organic papers
o862
Jinget al. C14H12N4O5 doi:10.1107/S160053680600287X Acta Cryst.(2006). E62, o862–o863
Acta Crystallographica Section E Structure Reports
Online
ISSN 1600-5368
N
-(2,4-Dinitrophenyl)-
N
000-(4-methoxybenzyl-idene)hydrazine
Zuo-Liang Jing,* Yu Liu, Xin Chen and Yu Ming
College of Sciences, Tianjin University of Science and Technology, Tianjin 300222, People’s Republic of China
Correspondence e-mail: jzl74@tust.edu.cn
Key indicators
Single-crystal X-ray study
T= 294 K
Mean(C–C) = 0.002 A˚
Rfactor = 0.034
wRfactor = 0.096
Data-to-parameter ratio = 11.6
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 20 January 2006 Accepted 24 January 2006
#2006 International Union of Crystallography All rights reserved
In the crystal structure of the title compound, C14H12N4O5,
molecules are connected viaweak intermolecular N—H O and C—H O hydrogen bonds, forming a three-dimensional network.
Comment
Metal complexes based on Schiff bases have attracted much attention because they can be utilized as model compounds of active centres in various proteins and enzymes (Kahwaet al., 1986; Santos et al., 2001). As part of an investigation of the coordination properties of Shiff bases functioning as ligands (Yuet al., 2005; Denget al., 2005; Jing, Fanet al., 2005a,b; Jing, Wanget al., 2005), we report the synthesis and structure of the title compound, (I).
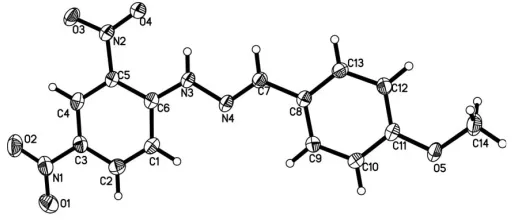
The molecular structure of (I) is approximately planar (Fig. 1); the central chromophore (C1–C6/N1–N4) and the 4-methoxybenzaldehyde group (C7—C13/O5) are planar with r.m.s. deviations of 0.0288 (2) and 0.0051 (5) A˚ , respectively; the dihedral angle between these two planes is 1.29 (7). An
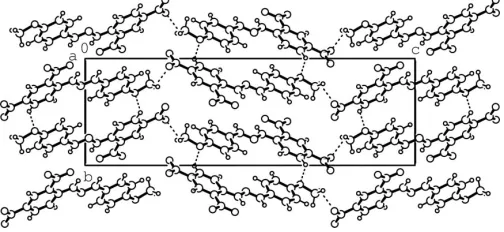
intramolecular N—H O hydrogen bond stabilizes the mol-ecular structure, while intermolmol-ecular N—H O and C— H O hydrogen bonds stabilize the crystal structure (Table 2 and Fig. 2).
Experimental
[image:1.610.204.460.609.721.2]An anhydrous ethanol solution (50 ml) of 4-methoxybenzaldehyde (1.36 g, 10 mmol) was added to an anhydrous ethanol solution
Figure 1
(50 ml) of (2,4-dinitrophenyl)hydrazine (2.03 g, 10 mmol) and the mixture was stirred at 330 K for 6 h under N2, whereupon a red
solution appeared. The solvent was removed and the residue was recrystallized from N,N-dimethylformamide. The product was isolated and then driedin vacuoto give the title compound in 82% yield. Red single crystals suitable for X-ray analysis were obtained by slow evaporation of anN,N-dimethylformamide solution of (I).
Crystal data
C14H12N4O5
Mr= 316.28
Monoclinic,P21=n a= 6.189 (2) A˚
b= 8.581 (3) A˚
c= 26.669 (11) A˚
= 96.392 (5) V= 1407.5 (9) A˚3
Z= 4
Dx= 1.492 Mg m 3
MoKradiation Cell parameters from 2340
reflections
= 3.1–26.1
= 0.12 mm1
T= 294 (2) K Block, red
0.240.200.18 mm
Data collection
Bruker SMART CCD area-detector diffractometer
’and!scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin= 0.963,Tmax= 0.979
7349 measured reflections
2481 independent reflections 1914 reflections withI> 2(I)
Rint= 0.017 max= 25.0
h=7!6
k=10!9
l=31!31
Refinement
Refinement onF2
R[F2> 2(F2)] = 0.034
wR(F2) = 0.096
S= 1.03 2481 reflections 213 parameters
H atoms treated by a mixture of independent and constrained refinement
w= 1/[2(F
o2) + (0.0536P)2
+ 0.1714P]
whereP= (Fo2+ 2Fc2)/3
(/)max< 0.001
max= 0.11 e A˚
3
min=0.21 e A˚
[image:2.610.46.298.74.190.2]3
Table 1
Selected geometric parameters (A˚ ,).
O5—C11 1.3614 (15) N1—C3 1.4500 (19) N2—C5 1.4462 (18)
N3—C6 1.3457 (18) N3—N4 1.3782 (15) N4—C7 1.2746 (18) C6—N3—N4 119.99 (12) C7—N4—N3 114.37 (12)
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
N3—H3 O4 0.869 (18) 1.984 (17) 2.6189 (17) 129.0 (14) N3—H3 O4i
0.869 (18) 2.620 (18) 3.4419 (19) 158.2 (14) C2—H2 O5ii
0.93 2.59 3.375 (2) 143 C14—H14B O2iii
0.96 2.56 3.188 (3) 123
Symmetry codes: (i)x;yþ2;z; (ii)xþ2;yþ1;z; (iii)xþ1 2;yþ
1 2;z
3 2.
The H atom attached to N atom was located in a difference Fourier map and refined freely. C-bound H atoms were included in calculated positions and refined using a riding model approximation, with C—H = 0.93 A˚ (aromatic) and 0.96 A˚ (methyl), and with Uiso(H) =
1.2Ueq(C) and 1.5Ueq(C), respectively.
Data collection:SMART(Bruker, 1999); cell refinement:SAINT
(Bruker, 1999); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics:
SHELXTL (Bruker, 1997); software used to prepare material for publication:SHELXTL.
References
Bruker (1997). SHELXTL (Version 5.10). Bruker AXS Inc., Madison, Wisconsin, USA.
Bruker (1999).SMART(version 5.0) andSAINT(Version 4.0). Bruker AXS Inc., Madison, Wisconsin, USA.
Deng, Q.-L., Yu, M., Chen, X., Diao, C.-H., Jing, Z.-L. & Fan, Z. (2005).Acta Cryst.E61, o2545–o2546.
Jing, Z.-L., Fan, Z., Yu, M., Chen, X. & Deng, Q.-L. (2005a).Acta Cryst.E61, o3208–o3209.
Jing, Z.-L., Fan, Z., Yu, M., Chen, X. & Deng, Q.-L. (2005b).Acta Cryst.E61, o3495–o3496.
Jing, Z.-L., Wang, X.-Y., Chen, X. & Deng, Q.-L. (2005).Acta Cryst.E61, o4316–o4317.
Kahwa, I. A., Selbin, J., Hsieh, T. C.-Y. & Laine, R. A. (1986).Inorg. Chim. Acta,118, 179–185.
Santos, M. L. P., Bagatin, I. A., Pereira, E. M. & Ferreira, A. M. D. C. (2001).J. Chem. Soc. Dalton Trans.838–844.
Sheldrick, G. M. (1996).SADABS. University of Go¨ttingen, Germany. Sheldrick, G. M. (1997).SHELXS97&SHELXL97. University of Go¨ttingen,
Germany.
Yu, M., Chen, X. & Jing, Z.-L. (2005).Acta Cryst.E61, o1345–o1346.
Figure 2
[image:2.610.315.566.93.146.2]supporting information
sup-1 Acta Cryst. (2006). E62, o862–o863
supporting information
Acta Cryst. (2006). E62, o862–o863 [https://doi.org/10.1107/S160053680600287X]
N
-(2,4-Dinitrophenyl)-
N
′
-(4-methoxybenzylidene)hydrazine
Zuo-Liang Jing, Yu Liu, Xin Chen and Yu Ming
(I)
Crystal data
C14H12N4O5
Mr = 316.28 Monoclinic, P21/n
a = 6.189 (2) Å
b = 8.581 (3) Å
c = 26.669 (11) Å
β = 96.392 (5)°
V = 1407.5 (9) Å3
Z = 4
F(000) = 656
Dx = 1.492 Mg m−3
Mo Kα radiation, λ = 0.71073 Å
Cell parameters from 2340 reflections
θ = 3.1–26.1°
µ = 0.12 mm−1
T = 294 K
Block, yellow
0.24 × 0.20 × 0.18 mm
Data collection
CCD area detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
φ and ω scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin = 0.963, Tmax = 0.979
7349 measured reflections 2481 independent reflections 1914 reflections with I > 2σ(I)
Rint = 0.017
θmax = 25.0°, θmin = 2.5°
h = −7→6
k = −10→9
l = −31→31
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.034
wR(F2) = 0.096
S = 1.03
2481 reflections 213 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H atoms treated by a mixture of independent and constrained refinement
w = 1/[σ2(F
o2) + (0.0536P)2 + 0.1714P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.11 e Å−3
Δρmin = −0.21 e Å−3
Special details
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
O1 0.3423 (2) 0.45260 (16) 0.24373 (4) 0.0786 (4)
O2 0.0374 (2) 0.57267 (15) 0.24395 (4) 0.0732 (4)
O3 −0.21341 (15) 0.91196 (13) 0.11194 (4) 0.0544 (3)
O4 −0.05330 (15) 0.93642 (13) 0.04464 (4) 0.0485 (3)
O5 1.16761 (16) 0.67880 (13) −0.15044 (4) 0.0534 (3)
N1 0.2021 (2) 0.54176 (16) 0.22460 (5) 0.0554 (4)
N2 −0.06467 (17) 0.88067 (13) 0.08692 (4) 0.0396 (3)
N3 0.31318 (19) 0.79195 (15) 0.03573 (4) 0.0410 (3)
H3 0.218 (3) 0.856 (2) 0.0209 (6) 0.063 (5)*
N4 0.49046 (17) 0.74551 (14) 0.01246 (4) 0.0407 (3)
C1 0.4396 (2) 0.63427 (17) 0.10686 (5) 0.0438 (4)
H1 0.5617 0.6076 0.0914 0.053*
C2 0.4153 (2) 0.57340 (18) 0.15287 (6) 0.0468 (4)
H2 0.5199 0.5063 0.1685 0.056*
C3 0.2329 (2) 0.61192 (16) 0.17645 (5) 0.0430 (3)
C4 0.0793 (2) 0.71209 (16) 0.15458 (5) 0.0413 (3)
H4 −0.0409 0.7378 0.1709 0.050*
C5 0.1041 (2) 0.77512 (15) 0.10779 (5) 0.0361 (3)
C6 0.2851 (2) 0.73685 (15) 0.08180 (5) 0.0369 (3)
C7 0.4991 (2) 0.80444 (17) −0.03114 (5) 0.0392 (3)
H7 0.3898 0.8727 −0.0439 0.047*
C8 0.6728 (2) 0.76941 (16) −0.06176 (5) 0.0366 (3)
C9 0.8453 (2) 0.66906 (16) −0.04617 (5) 0.0394 (3)
H9 0.8509 0.6203 −0.0149 0.047*
C10 1.0061 (2) 0.64189 (17) −0.07647 (5) 0.0407 (3)
H10 1.1199 0.5748 −0.0657 0.049*
C11 1.0002 (2) 0.71415 (16) −0.12336 (5) 0.0394 (3)
C12 0.8303 (2) 0.81308 (17) −0.13959 (5) 0.0405 (3)
H12 0.8245 0.8612 −0.1710 0.049*
C13 0.6694 (2) 0.83953 (17) −0.10867 (5) 0.0398 (3)
H13 0.5554 0.9063 −0.1196 0.048*
C14 1.1818 (3) 0.7624 (2) −0.19634 (6) 0.0601 (5)
H14A 1.0598 0.7361 −0.2203 0.090*
H14B 1.3141 0.7347 −0.2098 0.090*
H14C 1.1811 0.8723 −0.1897 0.090*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
supporting information
sup-3 Acta Cryst. (2006). E62, o862–o863
O2 0.0946 (9) 0.0761 (9) 0.0554 (7) 0.0086 (7) 0.0364 (7) 0.0042 (6)
O3 0.0427 (6) 0.0635 (7) 0.0608 (7) 0.0119 (5) 0.0231 (5) 0.0002 (5)
O4 0.0453 (6) 0.0573 (7) 0.0442 (6) 0.0096 (5) 0.0103 (5) 0.0060 (5)
O5 0.0477 (6) 0.0642 (7) 0.0528 (6) 0.0114 (5) 0.0257 (5) 0.0062 (5)
N1 0.0774 (10) 0.0484 (8) 0.0416 (7) −0.0004 (7) 0.0121 (7) −0.0024 (6)
N2 0.0341 (6) 0.0406 (7) 0.0454 (7) 0.0001 (5) 0.0098 (5) −0.0048 (5)
N3 0.0343 (6) 0.0494 (8) 0.0413 (7) 0.0062 (5) 0.0128 (5) 0.0004 (6)
N4 0.0339 (6) 0.0468 (7) 0.0435 (7) 0.0018 (5) 0.0133 (5) −0.0047 (5)
C1 0.0384 (7) 0.0446 (8) 0.0503 (9) 0.0047 (6) 0.0124 (6) −0.0024 (7)
C2 0.0481 (8) 0.0431 (9) 0.0494 (9) 0.0051 (6) 0.0055 (7) 0.0009 (7)
C3 0.0540 (9) 0.0389 (8) 0.0366 (7) −0.0022 (7) 0.0081 (6) −0.0023 (6)
C4 0.0450 (8) 0.0379 (8) 0.0430 (8) −0.0030 (6) 0.0142 (6) −0.0090 (6)
C5 0.0345 (7) 0.0360 (8) 0.0389 (7) −0.0009 (5) 0.0088 (6) −0.0056 (6)
C6 0.0346 (7) 0.0355 (7) 0.0415 (8) −0.0035 (6) 0.0080 (6) −0.0057 (6)
C7 0.0342 (7) 0.0437 (8) 0.0405 (8) 0.0021 (6) 0.0071 (6) −0.0024 (6)
C8 0.0332 (7) 0.0385 (8) 0.0389 (7) −0.0019 (6) 0.0073 (6) −0.0048 (6)
C9 0.0396 (7) 0.0437 (8) 0.0354 (7) 0.0006 (6) 0.0067 (6) 0.0015 (6)
C10 0.0358 (7) 0.0429 (8) 0.0439 (8) 0.0053 (6) 0.0058 (6) 0.0001 (6)
C11 0.0356 (7) 0.0415 (8) 0.0430 (8) −0.0015 (6) 0.0125 (6) −0.0058 (6)
C12 0.0422 (8) 0.0452 (8) 0.0347 (7) −0.0003 (6) 0.0067 (6) 0.0009 (6)
C13 0.0357 (7) 0.0429 (8) 0.0407 (8) 0.0049 (6) 0.0031 (6) −0.0018 (6)
C14 0.0614 (10) 0.0671 (11) 0.0573 (10) 0.0039 (8) 0.0313 (8) 0.0054 (9)
Geometric parameters (Å, º)
O1—N1 1.2252 (18) C4—C5 1.3841 (19)
O2—N1 1.2217 (17) C4—H4 0.9300
O3—N2 1.2254 (14) C5—C6 1.4199 (18)
O4—N2 1.2339 (15) C7—C8 1.4516 (18)
O5—C11 1.3614 (15) C7—H7 0.9300
O5—C14 1.4298 (18) C8—C13 1.3864 (19)
N1—C3 1.4500 (19) C8—C9 1.3984 (19)
N2—C5 1.4462 (18) C9—C10 1.3703 (18)
N3—C6 1.3457 (18) C9—H9 0.9300
N3—N4 1.3782 (15) C10—C11 1.393 (2)
N3—H3 0.869 (18) C10—H10 0.9300
N4—C7 1.2746 (18) C11—C12 1.383 (2)
C1—C2 1.357 (2) C12—C13 1.3809 (18)
C1—C6 1.412 (2) C12—H12 0.9300
C1—H1 0.9300 C13—H13 0.9300
C2—C3 1.392 (2) C14—H14A 0.9600
C2—H2 0.9300 C14—H14B 0.9600
C3—C4 1.364 (2) C14—H14C 0.9600
C11—O5—C14 117.74 (11) C1—C6—C5 116.28 (13)
O2—N1—O1 123.06 (14) N4—C7—C8 122.69 (13)
O2—N1—C3 118.84 (14) N4—C7—H7 118.7
O3—N2—O4 122.36 (12) C13—C8—C9 118.03 (12)
O3—N2—C5 118.80 (12) C13—C8—C7 118.68 (12)
O4—N2—C5 118.84 (10) C9—C8—C7 123.29 (12)
C6—N3—N4 119.99 (12) C10—C9—C8 120.69 (13)
C6—N3—H3 119.3 (10) C10—C9—H9 119.7
N4—N3—H3 120.7 (11) C8—C9—H9 119.7
C7—N4—N3 114.37 (12) C9—C10—C11 120.37 (13)
C2—C1—C6 122.04 (13) C9—C10—H10 119.8
C2—C1—H1 119.0 C11—C10—H10 119.8
C6—C1—H1 119.0 O5—C11—C12 124.60 (13)
C1—C2—C3 119.62 (14) O5—C11—C10 115.57 (12)
C1—C2—H2 120.2 C12—C11—C10 119.83 (12)
C3—C2—H2 120.2 C13—C12—C11 119.21 (13)
C4—C3—C2 121.22 (13) C13—C12—H12 120.4
C4—C3—N1 119.09 (13) C11—C12—H12 120.4
C2—C3—N1 119.67 (14) C12—C13—C8 121.87 (13)
C3—C4—C5 119.35 (12) C12—C13—H13 119.1
C3—C4—H4 120.3 C8—C13—H13 119.1
C5—C4—H4 120.3 O5—C14—H14A 109.5
C4—C5—C6 121.48 (13) O5—C14—H14B 109.5
C4—C5—N2 116.37 (11) H14A—C14—H14B 109.5
C6—C5—N2 122.15 (12) O5—C14—H14C 109.5
N3—C6—C1 120.10 (12) H14A—C14—H14C 109.5
N3—C6—C5 123.62 (13) H14B—C14—H14C 109.5
C6—N3—N4—C7 179.55 (12) C4—C5—C6—N3 178.22 (12)
C6—C1—C2—C3 0.2 (2) N2—C5—C6—N3 −1.4 (2)
C1—C2—C3—C4 −1.1 (2) C4—C5—C6—C1 −1.4 (2)
C1—C2—C3—N1 177.35 (13) N2—C5—C6—C1 179.02 (12)
O2—N1—C3—C4 1.0 (2) N3—N4—C7—C8 −179.74 (12)
O1—N1—C3—C4 −179.69 (14) N4—C7—C8—C13 −179.72 (13)
O2—N1—C3—C2 −177.49 (14) N4—C7—C8—C9 −0.3 (2)
O1—N1—C3—C2 1.8 (2) C13—C8—C9—C10 0.3 (2)
C2—C3—C4—C5 0.8 (2) C7—C8—C9—C10 −179.18 (13)
N1—C3—C4—C5 −177.72 (12) C8—C9—C10—C11 0.0 (2)
C3—C4—C5—C6 0.5 (2) C14—O5—C11—C12 7.1 (2)
C3—C4—C5—N2 −179.86 (12) C14—O5—C11—C10 −173.46 (13)
O3—N2—C5—C4 2.33 (18) C9—C10—C11—O5 −179.86 (12)
O4—N2—C5—C4 −177.93 (12) C9—C10—C11—C12 −0.4 (2)
O3—N2—C5—C6 −178.05 (12) O5—C11—C12—C13 179.85 (13)
O4—N2—C5—C6 1.69 (19) C10—C11—C12—C13 0.5 (2)
N4—N3—C6—C1 1.4 (2) C11—C12—C13—C8 −0.1 (2)
N4—N3—C6—C5 −178.13 (12) C9—C8—C13—C12 −0.2 (2)
C2—C1—C6—N3 −178.59 (14) C7—C8—C13—C12 179.25 (13)
supporting information
sup-5 Acta Cryst. (2006). E62, o862–o863
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
N3—H3···O4 0.869 (18) 1.984 (17) 2.6189 (17) 129.0 (14)
N3—H3···O4i 0.869 (18) 2.620 (18) 3.4419 (19) 158.2 (14)
C2—H2···O5ii 0.93 2.59 3.375 (2) 143
C14—H14B···O2iii 0.96 2.56 3.188 (3) 123