1025–1034.
41.Zhang, Y., B. Heym, B. Allen, D. Young, and S. T. Cole.1992. The catalase-peroxidase gene and isoniazid resistance ofMycobacterium tuberculosis. Na-ture358:501–503.