organic papers
Acta Cryst.(2005). E61, o2167–o2168 doi:10.1107/S160053680501682X Yanet al. C
12H13NO3
o2167
Acta Crystallographica Section E Structure Reports Online
ISSN 1600-5368
(5
S
,6
R
)-4,5-Dimethyl-6-phenylmorpholine-2,3-dione
Ji-Dan Yan, Ming Lu, Yu-Ping Guo, Chun-Bao Li* and Xiang-Yang Tang*
Department of Chemistry, Tianjin University, Tianjin 300072, People’s Republic of China
Correspondence e-mail:
hxyjd2002@yahoo.com.cn, txy@tju.edu.cn
Key indicators
Single-crystal X-ray study
T= 293 K
Mean(C–C) = 0.002 A˚
Rfactor = 0.031
wRfactor = 0.087 Data-to-parameter ratio = 9.9
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2005 International Union of Crystallography Printed in Great Britain – all rights reserved
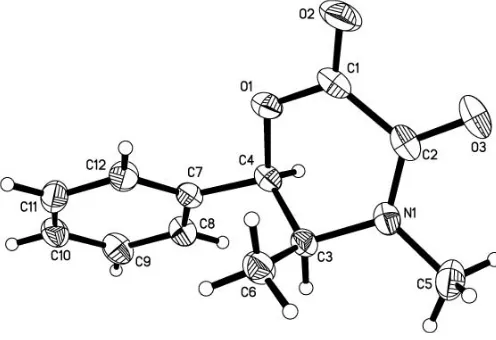
In the title compound, C12H13NO3, the angles at the Csp 3
atoms in the vicinity of the morpholine ring are slightly strained, as indicated by the values of 113.08 (13) and 115.18 (14) for C—C—C angles and 106.98 (13) for the N—C—C angle.
Comment
The title compound, (I), was prepared by the reaction of ephedrine hydrochloride and oxalyl chloride (Pansare et al., 2002). The present crystal structure determination of the compound has been carried out in order to elucidate the molecular conformation.
In the molecular structure of (I), the angles at atoms C3 and C4 indicate strain, compared with the expected value of 109.5 forsp3hydridization (Table 1). Similarly, the angles at thesp2 hybridized atom C2 are distorted from the ideal value of 120. In the six-membered morpholine ring, the atoms C1/C2/N1/C3 form the best least-squares plane, while atoms C4 and O1 deviate by 0.6838 (2) and 0.1179 (2) A˚ , respectively, from this plane. The conformation of the heterocyclic ring is twist–boat.
Experimental
To a stirred suspension of ephedrine hydrochloride (2 g, 9.9 mmol) and DMAP (dimethylpyridin-4-ylamine) (60 mg, 0.49 mmol) in dichloromethane (200 ml) at 273 K, triethylamine (5.5 ml, 39.6 mmol) was added. The mixture was stirred for 10 min and a solution of oxalyl chloride (1.3 ml, 14.9 mmol) in dichloromethane (100 ml) was added dropwise over a period of 4 h at 273 K. The mixture was further stirred at 273 K for 1 h and ice was added. The mixture was warmed to ambient temperature and the two phases were separated. The dichloromethane layer was washed with water (70 ml), dried (Na2SO4) and concentrated. The residue was purified by column
chromatography on silica gel to furnish (I) as a white solid (1.42 g, 65%). The title compound was crystallized from ethyl acetate (m.p.
455 K). Spectroscopic analysis: IR (KBr,, cm1): 3018, 1771, 1693, 1406, 1292, 1215, 1186, 1009;1H NMR (CDCl3,, p.p.m.): 7.50–7.28
(m, 5H), 5.90 (d, 1H), 3.77–3.66 (dp, 1H), 1.12 (d, 3H).
Crystal data
C12H13NO3
Mr= 219.23
Monoclinic,P21
a= 7.3829 (11) A˚
b= 6.7658 (10) A˚
c= 11.1074 (17) A˚
= 93.826 (2) V= 553.59 (14) A˚3
Z= 2
Dx= 1.315 Mg m
3 MoKradiation Cell parameters from 1600
reflections
= 3.4–26.6 = 0.10 mm1
T= 293 (2) K Prism, colourless 0.260.240.20 mm
Data collection
Bruker SMART CCD area-detector diffractometer
’and!scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin= 0.976,Tmax= 0.981 3758 measured reflections
1454 independent reflections 1263 reflections withI> 2(I)
Rint= 0.015
max= 28.1
h=9!7
k=8!8
l=14!14
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.031
wR(F2) = 0.087
S= 1.10 1454 reflections 147 parameters
H-atom parameters constrained
w= 1/[2
(Fo2) + (0.048P)2 + 0.0289P]
whereP= (Fo2+ 2Fc2)/3 (/)max= 0.002
max= 0.15 e A˚
3
min=0.14 e A˚
[image:2.610.45.293.72.242.2]3
Table 1
Selected geometric parameters (A˚ ,).
O1—C1 1.335 (2)
O1—C4 1.4564 (19)
C1—C2 1.539 (3)
C3—C4 1.514 (3)
C3—C6 1.519 (3)
C4—C7 1.508 (2)
O3—C2—N1 124.82 (19)
O3—C2—C1 118.51 (16)
N1—C2—C1 116.65 (14)
N1—C3—C4 106.98 (13)
N1—C3—C6 110.76 (15)
C4—C3—C6 115.18 (14)
O1—C4—C7 108.27 (12)
O1—C4—C3 109.22 (14)
C7—C4—C3 113.08 (13)
All H atoms were positioned geometrically and refined in a riding-model approximation (C—H = 0.93–0.98 A˚ ) with Uiso(H) =
1.2Ueq(carrier atom), or 1.5Ueq(carrier atom) for methyl H. In the
absence of anomalous dispersion effects, the Friedel pairs were merged. The absolute configuration was assigned on the basis of the known configuration of ephedrine hydrochloride.
Data collection:SMART(Bruker, 1997); cell refinement:SAINT
(Bruker, 1997); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics:
SHELXTL (Bruker, 1997); software used to prepare material for publication:SHELXTL.
The authors thank Tianjin University Young Teacher Foundation (grant No. W50501) for financial support.
References
Bruker (1997).SMART(Version 5.10),SAINT(Version 5.10) andSHELXTL
(Version 5.10). Bruker AXS Inc., Madison, Wisconsin, USA. Sheldrick, G. M. (1996).SADABS. University of Go¨ttingen, Germany. Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of
Go¨ttingen, Germany.
Pansare, S. V., Shinkre, B. A. & Bhattacharyya, A. (2002).Tetrahedron,58, 8985–8991.
Figure 1
supporting information
sup-1
Acta Cryst. (2005). E61, o2167–o2168
supporting information
Acta Cryst. (2005). E61, o2167–o2168 [https://doi.org/10.1107/S160053680501682X]
(5
S
,6
R
)-4,5-Dimethyl-6-phenylmorpholine-2,3-dione
Ji-Dan Yan, Ming Lu, Yu-Ping Guo, Chun-Bao Li and Xiang-Yang Tang
(5S,6R)-4,5-Dimethyl-6-phenylmorpholine-2,3-dione
Crystal data C12H13NO3
Mr = 219.23
Monoclinic, P21
Hall symbol: P 2yb a = 7.3829 (11) Å b = 6.7658 (10) Å c = 11.1074 (17) Å β = 93.826 (2)° V = 553.59 (14) Å3
Z = 2
F(000) = 232.0 Dx = 1.315 Mg m−3
Melting point: 455 K
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 1600 reflections θ = 3.4–26.6°
µ = 0.10 mm−1
T = 293 K Prism, colourless 0.26 × 0.24 × 0.20 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
φ and ω scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996) Tmin = 0.976, Tmax = 0.981
3758 measured reflections 1454 independent reflections 1263 reflections with I > 2σ(I) Rint = 0.015
θmax = 28.1°, θmin = 1.8°
h = −9→7 k = −8→8 l = −14→14
Refinement Refinement on F2
Least-squares matrix: full R[F2 > 2σ(F2)] = 0.031
wR(F2) = 0.087
S = 1.10 1454 reflections 147 parameters 1 restraint
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained w = 1/[σ2(F
o2) + (0.048P)2 + 0.0289P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max = 0.002
Δρmax = 0.15 e Å−3
Δρmin = −0.14 e Å−3
Special details
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
N1 0.72529 (19) 0.1308 (3) 0.94829 (11) 0.0455 (3) O1 0.46611 (15) 0.2184 (2) 0.75940 (11) 0.0493 (3) O2 0.25547 (17) 0.1225 (3) 0.87707 (14) 0.0725 (5) O3 0.4986 (2) 0.1340 (3) 1.07541 (11) 0.0706 (4) C1 0.4113 (2) 0.1611 (3) 0.86604 (17) 0.0498 (4) C2 0.5524 (3) 0.1420 (3) 0.97386 (15) 0.0488 (4) C3 0.7766 (2) 0.1324 (3) 0.82228 (13) 0.0406 (4)
H3 0.9016 0.1808 0.8220 0.049*
C4 0.6543 (2) 0.2808 (3) 0.75485 (15) 0.0403 (4)
H4 0.6693 0.4096 0.7946 0.048*
C5 0.8655 (3) 0.0976 (5) 1.04538 (16) 0.0656 (6)
H5A 0.8460 0.1850 1.1114 0.098*
H5B 0.9828 0.1229 1.0161 0.098*
H5C 0.8600 −0.0369 1.0724 0.098*
C6 0.7715 (3) −0.0753 (3) 0.77006 (17) 0.0508 (4)
H6A 0.8456 −0.1611 0.8215 0.076*
H6B 0.8169 −0.0731 0.6911 0.076*
H6C 0.6487 −0.1228 0.7646 0.076*
C7 0.6949 (2) 0.3030 (3) 0.62424 (14) 0.0387 (4) C8 0.8231 (2) 0.4407 (3) 0.59481 (16) 0.0484 (4)
H8 0.8784 0.5203 0.6548 0.058*
C9 0.8699 (3) 0.4612 (3) 0.47690 (17) 0.0567 (5)
H9 0.9563 0.5542 0.4580 0.068*
C10 0.7889 (3) 0.3446 (3) 0.38792 (16) 0.0531 (5)
H10 0.8201 0.3589 0.3086 0.064*
C11 0.6619 (3) 0.2068 (4) 0.41564 (16) 0.0532 (5)
H11 0.6082 0.1270 0.3552 0.064*
C12 0.6131 (2) 0.1859 (3) 0.53392 (15) 0.0474 (4)
H12 0.5258 0.0936 0.5522 0.057*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
supporting information
sup-3
Acta Cryst. (2005). E61, o2167–o2168
C4 0.0395 (8) 0.0412 (9) 0.0414 (8) −0.0009 (7) 0.0116 (6) −0.0036 (7) C5 0.0683 (12) 0.0859 (18) 0.0420 (9) −0.0145 (12) −0.0010 (8) 0.0061 (11) C6 0.0573 (11) 0.0484 (10) 0.0476 (9) 0.0110 (9) 0.0111 (8) 0.0002 (8) C7 0.0387 (8) 0.0382 (8) 0.0399 (8) 0.0038 (7) 0.0087 (6) 0.0014 (7) C8 0.0518 (10) 0.0447 (9) 0.0496 (9) −0.0067 (9) 0.0107 (8) −0.0045 (8) C9 0.0586 (11) 0.0545 (12) 0.0594 (11) −0.0094 (10) 0.0217 (9) 0.0047 (10) C10 0.0544 (10) 0.0654 (13) 0.0410 (8) 0.0056 (10) 0.0145 (7) 0.0064 (9) C11 0.0538 (10) 0.0640 (12) 0.0413 (8) −0.0045 (10) 0.0002 (8) −0.0032 (9) C12 0.0452 (9) 0.0522 (12) 0.0451 (8) −0.0080 (8) 0.0047 (7) 0.0016 (8)
Geometric parameters (Å, º)
N1—C2 1.328 (2) C5—H5C 0.9600
N1—C5 1.462 (2) C6—H6A 0.9600
N1—C3 1.4741 (18) C6—H6B 0.9600
O1—C1 1.335 (2) C6—H6C 0.9600
O1—C4 1.4564 (19) C7—C8 1.383 (2)
O2—C1 1.194 (2) C7—C12 1.385 (2)
O3—C2 1.2217 (19) C8—C9 1.383 (2)
C1—C2 1.539 (3) C8—H8 0.9300
C3—C4 1.514 (3) C9—C10 1.370 (3)
C3—C6 1.519 (3) C9—H9 0.9300
C3—H3 0.9800 C10—C11 1.372 (3)
C4—C7 1.508 (2) C10—H10 0.9300
C4—H4 0.9800 C11—C12 1.392 (2)
C5—H5A 0.9600 C11—H11 0.9300
C5—H5B 0.9600 C12—H12 0.9300
C2—N1—C5 119.65 (15) H5A—C5—H5C 109.5
C2—N1—C3 120.93 (14) H5B—C5—H5C 109.5
C5—N1—C3 119.17 (14) C3—C6—H6A 109.5
C1—O1—C4 117.50 (13) C3—C6—H6B 109.5
O2—C1—O1 120.41 (18) H6A—C6—H6B 109.5
O2—C1—C2 120.57 (16) C3—C6—H6C 109.5
O1—C1—C2 119.02 (14) H6A—C6—H6C 109.5
O3—C2—N1 124.82 (19) H6B—C6—H6C 109.5
O3—C2—C1 118.51 (16) C8—C7—C12 119.10 (15)
N1—C2—C1 116.65 (14) C8—C7—C4 118.53 (14)
N1—C3—C4 106.98 (13) C12—C7—C4 122.33 (15)
N1—C3—C6 110.76 (15) C7—C8—C9 120.62 (17)
C4—C3—C6 115.18 (14) C7—C8—H8 119.7
N1—C3—H3 107.9 C9—C8—H8 119.7
C4—C3—H3 107.9 C10—C9—C8 120.05 (18)
O1—C4—C7 108.27 (12) C10—C9—H9 120.0
O1—C4—C3 109.22 (14) C8—C9—H9 120.0
C6—C3—H3 107.9 C9—C10—C11 120.08 (17)
C7—C4—C3 113.08 (13) C9—C10—H10 120.0
C7—C4—H4 108.7 C10—C11—C12 120.29 (18)
C3—C4—H4 108.7 C10—C11—H11 119.9
N1—C5—H5A 109.5 C12—C11—H11 119.9
N1—C5—H5B 109.5 C7—C12—C11 119.86 (17)
H5A—C5—H5B 109.5 C7—C12—H12 120.1
N1—C5—H5C 109.5 C11—C12—H12 120.1