Rapid, Selective Digestion of Mitochondrial DNA in Accordance With the
matA
Hierarchy of Multiallelic Mating Types in the Mitochondrial Inheritance of
Physarum polycephalum
Y. Moriyama
1and S. Kawano
Laboratory of Plant Life System, Department of Integrated Biosciences, Graduate School of Frontier Sciences, University of Tokyo, Chiba 277-8562, Japan
Manuscript received December 16, 2002 Accepted for publication March 13, 2003
ABSTRACT
Although mitochondria are inherited uniparentally in nearly all eukaryotes, the mechanism for this is unclear. When zygotes of the isogamous protistPhysarum polycephalumwere stained with DAPI, the fluores-cence of mtDNA in half of the mitochondria decreased simultaneously to give small spots and then disappeared completelyⵑ1.5 hr after nuclear fusion, while the other mitochondrial nucleoids and all of the mitochondrial sheaths remained unchanged. PCR analysis of single zygote cells confirmed that the loss was limited to mtDNA from one parent. The vacant mitochondrial sheaths were gradually eliminated by 60 hr after mating. Using six mating types, the transmission patterns of mtDNA were examined in all possible crosses. In 39 of 60 crosses, strict uniparental inheritance was confirmed in accordance with a hierarchy of relative sexuality. In the other crosses, however, mtDNA from both parents was transmitted to plasmodia. The ratio of parental mtDNA was estimated to be from 1:1 to 1:10⫺4. Nevertheless, the matA hierarchy was followed. In these crosses, the mtDNA was incompletely digested, and mtDNA replicated during subsequent plasmodial development. We conclude that the rapid, selective digestion of mtDNA promotes the uniparental inheritance of mitochondria; when this fails, biparental inheritance occurs.
M
ITOCHONDRIA are inherited strictly maternally paternal mtDNA (0.01–0.1%) could be detected by sen-in many species. The maternal sen-inheritance of sitive PCR techniques (Gyllensten et al. 1991). This mitochondria was first reported in 1974 in horse-donkey promoted a reexamination, using PCR, of more com-hybrids (Hutchisonet al. 1974). Related studies reached mon intraspecific crosses between mammals from which the same conclusion in the rat (Hayashi et al. 1978; Kaneda et al. (1995) concluded that, in intraspecificKroon et al. 1978), the pocket gopher Geomys pinetis crosses (M.musculus), the paternal mtDNA was elimi-(Aviseet al. 1979), the frogXenopus laevis(Dawidand nated by the two-cell stage.
Blackler 1972), the fruit fly Drosophila melanogaster Uniparental inheritance of mitochondria has also (Reilly andThomas1980), and humans (Gileset al. been reported in the isogamous protist Physarum
poly-1980). Particularly in oogamous species, uniparental in- cephalum(Kawano et al. 1987;Kawano andKuroiwa heritance of mitochondria has been attributed to the 1989; Melandet al. 1991). The life cycle of Physarum small number of mitochondria in the male gamete. Al- includes two distinct vegetative forms: the haploid amoeba though fertilized eggs are heteroplasmic (i.e., they con- and the diploid plasmodium. The haploid myxamoebae tain mitochondria from both parents), a small popula- act as isogametes; individuals of different mating types tion of mitochondria derived from the male gamete is pair and fuse to form diploid zygotes that develop into segregated rapidly after repeated cell division. Conse- macroscopic, diploid plasmodia after repeated mitotic quently, most cells are thought to contain mitochondria cycles without cell division. Thus, the segregation of paren-from the female parent (Dawid and Blackler 1972; tal mtDNA is not involved in uniparental inheritance.
Hutchisonet al. 1974;Birky1995;Ankel-Simonsand There are more than just two mating types of Physarum;
Cummins1996). However, the idea of segregation of the mitochondria are transmitted uniparentally in ac-parental mitochondrial DNA (mtDNA) has recently cordance with the relative sexuality determined by the been challenged in several reports. Backcrosses between mating-type locus matA, which has at least 13 alleles.
Mus musculus and M. spretus (an interspecific cross) ThematAalleles can be ranked in a linear hierarchy to
yielded offspring in which a very small proportion of determine the loss of mtDNA (KawanoandKuroiwa 1989;Melandet al. 1991):matA7⬎ matA2⬎matA11⬎
matA12⬎matA1//matA15⬎matA6 (matA1 andmatA15
1Corresponding author:Laboratory of Plant Life System, Department
have not been tested against each other). The
mitochon-of Integrated Biosciences, Graduate School mitochon-of Frontier Sciences,
Uni-drial donor is generally the amoeba that possesses the
versity of Tokyo, Bldg. FSB-601, 5-1-5 Kashiwanoha, Kashiwa, Chiba
277-8562, Japan. E-mail: kk17527@mail.ecc.u-tokyo.ac.jp dominant matA allele, and in each mating pair, one
strain consistently acts as the mtDNA donor, although leles to demonstrate that digestion of mtDNA from one parent is highly selective and thorough, in accordance this strain does not always act as the donor when
com-bined in other mating pairs.Melandet al. (1991) sug- with the matA hierarchy of multiallelic mating types. In 21 of the 60 possible crosses, however, uniparental gested that the elimination of the mtDNA from one
parent is completed within 36 hr of mating. These facts mtDNA inheritance did not occur, and mtDNA from both parents was transmitted to plasmodia at varying suggest that the universal phenomenon of uniparental
inheritance of mitochondria requires a species-specific frequencies. Since the rapid, selective digestion of mtDNA in the recessive mitochondria was incomplete, recognition system by which the zygote cytoplasm
identi-fies and eliminates mitochondria or mtDNA from one leakage of paternal mtDNA occurred. parent.
Recently, several studies reported selective
destruc-MATERIALS AND METHODS
tion, rather than segregation, of sperm mitochondria
in the zygote, particularly in mammalian cells (Kaneda Strains and culture:The amoebal strains ofP. polycephalum
et al. 1995; Sutovskyet al. 1999, 2000). The possible used in this study are listed in Table 1. Myxamoebae were
involvement of ubiquitin in the destruction of sperm cultured on PGY plates (0.5% glucose, 0.05% yeast extract, 2 mmMgSO4, and 1.5% agar in 25 mmpotassium phosphate
mitochondria in fertilized cow and monkey eggs was
buffer, pH 6.6) at 23⬚ with live bacteria (Klebsiella aerogenes)
suggested. Conversely, in chloroplast inheritance, it has
for food. Zygote formation was induced on SM-30 mating
been demonstrated that fluorescent chloroplast
nucle-plates (30 mmcitrate buffer, pH 4.5, 10 mmMgSO4, and 1.5%
oids derived from the male (mt⫺) parent disappear after agar) at 23⬚. For efficient crossing, myxamoebae must carry zygote formation in the isogamous green algaeChlamy- different matA alleles, and in each mating pair, one strain consistently acts as the mitochondria donor. However, the
domonas reinhardtii(Kuroiwaet al. 1982;Nishimuraet al.
dominant strain does not necessarily act as the mitochondria
1999). Unfortunately, however, the behavior of mtDNA
donor in other combinations; the donor in each pair is
deter-before destruction is difficult to detect microscopically mined by the respectivematAalleles.
because of the small copy number and molecular size Plasmodium formation:About 3 days after zygote forma-of mammalian mtDNA. In our work, to investigate the tion, small agar blocks carrying young plasmodia were cut from the mating plates and transferred to malt extract agar
mechanism for eliminating mitochondria from one
par-(MEA) plates (Kawanoet al. 1987) for further growth at 23⬚.
ent, we observed the fate of mitochondria and
mt-nucle-Microscopic observation and fluorometry:To observe the
oids (complexes of mtDNA and proteins) throughout
mitochondria clearly, cells were fixed with 8% formaldehyde
the mating of Physarum. The mitochondria and mtDNA in 10⫻PBS (pH 11) containing 0.01% Tween 20 on SM-30 of Physarum are easily observed by phase-contrast and mating plates. DNA was stained with 4⬘,6-diamidino-2-phenyl-indole (DAPI), and a coverslip was placed over the stained
epifluorescence microscopy. The mitochondria are well
sample. Photographs were taken with a BX62 Olympus (Tokyo)
developed and contain 20–40 ⵑ63-kb mtDNA
mole-epifluorescence microscope equipped with a c4742 CCD
cam-cules, which are highly organized by proteins into a
era (Hamamatsu Photonics, Shizuoka, Japan) and an
Aqua-large rod-shaped mitochondrial nucleoid in each mito- cosmos system. The length of mt-nucleoids and the relative chondrion (Kuroiwa 1982; Takano et al. 2001). We mtDNA fluorescence were determined using the same system.
Electron microscopy:Samples were fixed with 1% osmium
observed the rapid, selective digestion of the mtDNA
tetroxide in PBS, pH 7.6 for 6 hr at 4⬚. They were then
dehy-from one parent during early zygote development of
drated in a graded ethanol series and embedded in Spurr’s
Physarum. The uniparental inheritance of
mitochon-resin (Spurr1969). Ultrathin sections (0.06–0.09m) were
dria seems to be promoted by this rapid, selective diges- cut with a glass knife on an ultramicrotome (Leica Ultracut tion of mtDNA. UCT; Leica Mikrosysteme, Vienna) and mounted on Formvar-Some articles have reported that biparental inheri- coated copper grids. The sections were stained with 3% uranyl acetate for 10 min at room temperature and lead citrate (0.13
tance of mtDNA does occur (Kondoet al. 1990;
Gyllen-mlead nitrate, 0.2mtrisodium citrate dehydrate) for 5 min
stenet al. 1991;Zouroset al. 1994;Kanedaet al. 1995;
at room temperature and then examined with an electron
Rawson et al. 1996). In particular, Gyllensten et al. microscope (H-7600; Hitachi, Tokyo).
(1991) detected paternal mtDNA by PCR in interspe- Isolation of single cells: A single amoeba or zygote was cific mitochondrial congenic mice. Since the paternal isolated from the SM-30 mating plate under a phase-contrast microscope (IMT-2; Olympus) using a capillary system that is
contribution was only 0.01–0.1%, these authors
sug-typically used for microinjection (MO-202; Narishige, Tokyo).
gested that earlier failures to detect paternal mtDNA
Single cells were transferred to individual microtubes
con-were due to the low sensitivity of the assays used. The
taining 10l 1⫻PCR buffer, 0.5% Tween 20, and 2g/ml
situation remains ambiguous, however, because many proteinase K. The samples were incubated overnight at 37⬚ reported cases of paternal transmission involve interspe- to digest proteins, and heated to 95⬚for 5 min to inactivate proteinase K. Each sample was divided into two tubes and
cific rather than intraspecific hybrids. Since matings in
used directly as template DNA for PCR.
nature by definition occur mostly within species, it is
DNA isolation:Approximately 20 mg of amoeba cells grown
important to examine whether mtDNA is also
biparen-on the PGY plates or plasmodium harvested biparen-on the MEA
tally transmitted in intraspecific hybrids. In this study, plates for 4 days was transferred to a microcentrifuge tube we used all possible crosses between 16 strains with and suspended in 500l of 10⫻Tris/saline EDTA (100 mm
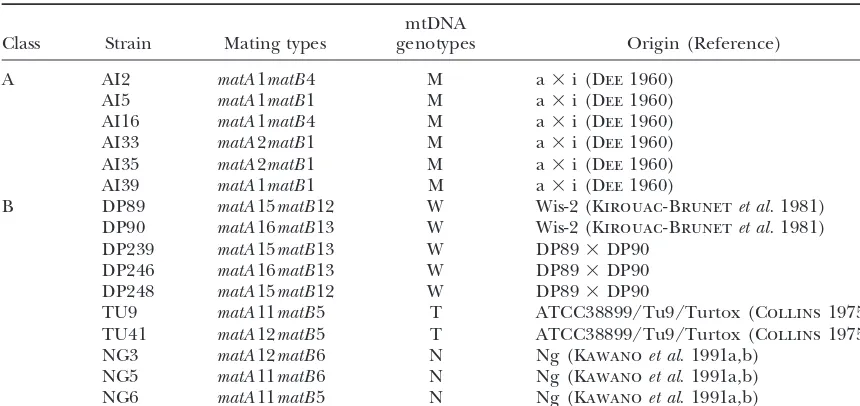
al-TABLE 1 Strains used in this study
mtDNA
Class Strain Mating types genotypes Origin (Reference)
A AI2 matA1matB4 M a⫻i (Dee1960)
AI5 matA1matB1 M a⫻i (Dee1960)
AI16 matA1matB4 M a⫻i (Dee1960)
AI33 matA2matB1 M a⫻i (Dee1960)
AI35 matA2matB1 M a⫻i (Dee1960)
AI39 matA1matB1 M a⫻i (Dee1960)
B DP89 matA15matB12 W Wis-2 (Kirouac-Brunetet al.1981) DP90 matA16matB13 W Wis-2 (Kirouac-Brunetet al.1981) DP239 matA15matB13 W DP89⫻DP90
DP246 matA16matB13 W DP89⫻DP90 DP248 matA15matB12 W DP89⫻DP90
TU9 matA11matB5 T ATCC38899/Tu9/Turtox (Collins1975) TU41 matA12matB5 T ATCC38899/Tu9/Turtox (Collins1975) NG3 matA12matB6 N Ng (Kawanoet al.1991a,b)
NG5 matA11matB6 N Ng (Kawanoet al.1991a,b) NG6 matA11matB5 N Ng (Kawanoet al.1991a,b)
SDS and 0.5 mg/ml proteinase K. After incubation of the bp, respectively. The T-type-specific primer also amplified a fragment of ⵑ3 kb from the mtDNA of M-type. However, suspension at 37⬚for 2 hr, 1 ml of saturated NaI with distilled
water was added, and the lysate was incubated at 0⬚for 30 min. preliminary experiments showed that this 3-kb fragment was not amplified from a mixture of mtDNA from two parents, The lysate was then centrifuged at 20,000⫻gfor 10 min at
4⬚, the surface debris was removed, and 5l of Glassmilk from due to competition with the 526-bp fragment from the T-type.
Estimation of the ratio of parental mtDNA with a PCR
a Gene Clean II kit (BIO 101, Vista, CA) was added. The
Glass-milk was washed and the DNA eluted according to the manu- matrix:To estimate the ratio of mtDNA from each parent in the plasmodium, a PCR matrix of different PCR cycles and facturer’s directions. The eluted DNA was used as the template
for PCR. different template ratios was made using purified M- and W-type mtDNA. The DNA was amplified by PCR with primer
Detection of parental mtDNA types:According to the
re-striction fragment length polymorphism analyses, the mtDNA sets i ⫹iii or i⫹iv from AI35 (M-type) or DP246 (W-type) and then purified with a GFX PCR DNA and gel band purifica-genotypes of the amoebae used in this study are classified into
M-, W-, T-, and N-types (see Table 1). Unlike the M-type, the tion kit (Amersham Pharmacia Biotech) as recommended by the manufacturer. The copy numbers of each product were mtDNA of the W-, T-, and N-types has a 2-kb deletion (Sakurai
et al. 2000). This difference was exploited to detect mtDNA estimated from the DNA concentration, and the two products were mixed at ratios of 1:109–109:1. Using such template mix-from single cells by semi-nested PCR (see Figure 3C). The
DNA from a single cell was separated into two subsamples and tures, PCR was carried out for 30 sec at 94⬚, 30 sec at 55⬚, and 1 min at 72⬚, for 20, 25, 30, and 35 cycles with primer sets each was amplified using the specific primers for either the
M-type or the other types (W, T, or N) of mtDNA. As one ii⫹iii or ii⫹iv. The products amplified with the two primer sets using the same template ratios were loaded in one lane complete round of PCR was insufficient to detect the mtDNA
from a single cell, a second round was performed with semi- and electrophoresed together. The electrophoresis patterns are shown in Figure 7. The parental mtDNA in the plasmo-nested primers. The primer sequences were as follows:
dium was detected by PCR using plasmodial DNA isolated i. F1, 5⬘-TACCCTGTATATGGAACAG-3⬘; with Glassmilk from a Gene Clean II kit as the template. For ii. F2, 5⬘-GAATTGATAGAAGAACTCAGAAGAGG-3⬘; PCR, 1l of 0.01⫻, 0.1⫻, and 1⫻template solution was used. iii. MR, 5⬘-GGTCCCCAAATATTTCTTATAGAATATGC-3⬘; PCR was performed with primer sets ii⫹iii or ii⫹iv for 20, iv. TR, 5⬘-TGCTTCCATAATTGCATCGT-3⬘. 25, 30, and 35 cycles of 30 sec at 94⬚, 30 sec at 55⬚, and 1 min at 72⬚. The ratio of mtDNA from the parents in the PCR reaction mixtures were prepared with ExTaq DNA
plasmodium was estimated by comparing this PCR pattern polymerase (Takara, Otsu, Japan) according to the
manufac-with the PCR matrix. turer’s instructions in a final volume of 50l. The first and
second rounds of PCR included 35 cycles at 94⬚for 0.5 min, at 54⬚for 0.5 min, and at 72⬚for 1 min. M-type mtDNA was
amplified from the sample in one of the paired tubes with RESULTS primers i and iii for the first round of PCR and with primers
ii and iii for the second round. A 1-l sample of the mixture Visualization of loss of mtDNA during zygote
forma-from the first round of PCR was used as template for the tion:Myxamoebae are uninucleate cells that act as isoga-second round. T-type mtDNA was amplified from the re- metes in crosses. In our serial observation of mating, maining tube with primers i and iv for the first round of PCR
syngamy occurred soon after mixing two myxamoebae
and with ii and iv for the second round. The lengths of the
strains with differentmatAalleles. Cell nuclei fusedⵑ2
fragments amplified with the second pairs of primers from
di-Figure1.—Loss of mt-nucleoids during zygote formation inP. polycephalum. (A–H) Merged im-ages from phase-contrast and fluorescence mi-croscopy. (A) Myxamoebae. (B) Zygote. (C–H) Timing of mt-nucleoid loss. (C and F) Fused cell. (D and G) Uninucleate zygote just after nuclear fusion. (E and H) Uninucleate zygote ⵑ1.5 hr after nuclear fusion. Bars: A–E, 5m; F–H, 1m.
vided repeatedly in the absence of cell division to form oid, although the shape of the mitochondria was consis-a multinucleconsis-ate, diploid plconsis-asmodium. Phconsis-ase-contrconsis-ast tent with the amoebal stage (Figure 1B). To investigate observations of myxamoebae clearly revealed the ellip- the loss of mt-nucleoids, the behavior of mitochondria tical mitochondria. After DAPI staining, cell nuclei and during zygote formation was analyzed throughout the mt-nucleoids emitted bright blue-white fluorescence. course of mating. Soon after two myxamoebae fused, a Each amoeba containedⵑ15 mitochondria before mat- full complement ofⵑ30 parental mitochondria mixed ing, and each mitochondrion contained a long rod- together. Each mitochondrion was characterized by the shaped mt-nucleoid at its center (Figure 1A). Zygote presence of a fluorescent mt-nucleoid (Figure 1, C and formation was induced on a mating plate by mixing two F). The fused cells formed a uninucleate zygote as a strains of different mating types. Surprisingly, about half result of nuclear fusion ⵑ2 hr after mating. The fluo-of the total mitochondria in zygotes lacked an mt-nucle- rescent mt-nucleoid persisted in every mitochondrion until this stage (Figure 1, D and G). However, 1 hr after nuclear fusion, mt-nucleoid fluorescence completely disappeared in about half of the mitochondria in the zygote (Figure 1, E and H).
The process of mtDNA loss: Rapid loss of
mt-nucleoids is expected to occur within an hour of nuclear fusion. We investigated this stage of mating in detail and found that mt-nucleoid fluorescence diminished synchronously to small spots, regardless of the position of the mitochondria in the zygote (Figure 2A). Within another 30 min, the mt-nucleoid fluorescence in these mitochondria disappeared completely. Mitochondria were arranged according to the time course of mt-nucleoid loss, as shown in Figure 2, B–E. In half of the mitochondria, the long rod-shaped mt-nucleoid present
Figure2.—Sequential images of mt-nucleoid loss. (A)
Fluo-in each mitochondrion just after nuclear fusion (Figure
rescent image showing the simultaneous loss of half the
mt-nucleoids in the zygote. (B–E) Representative images of mt- 2B) disappeared, starting from both ends of the
mt-nucleoid loss. Phase-contrast and fluorescent images are nucleoid ⵑ1 hr after nuclear fusion (Figure 2C). The merged. (B) Mitochondrion and its nucleoid in a uninucleate
fluorescence of each mt-nucleoid grew fainter and was
zygote just after nuclear fusion. (C and D) Beginning and end
rapidly reduced to a single small spot (Figure 2D). This
of mt-nucleoid loss. (E) Mitochondrion completely lacking
mito-Figure 3.—Detection of parental mt-DNA from a single cell by PCR. (A and B) Before and after isolation of a single zygote from a mating plate with a capillary tube. (C) Scheme for detecting parental mtDNA by semi-nested PCR. The primer sets used are marked by half arrows (seematerials and methods). (D) Inheritance of parental mtDNA, as detected in single zygotes at sev-eral developmental stages of AI35⫻U41. Three representative samples are shown for each stage of development. Bar, 30 m; arrows: (A) zygote; (B) after isolation of the zygote.
chondrion, and it disappeared completely without any division without cell division after mating, all of the mito-chondria derived from the parents are kept in a single other apparent major changes in the mitochondrion
(Figure 2E). These observations suggest the presence cell during zygote development. The total number of mito-chondria in a single cell was counted at 8, 12, 24, 36, of two types of mitochondria in the uninucleate zygote:
those in which the mt-nucleoid is lost and those in which 48, and 60 hr after crossing AI35 and TU41 (Figure 4). AI35 and TU41 hadⵑ14 and 16 mitochondria per cell, it persists. A mechanism for the uniparental inheritance
of mitochondria could be proposed if it is established respectively. There wereⵑ32 mitochondria in the zygote, 16 of which lost mtDNA by 8 hr after mating. As ex-that lost mt-nucleoids originate from a single parent.
Detection of parental mtDNA from a single cell by pected, the number of mitochondria with mtDNA
in-creased⬎25-fold to 430 by 60 hr after mating as a result
PCR:To determine the parental origin of the lost
mt-nucleoids, parental mtDNA in single cells was analyzed of repeated mitochondrial fission (Figure 4A). By con-trast, the number of mitochondria without mtDNA re-during zygote formation using semi-nested PCR. A
sin-gle gamete or zygote was isolated under a phase-contrast mained at 16 until 36 hr after mating (Figure 4B). Then, the number decreased toⵑ3 in a single cell by 48 hr, microscope using a microinjection capillary (Figure 3,
A and B), and its mtDNA was amplified by PCR. Two and all were lost by 60 hr after mating (Figure 4B). Since the decrease in mitochondria lacking mtDNA amoebal strains of different mating types were used,
AI35 (matA2; mtDNA, M-type) and TU41 (matA12; mtDNA, might have been due to mitochondrial sheath destruc-tion, including destruction of the outer and inner mem-T-type). Unlike M-type, the mtDNA of the T-type has a
2-kb mtDNA deletion, so that mtDNA specific for the branes and cristae, mitochondrial morphology was ex-amined during early zygote development. Morphological M- and T-types can be distinguished with PCR primers
(Figure 3C). The results from three representative sam- changes were examined by light and electron micros-copy during the early stages of zygote development to ples at each developmental stage are shown in Figure
3D. When AI35 and TU41 were crossed, the parental plasmodium. The fused, diploid nucleus divided to form a binucleate zygote (a small plasmodium). Atⵑ24 hr mtDNA coexisted in the fused cell and was detectable
in the uninucleate zygote just after nuclear fusion. How- after mating, no morphological changes were observed by phase-contrast microscopy in mitochondria lacking ever,ⵑ1.5 hr after nuclear fusion, no parental mtDNA
from TU41 was detected, and this was correlated with mtDNA (Figure 5A). The plasmodium becameⵑ50m in diameter and had many nucleiⵑ36 hr after mating the loss of mt-nucleoids. Thus, mt-nucleoid loss appears
to be the result of the selective digestion of mtDNA (Figure 5B). The vacant mitochondrial sheaths remained visible at this time, and no degraded mitochondrial from one parent. Such selective digestion may account
for uniparental inheritance of mitochondria. sheaths were observed. By 48 hr after mating, almost all of the vacant mitochondria had been eliminated
Fate of mitochondria lacking mtDNA during
plasmo-dial development:To investigate the fate of mitochon- (Figure 5C).
Although phase-contrast microscopy suggested that dria that lost mtDNA, mitochondria in the developing
no mt-nucleoid and has collapsed cristae. Almost the entire mitochondrion was an electron-transparent re-gion, although some parts retained double membranes and tubular cristae, enabling their detection using phase-contrast microscopy (Figure 5, B and D). Mito-chondrial degradation was observed by ⵑ36 hr after mating, and the elimination of the mitochondrial sheath was completed by 60 hr after mating. Compared with the rapid and selective digestion of mtDNA from one parent within 0.5 hr at 3 hr of mating, elimination of the vacant mitochondria derived from one parent was a lengthy procedure.
Mitochondrial inheritance according to thematA
hier-archy: In Physarum, there are multiple mating types,
and the mitochondrial inheritance mode is determined by allelesmatA1–matA16, which are ranked in a linear hierarchy with respect to mitochondrial inheritance (Kawano et al. 1987; Kawano and Kuroiwa 1989;
Melandet al. 1991). We investigated whether selective digestion of mtDNA occurred in accordance with the relative sexuality of matA. To determine whether the mtDNA from TU41 could survive when crossed with a
Figure 4.—Changes in the number of mitochondria de- second strain with a lower matA rank, AI16 (matA1;
rived from each parent in a single cell during early plasmodial mtDNA, M-type) was crossed with TU41. Since AI16 and development. The number of mitochondria containing AI35 have the same parent (Dee1960), they have the mtDNA in a cell (䊊) and the number of mitochondria that
same mitochondrial genotype (Table 1). Consequently,
lost mtDNA (䊉) are shown in A and B in different ranges of
crosses of these strains with TU41 also permitted
exami-they-axis. All measurements are shown as mean⫾SD. At-test
was used to estimate the significance of differences in the nation of the relationship between digestion selectivity
numbers of mitochondria between one stage and a previous and mtDNA molecules. In crosses of AI16 with TU41, stage (P⫽0.01).
mt-nucleoid loss occurred in the same manner as in the original AI35⫻TU41 cross. In the plasmodium of AI35⫻ TU41, mtDNA of TU41 was lost, as previously structural changes in the inner membrane by ⵑ36 hr
described (Figures 3D and 6A). In the plasmodium of after mating. Figure 5D represents two typical
mitochon-AI35⫻TU41, however, the TU41 mtDNA survived, and dria (left and right) at this stage. The left mitochondrion
the lost mt-nucleoids were of AI16 origin (Figure 6B). preserves an electron-dense mt-nucleoid at the center
The results show that the digestion of parental mtDNA of the matrix and has well-developed tubular cristae.
from one strain is highly selective and is in accord with Almost all of the mitochondria at this stage were of this
type. In contrast, the mitochondrion on the right has thematAhierarchy.
plasmodia (Figure 6D). Such biparental inheritance of mtDNA occurred only in the crossesmatA1⫻matA15,
matA1⫻matA16, andmatA2⫻matA15.
Ratio of parental mtDNA in biparental inheritance of
mtDNA:We examined the mtDNA type of every
plasmo-dium at 10 days after mating by PCR with 35 cycles, as shown (Figure 6). With this number of PCR cycles, it is possible to detect the presence/absence of either parental mtDNA, but it is not possible to estimate the ratio of parental mtDNA, because the PCR product is saturated under this condition. Therefore, to estimate the ratio of parental mtDNA in the 21 crosses that inher-ited mtDNA biparentally, the PCR efficiency of mixtures of the two different kinds of mtDNA (M- and W-types) in copy-number ratios of 1:109–109:1 was examined at
20, 25, 30, and 35 PCR cycles using different primer sets for the M- and W-types. The two PCR products using the different primer sets were loaded in one lane. The
Figure6.—Transmission pattern of mtDNA in four
repre-results were arranged in a matrix, as shown in Figure
sentative crosses. Parental mtDNA was detected from five
plas-7, to show the PCR efficiency with different numbers
modia in each cross of (A) AI35⫻TU41, (B) AI16⫻TU41,
(C) AI35⫻DP246, and (D) AI16⫻DP246. of cycles and different ratios of the two templates. This efficiency matrix can detect at least 1⫻ 105molecules
of mtDNA and can detect mtDNA from one genotype To confirm the strictly uniparental inheritance at the in a 105–100 times excess of mtDNA from the other.
PCR level in any combination of mating type, the trans- Therefore, using this PCR matrix, we estimated the ra-mission patterns of mtDNA were examined in all possi- tios of parental mtDNA in 1l of 0.01⫻, 0.1⫻, and 1⫻ ble crosses between the strains listed in classes A and B template DNA solution that was isolated from plasmo-in Table 1. The six straplasmo-ins ranked plasmo-in class A are progeny dium 10 days after mating for 20, 25, 30, and 35 cycles of of a⫻i and have eithermatA1 ormatA2. Their mtDNA PCR (Table 2). There were biased ratios in the parental is M-type. The strains ranked in class B have different mtDNA for the 21 crosses that inherited mtDNA bipa-origins and havematA11, matA12, matA15, or matA16, rentally. For example, the AI5 ⫻ DP246 plasmodium, depending on their origin. Since they have W-, T-, or which had inherited mtDNA biparentally, contained N-type mtDNA, the mtDNA transmission pattern can 10⫺3 as much mtDNA from AI5 as from DP246 (Table
be detected when they are crossed with class A strains. 2). Conversely, AI16⫻DP246 contained mtDNA from The mtDNA of parents was detected in plasmodia 10 both parents in equal amounts, according to the PCR days after mating by using PCR for 35 cycles. In 39 of matrix. Equal biparental inheritance occurred in 5 of 60 possible crosses, strict uniparental inheritance was the 21 crosses, including AI16⫻ DP246. The mtDNA confirmed (Table 2). For example, the mtDNA of AI35 genotypes of each plasmodium were abbreviated using (M-type) was transmitted in AI35 ⫻ TU41, while the Ⰶ,⬍, ⫽, or ⬎ according to the ratio of mtDNA from mtDNA of TU41 (T-type) was transmitted in AI16 ⫻ the parents (Table 2), and these results are arranged in TU41, as shown in Figure 6, A and B. These results are Table 3 according to mating types. The bias of parental arranged in Table 2. Strict uniparental inheritance of mtDNA in the crosses matA1 ⫻ matA15, matA1 ⫻ mtDNA occurred in all of the crosses ofmatA1 and -2 matA16, and matA2⫻ matA15 seems to obey thematA
strains with matA11 and -12 strains. The mtDNA of hierarchy. Furthermore, in matA1 ⫻ matA15, 5 of 12
matA11 and -12 strains (N-type) was transmitted in the crosses (AI5⫻DP89, AI39⫻DP89, AI2⫻DP248, AI5⫻ crosses withmatA1 strains, but was not transmitted in DP248, and AI39⫻DP248) showed exceptional unipa-the crosses withmatA2 strains. These results are in ac- rental inheritance of mtDNA, resulting in matA1 ⬎ cord with the matA hierarchy: matA2 ⬎ matA11 ⬎ matA15.
matA12⬎matA1. ThematA2 strains were mitochondrial Biased biparental inheritance caused by partial
Figure7.—PCR matrix of different PCR cycles from template solution containing different copy numbers of the two mitochon-drial DNA types. PCR was performed with strain-specific primers from template solution containing different copy numbers of the two mtDNA types for 20, 25, 30, and 35 cycles. Each mtDNA sample was amplified separately using different primer sets and applied to one lane. The upper fragment (673 kb) is the M-type product, and the lower fragment (526 kb) is the W-type.
mtDNA in the recessive mitochondria from AI16 was had well-developed mt-nucleoids (0.5–1.8 m, 0.4–2.2 rfiu), but some mitochondria contained small mt-nucle-not complete 24 hr after mating; very faint, small spots,
representing fluorescent mt-nucleoids, persisted in some oids that emitted very faint fluorescence (0.1–0.25m, 0–0.5 rfiu). These very small mt-nucleoids were gener-mitochondria (Figure 8, A–C). Surprisingly, 24–36 hr
after mating, the persistent mt-nucleoids seemed to in- ated by incomplete mtDNA digestion. Such small mt-nucleoids were rarely observed 36 hr after mating crease gradually in size due to mtDNA replication
(Fig-ure 8, D–F). ure 8, D–F), because they appear to become larger (0.25⫹ m, 0.5⫹rfiu). During plasmodial development, We measured the length and fluorescence intensity
of mt-nucleoids stained with DAPI in cells 24 and 36 mtDNA is replicated, and the size of these surviving mt-nucleoids increases, as shown in Figure 9.
hr after mating and in mature plasmodium, using an
epifluorescence microscope equipped with a CCD cam- At 36 hr after mating, the vacant mitochondria that had lost mt-nucleoids completely remained at the points era and fluorometric software. The values of their major
axes (in micrometers) and fluorescence intensities (rfiu; of origin. In the plasmodia, the vacant mitochondria were completely eliminated, and the surviving mt-nucle-relative fluorescent intensity unit) are arranged in a
scatter plot in Figure 9. At 24 hr after mating, 18% of oids and mitochondria were indistinguishable from each other (0.4–1.8m, 0.3–1.9 rfiu). However, the biased the mitochondria were vacant (0m, 0 rfiu), and 69%
TABLE 3
The relation between mating type and the inheritance mode of mtDNA
matA11 matA12 matA15 matA16
Mating mtDNA N N T N T W W W W W
type genotype Strain NG5 NG6 TU9 NG3 TU41 DP89 DP239 DP248 DP90 DP246
matA1 M AI2 N N T N T mⰆW m⬍W W m⬍W M⫽W
M AI5 N N T N T W m⬍W W mⰆW mⰆW
M AI16 N N T N T mⰆW m⬍W mⰆW M⫽W M⫽W
M AI39 N N T N T W m⬍W W m⬍W m⬍W
matA2 M AI33 M M M M M M⬎w M⬎w M⫽W M M
Figure8.—Incomplete digestion of mtDNA in a plasmodium showing biparental inheritance of mtDNA. (A–F) Merged images from phase-con-trast and DAPI fluorescence microscopy of whole cells. (A–C) Binucleate zygoteⵑ24 hr after mat-ing. (D–F) Multinucleate zygoteⵑ36 hr after mat-ing. B and E are enlargements of areas in A and D, respectively. Bars: A and D, 10m; B and E, 5m.
biparental inheritance of mtDNA suggests that the in- suggest that complete digestion of each mt-nucleoid in mitochondria is needed to destroy and eliminate the completely digested mtDNA was of uniparental origin
and that the normal copy number in mitochondria was mitochondrial sheath. Incomplete digestion of mtDNA enables biased biparental inheritance.
restored during plasmodial development. These results
DISCUSSION
Rapid, selective digestion of mtDNA causes
uniparen-tal inheritance of mitochondria:Mitochondrial
inheri-tance is thought to be predominantly uniparental in nearly all eukaryotes. The combination of mainly unipa-rental inheritance and frequent mutation invites great interest in mtDNA as an indicator of evolutionary rela-tionships (Ingmanet al. 2000). However, the mechanism behind uniparental inheritance has been unclear. In this study, we observed the rapid and simultaneous loss of mt-nucleoids in about half of the mitochondria dur-ing an early stage of zygote maturation (Figures 1 and 2). Molecular analysis of a single cell showed that mt-nucleoid loss coincided with the uniparental inheri-tance of mitochondria (Figure 3); after mt-nucleoid loss, mtDNA from only one of the two parents was detected by PCR. These results indicate that the lost mt-nucleoids were of uniparental origin. The loss of mtDNA has also been shown in higher plants and algae (Kuroiwaand
Hori1986;CorriveauandColeman1991;Nagataet al. 1999). This loss of mtDNA organized in mt-nucleoids occurs before fertilization in the mature generative cell inside a pollen grain or in the male gamete before fertilization. In Physarum, however, mt-nucleoid loss
Figure 9.—Scatter plots of fluorescence intensity vs.mt- occurs in the zygote after mating. Although mitochon-nucleoid length at 24 and 36 hr after mating and in plasmodia dria from both parents were well mixed in the zygote, in AI5 ⫻ DP246. The relative fluorescence intensity of
mt-mtDNA from the mitochondrial recipient strain was
nucleoids stained with DAPI was plotted against the length of
digested synchronously and completely, while that from
the major axis. Vacant mitochondria that had lost their
pro-tected from digestion. The mt-nucleoid loss progressed reasonable to assume that modification and destruction of the mitochondrial membrane plays an important role synchronously after nuclear fusion (Figure 2). This
sug-gests that a nuclease, or nuclease activation signal, is in mitochondrial inheritance. In Physarum, however, such ubiquitination before gamete fusion is unlikely, synchronously transported from the cytoplasm to
tar-geted mitochondria within a very limited period. The since the gamete has only relative sexuality, which is determined by thematA allele of the mating partner. digestion of mtDNA seemed to be independent of the
mtDNA sequence, since mtDNA with the same sequence As the choice of a fusion partner is random, the mito-chondria or mtDNA cannot be primed before mating. were digested in the AI35⫻TU41 cross, but not in the
AI16⫻TU41 cross (Figure 6). The results of reciprocal At least the integrity of the mitochondrial membrane is conserved just before the digestion of mtDNA, as the crosses in which one strain (TU41) played a dual role
in uniparental inheritance, acting as a recipient in one digestion of mtDNA in dividing mitochondria occurs (Figure 1B). We believe that prohibitin ubiquitination cross (AI35⫻TU41) and a donor in another (AI16⫻
TU41), confirmed that mtDNA of uniparental origin may perform an important role in the destruction of the inner mitochondrial membrane after mtDNA digestion. are not destined to be digested before mating. The
uniparental inheritance of mitochondria seems to in- Incomplete digestion of mtDNA causes biased
bipa-rental inheritance of mitochondria: In Physarum, the
volve mechanisms that recognize the origin of mtDNA
and promote the selective digestion of mtDNA from inheritance mode of mtDNA is determined by the mat-ing-type locus matA, which has at least 13 alleles. We one parent in the zygote.
After the complete digestion of mtDNA, the number examined the recognition and digestion of mtDNA among multiple mating types, using PCR for 35 cycles of DNA of mitochondria that contained mtDNA increased
greatly, whereas the number of mitochondria that lost from plasmodia 10 days after mating. Of the eight possi-ble combinations of six mating types, five (39 of possipossi-ble mtDNA remained unchanged untilⵑ36 hr after mating.
After another 24 hr, they were lost completely (Figure 60 crosses) showed strict uniparental inheritance of mi-tochondria in accordance with the relative sexuality of 4). Degraded or disintegrated mitochondria were not
observed directly at these stages by phase-contrast mi- matA(Figure 6, Table 3). Conversely, mtDNA from both parents was transmitted in three combinations ofmatA
croscopy (Figure 5, A–C). InSaccharomyces cerevisiaeand
C. reinhardtii, it is well known that organelles derived (21 crosses).
To estimate the ratio of parental mtDNA readily, we from both parents fuse. However, the decrease in the
number of mitochondria that lost mtDNA was not due made PCR matrices that showed the efficiency of the PCR reaction depending on the ratio of parental mt-to fusion with mimt-tochondria that contained mtDNA,
since no fused mitochondria were observed at these DNA. Our PCR matrix method is very useful for treating many samples to estimate the ratio of parental mtDNA. stages in Physarum. Moreover, the strains used here do
not have an mF plasmid, which is known to promote The ratio ranged from 1:10⫺4to equal amounts (Tables
2 and 3), and one of the parental mtDNA genotypes mitochondrial fusion inP.polycephalum(Kawanoet al.
1993). always dominated in accordance with thematA hierar-chy. In the zygotes of these crosses, the digestion of More detailed observation by electron microscopy
re-vealed that degradation of the mitochondrial inner mtDNA was incomplete (Figure 8, A–C). The ratios of mtDNA from parents, listed in Table 2, may depend on membrane occurred without any changes in size of
mito-chondriaⵑ36 hr after mating (Figure 5D). Morphologi- the effectiveness of the digestion of mtDNA. The aberrant mitochondria with small mt-nucleoids (Figures 8, C and cal changes that do not result in changes in the overall
size of mitochondria have also been noted during apop- F) become normal sized by replicating their mtDNA (Figure 9). The temporary reduction and subsequent tosis and necrosis (Lemasterset al. 1998;Scorranoet
al. 2002). In Physarum, it is likely that the inner mem- replication of mtDNA can explain the biased biparental inheritance of mtDNA from parents in Physarum. brane of mitochondria that lose mtDNA is disrupted,
and then the empty, nonfunctional mitochondria are The presence of leaked paternal mtDNA, particularly in interspecific hybrids between closely related species, removed, probably by lysosomes. In hamsters, rats, and
bovines, degradation of sperm mitochondria has been occurs in several genera, such as Mytilus (Zouroset al. 1994; Rawson et al. 1996), Drosophila (Kondo et al. observed during the early stage of embryonic
develop-ment (Szollosi 1965; Hiraoka and Hirao 1988; 1990), and Mus (Gyllensten et al. 1991; Kaneda et al. 1995), and in humans (Schwartz and Vissing Kanedaet al. 1995;Sutovskyet al. 1999, 2000).
Sutov-skyet al. (1999, 2000) reported that sperm mitochon- 2002). In such cases, the selection or digestion of
mt-DNA from one parent in the zygote might fail. The dria were ubiquitinated before fertilization and
subse-quently destroyed in the mammalian egg. They insisted biased biparental inheritance of mtDNA in Physarum suggests that the destruction of mitochondria from a that the destruction of mitochondria invites uniparental
inheritance of mitochondria. The candidate ubiquitin strain lower in the hierarchy never occurs unless the mtDNA is digested completely. If the destruction of substrate was proposed to be prohibitin, an integral
cific crosses during early mouse embryogenesis. Proc. Natl. Acad.
the critical mechanism of mitochondrial inheritance,
Sci. USA92:4542–4546.
strict uniparental inheritance of mitochondria would Kawano, S., andT. Kuroiwa, 1989 Transmission pattern of mito-occur even though complete digestion of mtDNA failed. chondrial DNA during plasmodium formation inPhysarum
poly-cephalum.J. Gen. Microbiol.135:1559–1566.
Our results suggest that complete digestion of mtDNA
Kawano, S., R. W. Anderson, T. NanbaandT. Kuroiwa, 1987
Poly-is needed for the destruction of mitochondria and that morphism and uniparental inheritance of mitochondrial DNA uniparental inheritance of mitochondria is directly inPhysarum polycephalum.J. Gen. Microbiol.133:3175–3182.
Kawano, S., H. Takano, K. MoriandT. Kuroiwa, 1991a A
mito-caused by the rapid and selective digestion of the
mt-chondrial plasmid that promotes mitomt-chondrial fusion in
Phy-DNA from one parent. The mechanisms for recognizing sarum polycephalum. Protoplasma160:167–169.
the parental origin of mtDNA and for promoting the Kawano, S., H. Takano, K. MoriandT. Kuroiwa, 1991b The oldest laboratory strain ofPhysarum polycephalum. Physarum Newslett.
selective digestion of mtDNA are unknown. We are now
22:70–75.
investigating mitochondrial nuclease(s) that are nuclear Kawano, S., H. Takano, J. Imai, K. Mori andT. Kuroiwa, 1993 DNA coded and involved in the recognition and diges- A genetic system controlling mitochondrial fusion in the slime
mould,Physarum polycephalum. Genetics133:213–224.
tion of mtDNA from one parent. The isolation and
Kirouac-Brumet, J., S. ManssonandD. Pallota, 1981 Multiple
characterization of nuclease(s) should explain the
leak-allelism at thematBlocus inPhysarum polycephalum. Can. J. Genet.
age of paternal mtDNA to subsequent generations. Cytol.23:9–16.
Kondo, R., Y. Satta, E. T. Matsuura, H. Ishiwa, N. Takahataet
We thank T. Kuroiwa (Graduate School of Science, University of
al., 1990 Incomplete maternal transmission of mitochondrial Tokyo) for helpful discussions and A. Hirata (Graduate School of DNA in Drosophila. Genetics126:657–663.
Frontier Science, University of Tokyo) for advice on electron micros- Kroon, A. M., W. M. de VosandH. Bakker, 1978 The heterogeneity copy. We also thank S. Matsunaga (Graduate School of Engineering, of rat-liver mitochondrial DNA. Biochim. Biophys. Acta519:269– University of Osaka) for helpful technical advice. This study was sup- 273.
ported by grants for Scientific Research in Priority Areas (no. Kuroiwa, T., 1982 Mitochondrial nuclei. Int. Rev. Cytol.75:1–59.
Kuroiwa, T., andT. Hori, 1986 Preferential digestion of male chlo-13440246 to S.K.) from the Ministry of Education, Culture, Sports,
roplast nuclei and mitochondrial nuclei during gametogenesis Science, and Technology of Japan.
ofBryopsis maximaOkamura. Protoplasma133:85–87.
Kuroiwa, T., S. Kawano, S. Nishibayashi and C. Sato, 1982 Epifluorescent microscopic evidence for maternal inheri-tance of chloroplast DNA. Nature298:481–483.
LITERATURE CITED Lemasters, J. J., A. L. Nieminen, T. Qian, L. C. Trost, S. P. Elmore
et al., 1998 The mitochondrial permeability transition in cell
Ankel-Simons, F., andJ. M. Cummins, 1996 Misconceptions about
death: a common mechanism in necrosis, apoptosis and autoph-mitochondria and mammalian fertilization: implications for
theo-agy. Biochim. Biophys. Acta1366:177–196. ries on human evolution. Proc. Natl. Acad. Sci. USA93:13859–
Meland, S., S. Johansen, T. Johansen, K. HaugliandF. Haugli, 13863.
1991 Rapid disappearance of one parental mitochondrial
geno-Avise, J. C., C. Giblin-Davidson, J. Laerm, J. C. PattonandR. A.
type after isogamous mating in the myxomycetePhysarum polyceph-Lansman, 1979 Mitochondrial DNA clones and matriarchal
alum.Curr. Genet.19:55–59. phylogeny within and among geographic populations of the
Nagata, N., C. Saito, A. Sakai, H. KuroiwaandT. Kuroiwa, 1999 pocket gopher,Geomys pinetis. Proc. Natl. Acad. Sci. USA 76:
The selective increase or decrease of organellar DNA in genera-6694–6698.
tive cells just after pollen mitosis one controls cytoplasmic
inheri-Birky, C. W., Jr., 1995 Uniparental inheritance of mitochondrial
tance. Planta209:53–65. and chloroplast genes: mechanisms and evolution. Proc. Natl.
Nishimura, Y., O. Misumi, S. Matsunaga, T. Higashiyama, A.
Acad. Sci. USA92:11331–11338.
Yokotaet al., 1999 The active digestion of uniparental
chloro-Collins, O.R., 1975 Mating types in five isolates ofPhysarum
polyceha-plast DNA in a single zygote ofChlamydomonas reinhardtiiis
re-lum. Mycologia67:98–107.
vealed by using the optical tweezers. Proc. Natl. Acad. Sci. USA
Corriveau, J. L., andA. W. Coleman, 1991 Monitoring by
epifluor-96:12577–12582. escence microscopy of organelle DNA fate during pollen
develop-Rawson, P. D., C. L. SecorandT. J. Hilbish, 1996 The effects of ment in five angiosperm species. Dev. Biol.147:271–280.
natural hybridization on the regulation of doubly uniparental
Dawid, I. B., andA. W. Blackler, 1972 Maternal and cytoplasmic
mtDNA inheritance in blue mussels (Mytilusspp.). Genetics144:
inheritance of mitochondrial DNA in Xenopus. Dev. Biol. 29:
241–248. 152–162.
Reilly, J. G., andC. A. Thomas, Jr., 1980 Length polymorphisms,
Dee, J., 1960 A mating type system in an acellular slime-mould.
restriction site variation, and maternal inheritance of mitochon-Nature185:780–781.
drial DNA ofDrosophila melanogaster.Plasmid3:109–115.
Giles, R. E., H. Blanc, H. M. CannandD. C. Wallace, 1980
Mater-Sakurai, R., N. Sasaki, H. Takano, T. AbeandS. Kawano, 2000 In
nal inheritance of human mitochondrial DNA. Proc. Natl. Acad.
vivoconformation of mitochondrial DNA revealed by pulsed-field Sci. USA77:6715–6719.
gel electrophoresis in the true slime mold,Physarum polycephalum. Gyllensten, U., D. Wharton, A. JosefssonandA. C. Wilson, 1991
DNA Res.7:83–91. Maternal inheritance of mitochondrial DNA during backcrossing
Schwartz, M., andJ. Vissing, 2002 Paternal inheritance of mito-of two species mito-of mice. Nature352:255–257.
chondrial DNA. N. Engl. J. Med.22:576–580.
Hayashi, J., H. Yonekawa, O. Gotoh, J. MotohashiandY.
Taga-Scorrano, L., M. Ashiya, K. Buttle, S. Weiler, S. A. Oakeset al.,
shira, 1978 Two different molecular types of rat mitochondrial
2002 A distinct pathway remodels mitochondrial cristae and DNAs. Biochem. Biophys. Res. Commun.81:871–877.
mobilizes cytochrome c during apoptosis. J. Dev. Cell2:55–67.
Hiraoka, J., andY. Hirao, 1988 Fate of sperm tail components after
Spurr, A. R., 1969 A low-viscosity epoxy resin embedding medium incorporation into the hamster egg. Gamete Res.19:369–380.
for electron microscopy. J. Ultrastruct. Res.26:31–43.
Hutchison, C. A., III,J. E. Newbold, S. S. PotterandM. H. Edgell,
Sutovsky, P., R. D. Moreno, J. Romalho-Santos, T. Dominko, C.
1974 Maternal inheritance of mammalian mitochondrial DNA.
Simerlyet al., 1999 Ubiquitin tag for sperm mitochondria. Na-Nature251:536–538.
ture402:371–372.
Ingman, M., H. Kaessmann, S. Paaboand U. Gyllensten, 2000
Sutovsky, P., R. D. Moreno, J. Romalho-Santos, T. Dominko, C.
Mitochondrial genome variation and the origin of modern
hu-Simerlyet al., 2000 Ubiquitinated sperm mitochondria, selec-mans. Nature408:708–713.
tive proteolysis, and the regulation of mitochondrial inheritance
Kaneda, H., J. Hayashi, S. Takahama, C. Taya, K. F. Lindahlet al.,
Szollosi, D. J., 1965 The fate of sperm middle-piece mitochondria Zouros, E., B. A. Oberhauser, C. SaavedraandK. R. Freeman, 1994 An unusual type of mitochondrial DNA inheritance in the in the rat egg. Exp. Zool.159:366–377.
Takano, H., T. Abe, R. Sakurai, Y. Moriyama, Y. Miyazawaet al., blue musselMytilus.Proc. Natl. Acad. Sci. USA91:7463–7467. 2001 The complete DNA sequence of the mitochondrial
ge-Communicating editor: N.Takahata