0095-1137/95/$04.0010
Copyrightq1995, American Society for Microbiology
Genomic and Proteinic Characterization of Strain S, a Rickettsia
Isolated from Rhipicephalus sanguineus Ticks in Armenia
M. EREMEEVA,
1† N. BALAYEVA,
1,2V. ROUX,
1V. IGNATOVICH,
2M. KOTSINJAN,
3AND
D. RAOULT
1*
Unite´ des Rickettsies, Centre National de la Recherche Scientifique EP J0054, Faculte´ de Me´decine,
13385 Marseille Ce´dex 5, France
1; Gamaleya Research Institute of Epidemiology
and Microbiology, Academy of Medical Sciences of Russia, 123098 Moscow,
Russia
2; and Institute of Epidemiology, Microbiology and
Medical Parasitology, Erevan, Armenia
3Received 17 April 1995/Returned for modification 8 May 1995/Accepted 17 July 1995
Strain S, a spotted fever group (SFG) rickettsia isolated from
Rhipicephalus sanguineus
ticks collected in
Armenia, was identified. Microimmunofluorescence, sodium dodecyl sulfate-polyacrylamide gel protein
elec-trophoresis and Western immunoblotting, PCR and then restriction fragment length polymorphism analysis,
pulsed-field gel electrophoresis, and 16S rRNA gene sequencing were used to compare strain S with reference
isolates. Strain S was found to possess proteinic, antigenic, and genomic patterns which were unique among
SFG rickettsiae. Strain S is characterized by its high degree of pathogenicity for experimental animals, but its
role as a potential human pathogen should be determined. The role of
R. sanguineus
ticks in the epidemiology
of SFG rickettsiae is discussed.
Spotted fever group (SFG) rickettsiae are obligate
intracel-lular bacteria that are widely distributed throughout the world.
These bacteria are associated with acarian vectors which, while
feeding, can transmit rickettsiae to humans and animals (27,
55). Resultant human diseases have characteristic clinical
fea-tures, including fever, headache, maculopapular eruption, and
eschar formation (tache noire).
Three SFG rickettsioses are prevalent in the former USSR:
Boutonneuse fever on the Black Sea Coast (43, 56), Astrakhan
fever on the north part of the Caspian Sea Coast (50), and
Siberian tick spotted fever typhus in a wide area extending
from the Altai Region and Kazakhstan to eastern Siberia and
the Far East of Russia (43, 56). Since 1950, human cases of
spotted fever-like infection have been reported in Armenia,
and the seroprevalence of specific antibodies to SFG
rickett-siae have been determined in sera from humans and domestic
and wild animals (51). Several strains of rickettsiae were
iso-lated from Dermacentor marginatus and Rhipicephalus
san-guineus ticks collected from sheep, cattle, and dogs and from
the brains of small rodents. These isolates have a broad
spec-trum of virulence in laboratory animals and are serologically
distinct in complement fixation tests (33, 43, 51).
Recently, genetic identification of several Armenian isolates
has been performed (4, 17). Restriction fragment length
poly-morphism (RFLP) analysis of PCR-amplified DNA fragments
(PCR-RFLP) of five isolates from D. marginatus ticks showed
one isolate to be indistinguishable from Rickettsia sibirica and
four isolates to be indistinguishable from Rickettsia slovaca.
These findings were confirmed by the analysis of the protein
and chromosomal DNA profiles of the isolates (17).
The purpose of this report is to describe the antigenic and
genomic identifications of strain S isolated from R. sanguineus
ticks in Armenia. When it was first isolated, this strain was
considered to be an atypical strain since it could be
differen-tiated serologically from R. sibirica, Rickettsia conorii, and
Rickettsia akari by the complement fixation test.
MATERIALS AND METHODS
Isolation of the strain.R. sanguineus ticks identified by standard taxonomic characteristics (38) were collected in May 1955 from sheep in Kutschak Village of the Aparanskyi Region (Armenia). Nineteen ticks were triturated and were suspended in a physiological saline buffer (0.15 M NaCl [pH 7.0]) and were inoculated intraperitoneally into two guinea pigs. One guinea pig died 2 days later, while the other developed a fever on the fourth day after inoculation. On day 6, the guinea pig was euthanized and dissected. Splenomegaly, periorchitis, and a peritoneal exudate were found. Rickettsia-like bacteria were observed in smears from the peritoneum and the tunica vaginalis by Zdrodovskyi staining (56).
Samples of brain, spleen, and testicles were homogenized in physiological saline buffer and were inoculated individually or as mixtures into a guinea pig previously unexposed to SFG rickettsiae. From 3 to 8 days after inoculation, all guinea pigs developed periorchitis and a fever (39.6 to 40.88C) which persisted for 3 to 5 days.
After several passages in guinea pigs, chicken embryos were inoculated and a rickettsial culture was established. The isolated rickettsial strain was named strain S.
The biological characteristics of strain S were determined by experimental inoculations of outbread male guinea pigs, white rats and white mice, and chicken embryos as described elsewhere (56).
Reference strains of rickettsiae.The following reference strains of rickettsiae were used. R. conorii Moroccan was obtained from the American Type Culture Collection (VR-141); R. sibirica K-1 (strain 246 of the American Type Culture Collection; VR-151) and Astrakhan spotted fever rickettsia (human isolate) were from the collection of the Gamaleya Research Institute of Epidemiology and Microbiology (Moscow, Russia). Rickettsia slovaca 13-B, Israeli tick typhus (IsTT) rickettsia strain ISTT CDC-1, and Rickettsia africae ESF 2500-1 were kindly supplied by G. A. Dasch (Naval Medical Research Institute, Bethesda, Md.). Comparison with other known SFG rickettsiae was performed by using the data bank of the Unite´ des Rickettsies (Marseille, France) and the Computer Q-Gel-1 system (Appligene; Madison Corp.).
Growth and purification of rickettsiae.The rickettsiae were grown in Vero cell monolayers in Eagle’s minimum essential medium (Seromed, Berlin, Germany) supplemented with 4% fetal calf serum (Seromed)–1%L-glutamine at 328C in a 5% CO2atmosphere (6, 17). Five to 7 days after inoculation, heavily infected cells were harvested and sonicated, and the rickettsiae were purified by differ-ential centrifugation and centrifugation through a 25% sucrose cushion; this was followed by Renografin (Radioselectan; Schering, Lys-Lez-Lannoy, France) den-sity gradient centrifugation (1, 17).
The protein concentration of the purified rickettsial suspension was deter-mined by the Lowry method in 5% sodium dodecyl sulfate (SDS) solution (35). Preparation of mouse antisera.Mouse polyclonal antisera to the rickettsiae * Corresponding author. Mailing address: Unite´ des Rickettsies,
Centre National de Recherche Scientifique EP J0054, Faculte´ de Me ´-decine, 27 Boulevard Jean Moulin, 13385 Marseille Ce´dex 5, France. Phone: (33) 91 83 43 75. Fax: (33) 91 83 03 90.
† Present address: School of Medicine, University of Maryland at Baltimore, Baltimore, MD 21201-1559.
2738
on May 15, 2020 by guest
http://jcm.asm.org/
were obtained by the method of Philip et al. (36). On days 0 and 7, four male Swiss Webster mice were injected via the tail vein with 0.1 ml of infected cell suspension containing about 103to 104PFU of rickettsiae. On day 10 the mice were exsanguinated, and sera from each group of mice were pooled and stored at2208C.
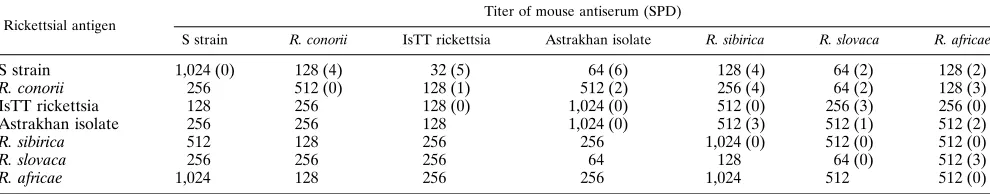
MIF test.The microimmunofluorescence (MIF) test with mouse polyclonal antisera was performed by the method of Philip et al. (36) with fluorescein isothiocyanate-labeled goat anti-mouse immunoglobulin M (IgM) and IgG (di-lution, 1:200; Immunotech, Marseille, France). Rickettsial suspensions partially purified from Vero cells by differential centrifugation (1) were used as antigens. Noninfected Vero cells were included as a negative control in all tests. The antibody titer in serum was determined as the endpoint of twofold serum dilu-tions at which rickettsiae could still be detected by the MIF test. The specificity difference (SPD) between each pair of strains was calculated by the formula SPD 5(Aa1Bb)2(Ab1Ba), where Aa (or Bb) is the antibody titer of serum sample A (or B) reacting with the homologous antigen a (or b), and Ab (or Ba) is the antibody titers in serum in a reaction with the heterologous antigen b (or a). Antibody titers were expressed as log2of the endpoint titer. It was concluded that two isolates were distinct species if the SPD was.3 (P,0.01) (6, 36).
SDS-PAGE and immunoblotting. SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting with mouse antisera were performed by previously described methods (17). High-range prestained SDS-PAGE standards (Bio-Rad, Hercules, Calif.) were used to estimate the molecular masses of the rickettsial proteins.
Peroxidase-labeled goat anti-mouse IgG and IgM antibodies (dilution, 1:100; Immunotech) and 4-chloro-1-naphthol (Sigma, St. Louis, Mo.) with 0.015% H2O2were used to detect rickettsial antigens in the immunoblots.
PCR amplification and RFLP analysis.PCR amplification, DNA digestion, and electrophoresis were carried out as described previously (18, 42). Thirty-five cycles of denaturation (958C for 20 s), annealing (488C for 30 s), and sequence extension (608C for 2 min) were carried out in a thermal cycler (PREM III; Lep Scientific, Flobio, Courbevoie, France). The Rr 190.70p and Rr 190.602n oligo-nucleotide primer pairs encoding for a 532-bp sequence of the 190-kDa protein gene of R. rickettsii (Bioprobe Systems, Montreuil-sous-Bois, France) were used (42). Amplified DNA samples were digested with RsaI and PstI restriction endonucleases (New England Biolabs, Beverly, Mass.), and the digested products were separated on an 8% polyacrylamide gel for 4 h at 100 V. DNA standard marker V (Boehringer Mannheim GmbH, Mannheim, Germany) was run simul-taneously with the samples to determine the DNA fragment sizes.
PFGE.The preparation of DNA-embedded agarose blocks; digestion with SmaI, EagI, and BssHII restriction endonucleases (New England BioLabs); and migration in a 1% agarose gel were carried out as described previously (17, 45). A bacteriophage lambda ladder pulsed-field gel marker of 48.5 to 1,018 kb and a low-range pulsed-field gel marker of 0.1 to 200 kb (New England BioLabs) were used to determine the band sizes.
A dendrogram of the genetic relationships between the SFG rickettsial isolates was established from the three restriction profiles obtained after pulsed-field gel electrophoresis (PFGE) migration as described previously (45). DNA divergence between rickettsial genomes was estimated by using the Dice coefficient (20), and the dendrogram was built by the unweighted pair-group method with arithmetic means (UPGMA) and version 3.4 of the PHYLIP software package, a commer-cial computer programming package (supplied by J. Felsenstein, Seattle, Wash.). 16S rRNA gene sequencing.Sequencing of PCR-derived fragments of the 16S rRNA gene was performed by using the AutoRead sequencing kit (Pharmacia, LKB, Uppsala, Sweden). Oligonucleotide primer sequences (Eurogentec, Sera-ing, Belgium), amplification conditions, and electrophoresis of the sequenced products in the A.L.F. DNA Sequencer (Pharmacia) have been described pre-viously (46).
The 16S rRNA gene sequences were aligned by using the multisequence alignment program CLUSTAL, which is a part of the BISANCE software pack-age (13). Phylogenetic relationships were inferred with version 3.5 of the PHYLIP software package. The evolutionary distance values were determined by the method of Jukes and Cantor (28) and were used to construct a dendrogram by the neighbor-joining method (47). Bootstrap analysis was performed to
esti-mate the stability of the tree that was obtained. Values from the bootstrap analysis were obtained from a consensus tree based on 100 randomly generated trees by using SEQBOOT and CONSENSE in the same software package.
Nucleotide sequence accession numbers.The nucleotide sequences of the 16S rRNA genes of the following reference rickettsial strains were obtained from the GenBank data base: R. prowazekii Breinl, M21789; R. helvetica C9P9, L36212; R. japonica YH, L36213; R. massiliae Mtu1, L36214; R. montana tick strain, L36215; R. rhipicephali 3-7-6, L36216; R. rickettsii Sheila Smith, L36217; R. sibirica K-1, L36218; Thai tick typhus rickettsia strain TT-118, L36220; R. typhi Wilmington, L36221; R. slovaca 13-B, L36224; R. conorii Moroccan, L36105; R. africae ESF 2500-1, L36098; IsTT rickettsia ISTT CDC-1, L36223; Astrakhan spotted fever rickettsia A-167, L36100; R. bellii 369L42-1, L36103; R. canada 2678, L36104; R. akari MK, L36099; R. australis Phillips, L36101; R. parkeri maculatum 20, L36673; strain Bar 29, L36102; ELB bacterium, L28944; and AB bacterium, U04163. The nucleotide sequence of the 16S rRNA gene of strain S will appear in GenBank under accession number U25042.
RESULTS
Biological characteristics of strain S.
Although no mortality
occurred in guinea pigs inoculated with strain S (1 ml of 10
23diluted suspension with an ID
50of 5.5 for chicken embryos),
the animals developed a fever (39.6 to 40.0
8
C) that lasted for 3
to 8 days and had periorchitis for 2 to 4 days, depending on the
initial number of inoculated rickettsiae. Thin bacillary and long
chain-form rickettsiae were detected in the cytoplasms and
nuclei of cells from the peritoneum and tunica vaginalis of
infected animals.
Intraperitoneal infection of white mice with the suspension
of strain S described above resulted in splenomegaly and the
formation of a peritoneal exudate in which rickettsiae could be
detected microscopically. Intravenous inoculation of 0.1 ml of
strain S-infected cell culture (about 10
4PFU of rickettsiae)
caused the death of mice weighing 12 to 14 g on days 4 to 6.
Intraperitoneal inoculation of white rats resulted in fever and
splenomegaly, and scrotal swelling was observed in animals
weighing less than 100 g.
Strain S was characterized by moderate growth in chicken
embryos (ID
50CE5
5.5) but grew well in Vero cell cultures, in
which it had a cytopathic effect.
Serological characterization.
The results of serological
typ-ing of strain S are given in Table 1. Mouse antisera to strain S
cross-reacted with all of the reference SFG rickettsiae. The
lowest SPDs (SPDs
5
2) were found between strain S and R.
slovaca and R. africae. Strain S had less serological
cross-reac-tivity with R. conorii, R. sibirica, the Astrakhan isolate, and the
IsTT rickettsia, for which the SPDs were
$
3.
SDS-PAGE and immunoblotting.
Strain S had a
low-molec-ular-mass protein (
,
106-kDa) profile which was very similar to
those of the reference SFG rickettsiae (Fig. 1). However, the
high-molecular-mass proteins (
.
106 kDa) of strain S had
mo-lecular masses different from those of the reference rickettsiae.
The four proteins in strain S (165, 151, 123, and 112 kDa) were
clearly different from those of R. slovaca (151, 146, 130, and
R. slovaca 256 256 256 64 128 64 (0) 512 (3)
R. africae 1,024 128 256 256 1,024 512 512 (0)
on May 15, 2020 by guest
http://jcm.asm.org/
[image:2.612.59.554.94.191.2]120 kDa), R. africae (176, 142, 121 to 123, and 115 kDa), R.
conorii (143, 140, and 128 kDa), R. sibirica (176, 144, 120, and
114 kDa), the IsTT rickettsia (168, 153, 146, and 119 kDa), and
the Astrakhan isolate (159, 150, 140, and 125 kDa).
Immunoblotting with polyclonal mouse sera against R.
sibirica demonstrated (Fig. 2) that the immunodominant
pro-teins of strain S (165 and 112 kDa) were clearly different from
those of the reference SFG rickettsiae, mainly R. slovaca (151
and 121 kDa), R. sibirica (144 and 114 kDa), R. africae (142
kDa), the IsTT rickettsia (119 kDa), R. conorii (128 kDa), and
the Astrakhan isolate (140 and 125 kDa). The same patterns
were detected when anti-R. conorii, anti-IsTT rickettsia, and
anti-Astrakhan strain sera were used (data are not shown).
Identification by PCR-RFLP analysis.
Results of the
PCR-RFLP analysis are given in Fig. 3. Comparison of the
amplifi-cation products with the Rr 190.70p and Rr 190.602n primer
pair showed that strain S had two RsaI restriction fragments
similar to those of R. africae (Fig. 3B). Its PstI restriction
profile consisted of three bands identical to those identified for
R. sibirica and R. africae and similar to those of the IsTT
rickettsia and the Astrakhan isolate (Fig. 3A).
Comparison of chromosomal DNAs by PFGE.
Restriction
profiles of entire chromosomal DNAs obtained by PFGE are
presented in Fig. 4. Eighteen to 20 fragments were identified
after SmaI restriction endonuclease digestion (Fig. 4B), 13 to
17 fragments were identified after EagI digestion (Fig. 4A),
and 3 to 6 fragments were identified after BssHII digestion
(Fig. 4C). SmaI digestion generated closely related patterns;
however, specific patterns were determined for each isolate
compared. EagI and BssHII profiles were more specific and
allowed for the easy differentiation of strain S from the other
strains.
The genetic divergence of the rickettsial isolates that were
compared was determined by numerical analysis. The
UP-GMA method that was used grouped strain S with R. africae on
the basis of the fact that they had 81% genetic similarity (Fig.
5). This cluster had 26 to 29% genetic divergence with IsTT
rickettsiae, R. conorii, and R. sibirica and 33 and 36% genetic
divergence with the Astrakhan isolate and R. slovaca,
respec-tively.
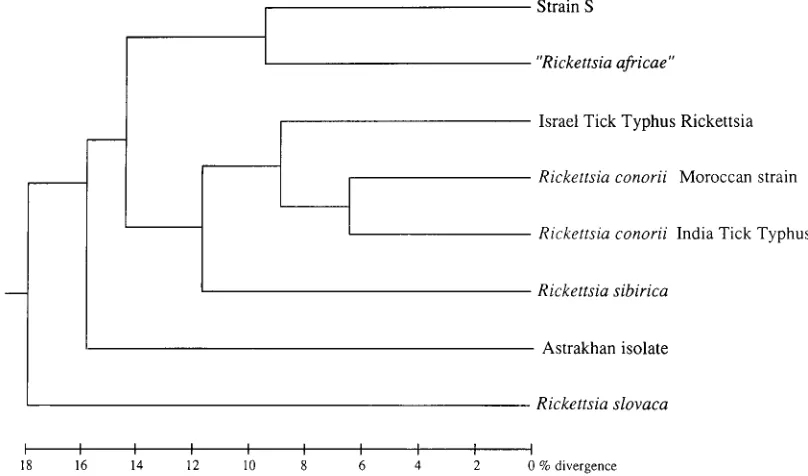
16S rRNA gene sequencing.
The 16S rRNA gene of strain S
was sequenced, and its specific differences from the reference
SFG rickettsiae were established. The sequence of strain S
differed in three nucleotides from those of R. africae and R.
sibirica and differed in four nucleotides from that of R. parkeri.
When a numerical analysis was performed, strain S was
clustered with R. africae, and both of them were grouped with
R. sibirica and R. parkeri (Fig. 6). A bootstrap analysis was done
to estimate the reliability of the phylogenetic analysis. A
boot-strap analysis probability of 100% was determined for the
cluster of R. sibirica, R. parkeri, R. africae, and strain S within
the SFG rickettsiae. However, the phylogenetic relatedness of
strain S to R. africae has been found to have a probability of
only 43%.
DISCUSSION
The SFG rickettsiae include 14 different species and several
isolates which have no official name but which differ from the
other isolates (6, 7, 44, 55). Until recently, the species
defini-tion of SFG rickettsiae has been based on their serotype
de-termined by the complement fixation test (37), toxin
neutral-ization test in mice (9), cross-immunity test in guinea pigs (11),
or MIF test (36). However, SFG rickettsiae express significant
cross-reactivity between recognized species, which makes the
serological differentiation of newly isolated strains difficult
(36). It led several tick isolates that originated from areas
FIG. 1. Coomassie R-250-stained SDS-PAGE profiles of the strains of SFGrickettsiae examined. Lanes: 1, R. slovaca; 2, R. sibirica; 3, strain S; 4, R. conorii; 5, Astrakhan isolate; 6, R. africae; 7, IsTT rickettsia. S, standard proteins, with molecular masses (in kilodaltons) indicated on the right side.
FIG. 2. Western immunoblotting of SDS-PAGE-separated high-molecular-mass rickettsial proteins reacted with mouse antisera to R. sibirica. Lanes: 1, strain S; 2, R. slovaca; 3, R. sibirica; 4, R. africae; 5, IsTT rickettsia; 6, R. conorii; 7, Astrakhan isolate. The positions of standard proteins are indicated on the right side (numbers are in kilodaltons).
FIG. 3. Ethidium bromide-stained polyacrylamide gel electrophoretograms of restriction endonuclease PstI-digested (A) and RsaI-digested (B) rickettsial DNA amplified by PCR with the Rr 190.70p and Rr 190.602n primer pair. Lanes: 1, strain S; 2, R. conorii; 3, R. slovaca; 4, R. sibirica; 5, IsTT rickettsia; 6, R. africae; 7, Astrakhan isolate. S, molecular size markers with sizes indicated on the right side (in base pairs).
on May 15, 2020 by guest
http://jcm.asm.org/
where recognized SFG rickettsioses are endemic or areas
where they are not endemic to be characterized as atypical
strains (33, 44, 51, 52).
Genomic identification of rickettsiae on the basis of RFLP
analysis of surface protein gene amplicons has recently been
proposed as an alternative to classical serological methods (6,
18, 41, 42). This new method has been shown to have high
degrees of reproducibility and specificity and correlates well
with the current classifications by the MIF test. The use of
RFLP analysis of surface protein gene amplicons together with
PFGE and SDS-PAGE analyses was shown to be of value for
the classification of known rickettsiae and the identification of
new isolates (17).
In the present study we used PCR-RFLP, the MIF test,
SDS-PAGE, PFGE, and 16S rRNA gene sequencing to
deter-mine the relationship of strain S (17) to recognized SFG
rick-ettsiae. PCR-RFLP analysis with RsaI showed the profile of
strain S to be similar to that of R. africae, while PCR-RFLP
analysis with PstI showed that the profile was identical to those
of R. sibirica and R. africae. This similarity between the
PCR-RFLP profiles of strain S and R. africae has been reported
previously (17). Our MIF studies in which the SPD between
strain S and R. africae was 2 provides further evidence of the
relatedness of these organisms. Strain S, however, had specific
high-molecular-mass proteins, detected by SDS-PAGE, which
were distinct from those of R. africae, those of the five other
reference SFG rickettsiae tested, and those in our data bank.
These findings suggest that strain S is a new type of SFG
rickettsia.
[image:4.612.80.535.74.213.2]To confirm the identification of strain S as a new type of
SFG rickettsia we studied the entire chromosomal DNAs using
PFGE, a proposed method for establishing the correct
taxo-nomic position of bacterial isolates (2, 10, 32, 45). Strain S was
found to have specific genomic polymorphism patterns which
were distinct from those of the known SFG rickettsiae. A close
similarity was found, however, to the patterns of R. africae, the
FIG. 4. EagI (A), SmaI (B), and BssHII (C) restriction endonuclease profiles of SFG rickettsiae obtained by PFGE. Lanes: 1, R. slovaca; 2, R. sibirica; 3, R. conorii; 4, Astrakhan isolate; 5, strain S; 6, R. africae; 7, IsTT rickettsia. Lanes S, DNA size markers (in kilobases).FIG. 5. Dengrogram of genetic relatedness among strain S and reference SFG rickettsial strains established on the basis of the PFGE restriction profile comparison.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:4.612.103.507.471.709.2]etiological agent of African tick-bite fever (29, 30), which is
transmitted by Amblyomma ticks (31).
Strain S was also found to be related to R. africae on the
basis of 16S rRNA gene sequence analysis, and both of them
have been grouped with R. sibirica and R. parkeri. The 16S
rRNA gene provides information about the phylogenetic
ori-gin of the isolates examined (19, 48, 49). However, the close
phylogenetic relatedness of strain S and R. africae is difficult to
consider because the confidence for this cluster is very low,
corresponding to a probability of only 43% by bootstrap
anal-ysis. This value might be related to a few differences in the 16S
rRNA gene among SFG rickettsiae which do not allow us to
obtain a precise phylogenetic analysis of these organisms (23,
46).
Genetic homology determined by DNA-DNA hybridization
is the reference method of species designation used today (19,
48, 54). Classical DNA relatedness criteria have been
estab-lished for human-adapted members of the family
Enterobacte-riaceae. Two strains are considered to belong to one species if
there is 70% or greater DNA homology (54). Accordingly, the
data available for SFG rickettsial DNA-DNA relatedness
sug-gest that there are only three species within the group (53). On
the basis of 91 to 73% DNA homology, R. rickettsii, R. conorii,
R. sibirica, and R. montana, organisms with distinct phenotypic
characteristics, form one species. R. akari and R. australis,
which share 46 and 53% homology, respectively, with R.
rick-ettsii are the other two species. The use of DNA homology for
the identification of SFG rickettsial isolates is not widely used,
however, since it requires large numbers of organisms for such
analysis. Moreover, rickettsiae differ from free-living bacteria
by the absence of genetic recombination and have a low level
of genetic variability (21), and the applicability of DNA
ho-mology studies with these organisms can therefore be
ques-tioned. The limits of variability in the described rickettsial
species is being examined in a number of studies (21–23, 39, 45,
46). Until these studies are completed and additional
charac-teristics of these isolates are described, it will be hard to know
precisely what comprises a meaningful rickettsial species. Other
taxonomic criteria, such as physiological or virulence
charac-teristics combined with ecoepidemiological and chemotaxic
characteristics, might be the most appropriate criteria for use
in the differentiation of rickettsiae at the species level. For
practical reasons it might be useful to give different names to
rickettsial isolates that cause clinically different spotted fevers.
Although it has been proposed that strain S causes human
SFG rickettsiosis in Armenia (27), the organism has not been
isolated from humans but has only been isolated from R.
san-guineus. Accordingly, this organism might best be called a new
[image:5.612.125.488.68.433.2]type of SFG rickettsia. If the organism is found to be a human
pathogen, this might justify the allocation of a species name.
FIG. 6. Phylogenetic position of strain S among SFG rickettsiae determined by 16S rRNA gene nucleotide sequence analysis. The dendrogram was constructed by the neighbor-joining method. The scale bar represents 0.5% difference in nucleotide sequences.on May 15, 2020 by guest
http://jcm.asm.org/
strains were isolated from ticks and were genetically identified,
and correlation between the number of rickettsia-infected ticks
and the seroprevalence in humans was determined (25, 34, 40,
43). R. sanguineus has been implicated in Astrakhan fever
rickettsiosis (14), since the rickettsial agent of this disease has
been identified by PCR-RFLP analysis in 60% of R. sanguineus
ticks collected in the area where Astrakhan fever rickettsiosis
is endemic. The rickettsial strain, however, has yet to be
iso-lated. It is possible that R. sanguineus could also be a vector of
R. sibirica in west Pakistan (44) and R. rickettsii in Mexico (12),
where rickettsiae have been detected in ticks by the MIF test.
Several new rickettsial isolates with undetermined
pathogenic-ity for humans have recently been found to infect R. sanguineus
ticks, including R. rhipicephali in the United States (12) and
France (15), R. massiliae in France (5) and its genomic variant,
strain GS, in Greece (3), and an Mtu 5-like strain in Spain (8).
The results of our study extend the present knowledge of the
geographical distribution and genetic diversity of SFG
rickett-siae transmitted by R. sanguineus ticks. They also provide
ev-idence for a larger role of this tick in the epidemiology of
rickettsial diseases. Our finding that strain S is pathogenic in
laboratory animals suggests that further investigations are
war-ranted to determine if this organism causes disease in humans.
ACKNOWLEDGMENTS
We thank P. Kelly for the review of the manuscript.
The research described in this report was made possible in part by grant M28000 from the International Science Foundation.
REFERENCES
1. Aniskovich, L. P., M. E. Eremeeva, N. M. Balayeva, V. F. Ignatovich, M. I. Artemiev, V. V. Emelyanov, and N. S. Smirnova.1989. Methods for purifi-cation of Rickettsia prowazekii separated from the host tissue: a step-by-step comparison. Acta Virol. 33:361–370.
2. Arbeit, R. D., M. Arthur, R. Dunn, C. Kim, R. K. Selander, and R. Goldstein. 1990. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J. Infect. Dis. 161:230–235.
3. Babalis, T., Y. Tselentis, V. Roux, A. Psaroulaki, and D. Raoult. 1994. Isolation and identification of a rickettsial strain related to Rickettsia mas-siliae in greek ticks. Am. J. Trop. Med. 50:365–372.
4. Balayeva, N. M., M. E. Eremeeva, and D. Raoult. 1994. Genotypic identifi-cation of Rickettsia slovaca among spotted fever group rickettsia isolates from Dermacentor marginatus in Armenia. Acta Virol. 6:321–325. 5. Beati, L. Personal communication.
6. Beati, L., J. P. Finidori, B. Gilot, and D. Raoult. 1992. Comparison of serologic typing, sodium dodecyl sulfate-polyacrylamide gel electrophoresis protein analysis, and genetic restriction fragment length polymorphism anal-ysis for identification of rickettsiae: characterization of two new rickettsial strains. J. Clin. Microbiol. 30:1922–1930.
7. Beati, L., and D. Raoult. 1993. Nouvelles rickettsies du groupe boutonneux en France et dans le monde. Med. Mal Infect. 23(Spe´cial):491–498. 8. Beati, L., V. Roux, A. Ortuno, J. Castella, F. Segura-Porta, and D. Raoult.
Phenotypic and genotypic characterization of SFG rickettsiae isolated from Catalan Rhipicephalus sanguineus ticks. Submitted for publication. 9. Bell, E. J., and H. G. Stoenner. 1960. Immunologic relationships of the
spotted fever group rickettsias determined by toxin neutralisation tests in mice with convalescent animal serums. J. Immunol. 84:171–182.
10. Beyazova, M., and M. P. Lechevalier. 1993. Taxonomic utility of restriction endonuclease fingerprinting of large DNA fragments from Streptomyces strains. Int. J. Syst. Bacteriol. 43:674–682.
11. Bozeman, M. F., J. W. Humphries, J. M. Campbell, and P. L. O’Hara. 1960.
15. Drancourt, M., R. Regnery, and D. Raoult. 1991. Identification of tick-isolates by centrifugation-shell vial assay followed by polymerase chain re-action and restriction endonuclease length polymorphism analysis, p. 232– 238. In J. Kazar and D. Raoult (ed.), Rickettsiae and rickettsial diseases. Proceedings of the Fourth International Symposium on Rickettsiae and Rickettsial Diseases. Veda Publishing House of the Slovak Academy of Sciences, Bratislava, Czechoslovakia.
16. Durand, P., and E. Conseil. 1930. Transmission expe´rimentale de la fievre boutonneuse par Rhipicephalus sanguineus. C. R. Acad. Sci. Ser. D 190:1244. 17. Eremeeva, M. E., N. M. Balayeva, V. F. Ignatovich, and D. Raoult. 1993. Proteinic and genomic identification of spotted fever group rickettsiae iso-lated in the former USSR. J. Clin. Microbiol. 31:2625–2633.
18. Eremeeva, M., X. Yu, and D. Raoult. 1994. Differentiation among spotted fever group rickettsiae species by analysis of restriction fragment length polymorphism of polymerase chain reaction amplified DNA. J. Clin. Micro-biol. 32:803–810.
19. Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to quarantee species identity. Int. J. Syst. Bacteriol. 42:166–170.
20. Freney, J., F. Renaud, W. Hansen, and C. Bollet. 1992. Manuel de bacte ´ri-ologie clinique. Elsevier, Paris.
21. Fuerst, P., K. P. Poetter, C. Pretzman, and P. S. Perlman. 1990. Molecular genetics of populations of intracellular bacteria: the spotted fever group rickettsiae. Ann. N. Y. Acad. Sci. 590:430–438.
22. Fuerst, P. A., and K. P. Poetter. 1991. DNA sequence differentiation in North American spotted fever group species of Rickettsia, p. 162–169. In J. Kazar and D. Raoult (ed.), Rickettsiae and rickettsial diseases. Proceedings of the Fourth International Symposium on Rickettsiae and Rickettsial Dis-eases. Veda Publishing House of the Slovak Academy of Sciences, Bratislava, Slovakia.
23. Fuerst, P. A., and D. R. Stothard. 1994. Relative phylogenetic information in the 16S and 23S rRNA and 17 kDa antigens genes for evolutionary studies of Rickettsia. 11th sesqui-annual meeting of the American Society for Rick-ettsiology and Rickettsial Diseases 1994. American Society for Rickettsiol-ogy and Rickettsial Diseases, St. Simons Island, Ga.
24. Gilot, B., M. L. Laforge, J. Pichot, and D. Raoult. 1990. Relationships between the Rhipicephalus sanguineus compex ecology and Mediterranean spotted fever in France. Eur. J. Epidemiol. 6:357–362.
25. Goldwasser, R. A., Y. Steiman, W. Klinberg, T. A. Swartz, and M. A. Klin-berg.1974. The isolation of strains of rickettsiae of the spotted fever group in Israel and their differentiation from other members of the group by immunofluorescence methods. Scand. J. Infect. Dis. 6:53–62.
26. Herrero-Herrero, J. I., R. Ruiz-Beltran, A. M. Martin-Sanchez, and E. J. Garcia.1989. Mediterranean spotted fever in Salamanca, Spain. Epidemio-logical study in patients and serosurvey in animals and healthy human pop-ulation. Acta Trop. (Basel) 46:335–350.
27. Hoogstraal, H. 1967. Ticks in relation to human diseases caused by Rickettsia species. Annu. Rev. Entomol. 12:377–420.
28. Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21–132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York.
29. Kelly, P. J., L. Beati, P. Mason, R. Makombe, L. Matthewman, D. Raoult, and M. Dreary.1992. African tick-bite fever—a new spotted fever group rickettsiosis under an old name. Lancet 340:982–983.
30. Kelly, P. J., L. Beati, L. A. Matthewman, P. R. Mason, and D. Raoult. 1994. A new spotted fever group rickettsia from Africa. J. Trop. Med. Hyg. 97: 129–137.
31. Kelly, P. J., and P. R. Mason. 1991. Transmission of a spotted fever group rickettsia by Amblyomma hebraeum (Acari: Ixodidae). J. Med. Entomol. 28:598–600.
32. Knattak, M. N., and R. C. Matthews. 1993. Genetic relatedness of Bordetella species as determined by macrorestriction digests resolved by pulsed-field gel electrophoresis. Int. J. Syst. Bacteriol. 43:659–664.
33. Makarova, V. A., I. V. Tarasevich, and L. F. Plotnikova. 1978. Antigenic structure of Rickettsiae isolates in Czechoslovakia and USSR and their po-sition in the spotted fever group, p. 293–297. In J. Kazar, R. A. Ormsbee, and I. V. Tarasevich (ed.), Rickettsiae and rickettsial diseases. Proceedings of the Second International Symposium on Rickettsiae and Rickettsial Diseases. Veda Publishing House of the Slovak Academy of Sciences, Bratislava, Czechoslovakia.
on May 15, 2020 by guest
http://jcm.asm.org/
34. Manor, E., J. Ighbarieh, B. Sarov, I. Kassis, and R. Regnery. 1992. Human and tick spotted fever group rickettsia isolates from Israel: a genotypic analysis. J. Clin. Microbiol. 30:2656–2657.
35. Peterson, G. L. 1983. Determination of total protein. Methods Enzymol. 91:95–119.
36. Philip, R. N., E. A. Casper, W. Burgdorfer, R. K. Gerloff, L. E. Hugues, and J. Bell.1978. Serologic typing of Rickettsiae of the spotted fever group by micro immunofluorescence. J. Immunol. 121:1961–1968.
37. Plotz, H., R. L. Reagan, and K. Wertman. 1944. Differentiation between Fie`vre Boutonneuse and Rocky Mountain Spotted Fever by means of com-plement fixation. Proc. Soc. Exp. Biol. Med. 55:173–176.
38. Pomerantzev, B. I. 1959. Fauna of USSR. ARACHIDA. Ixodid ticks (Ixo-didae). The American Institute of Biological Sciences, Washington, D.C. 39. Ralph, D., C. Pretzman, N. Daugherty, and K. Poeter. 1990. Genetic
rela-tionships among the members of the family Rickettsiaceae as shown by DNA restriction polymorphism analysis. Ann. N. Y. Acad. Sci. 590:541–552. 40. Raoult, D., H. Tissot Dupont, C. Chicheportiche, O. Peter, B. Gilot, and M.
Drancourt.1993. Mediterranean spotted fever in Marseille, France: corre-lation between prevalence of hospitalized patients, seroepidemiology, and prevalence of infected ticks in three different areas. Am. J. Trop. Med. 48:249–256.
41. Regnery, R. L. 1990. Use of DNA probes for differentiation of spotted fever group and other rickettsiae. Ann. N. Y. Acad. Sci. 590:422–429. 42. Regnery, R. L., C. L. Spruill, and B. D. Plikaytis. 1991. Genotypic
identifi-cation of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J. Bacteriol. 173:1576–1589.
43. Rehacek, J., and I. V. Tarasevich. 1988. Acari-borne rickettsiae and rickett-sioses in Eurasia. Veda Publishing House of the Slovak Academy of Sci-ences, Bratislava, Czechoslovakia.
44. Robertson, R. G., and C. L. Wisseman, Jr. 1973. Tick-borne rickettsiae of spotted fever group in West Pakistan. II. Serological classification of isolates from west Pakistan and Thialand: evidence for two new species. Am. J. Epidemiol. 97:55–64.
45. Roux, V., and D. Raoult. 1993. Genotypic identification and phylogenetic analysis of the spotted fever group rickettsiae using pulsed field gel
electro-phoresis. J. Bacteriol. 175:4895–4904.
46. Roux, V., and D. Raoult. 1995. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res. Microbiol. 146:385–396.
47. Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. 48. Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for
DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846–849. 49. Stothard, D. R., J. B. Clark, and P. A. Fuerst. 1994. Ancestral divergence of
Rickettsia bellii from the spotted and typhus groups of Rickettsia and antiq-uity of the genus Rickettsia. Int. J. Syst. Bacteriol. 44:798–804.
50. Tarasevich, I. V., V. A. Makarova, N. F. Fetisova, A. Stepanov, E. D. Miskarova, N. M. Balayeva, and D. Raoult.1991. Astrakhan fever: new spotted fever group rickettsiosis. Lancet 337:172–173.
51. Tarasevich, I. V., L. F. Plotnikova, N. F. Fetisova, V. A. Makarova, V. A. Jablonskaya, J. Rehacek, M. Zupancicova, E. Kovacova, J. Urvo¨lgyi, R. Brezina, A. V. Zakarjan, and M. E. Kocinjan.1976. Rickettsioses studies. 1. Natural foci of rickettsioses in the Armenian Soviet Socialist Republic. Bull. W. H. O.53:25–30.
52. Vorontzova, T. A., A. A. Pchelkina, I. I. Seledtzov, and V. J. Malyshev. 1980. Use of the antibody neutralization test for investigation of Rickettsia sibirica circulation in natural foci. Med. Parasitol. 49:62–73. (In Russian.) 53. Walker, D. H. 1989. Rocky Mountain spotted fever: a disease in need of
microbiological concern. Clin. Microbiol. Rev. 2:227–240.
54. Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackenbrandt, M. P. Starr, and H. G. Truper.1987. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37:463–464.
55. Weiss, E., and J. W. Moulder. 1984. Order I Rickettsiales, Gieszczkiewicz 1939, 25AL
, p. 687–701. In N. R. Kreig and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore. 56. Zdrodowsky, P. F., and E. M. Golinevitch. 1972. Rickettsiae and rickettsial
diseases. Meditzina, Moscow. (In Russian.)