0022-538X/99/$04.0010
Copyright © 1999, American Society for Microbiology. All Rights Reserved.
Recombination between Two Identical Sequences
within the Same Retroviral RNA Molecule
JIAYOU ZHANG*ANDCHRISTY M. SAPP
Department of Microbiology and Immunology and Markey Cancer Center, University of Kentucky, Lexington, Kentucky 40536-0096
Received 10 November 1998/Accepted 12 March 1999
As a consequence of being diploid viruses, members of theRetroviridaehave a high recombination rate. To measure recombination between two identical sequences within the same RNA molecule per round of retroviral replication cycle, a murine leukemia virus based vector (JZ44213*Hyg) has been constructed. It carries a drug resistance gene,hyg, and a 290-bp repeat sequence of the 3*hyggene inserted into the 3*untranslated region of the green fluorescent protein gene (gfp). Under fluorescence microscopy, Hygrcells containing the
recombinant proviruses were clear, while a green color was observed in the drug-resistant cells carrying the parental proviruses. The rate of recombination was determined by the ratio of the number of clear colonies to the total number of Hygrcolonies (green and clear colonies). The rate of recombination was found to be 62%
by this method. The intermolecular recombination rate between an infectious virus bearing two copies of the 290-bp segment and a noninfectious chimeric RNA virus containing only a single copy of this sequence was also measured.
The presence of two genomic RNA molecules in retroviral virions results in a high rate of recombination (4). To study recombination, two different proviruses were introduced into one retroviral helper cell line. The recombinants were then judged by the expression of markers from both parental pro-viruses. Evidence shows that intermolecular recombinations occur frequently (8, 17). Previously, it was deduced that re-combination may occur between two sequences within the same RNA molecule (intramolecular) as well as between se-quences within two separate RNA molecules (intermolecular) (15). If intramolecular recombination occurs, it will result in a deletion within the progeny provirus. However, there is no direct evidence indicating that intramolecular recombination occurs. This is because, in the experiments described above, retroviral particles were able to package either two different RNA molecules or two identical RNA molecules. The deletion can be the result of an intramolecular event or the recombi-nation of an upstream sequence from one RNA molecule with a downstream sequence from the other RNA molecule. Since current technology cannot separate viral particles containing two identical molecules (homogeneous) from those containing two different molecules (heterogeneous), intermolecular re-combination cannot be excluded as the source of deletion recombinants.
Previously, retroviral vectors containing two identical se-quences separated by a packaging signal within the same RNA molecule were constructed and used to infect cells to examine deletion rates (5, 9). The investigators found that the rate of deletion in progeny proviruses was 57 to 93% after one round of replication. This result does not exclude the possibility that the high rate of deletion resulted from the packaging signal being a hot spot for reverse transcriptase template switching events (9). In addition, because the viruses analyzed were from a pool of transfected cells (5), it could not be determined if the
deletion occurred during reverse transcription or occurred dur-ing transfection with the proviral DNA.
To further study recombination between two identical se-quences within the same RNA molecule, murine leukemia virus (MLV)-based vector (JZ442 1 39 Hyg) has been con-structed. This vector contains a wholehyggene and an unse-lected color marker reporter gene, the green fluorescent pro-tein gene (gfp). Additionally, it carries an extra 39 hyg gene segment of 290 bp inserted into the 39untranslated region of thegfpgene. This construct allows us to demonstrate that a high rate of deletion does not relate to the packaging signal sequence. The intermolecular recombination rate between an infectious virus bearing two copies of the 290-bp segment and a chimeric RNA virus containing a single copy of this sequence was also measured. The rate of intermolecular recombination in the presence of two copies of identical sequences on the infectious RNA molecule did not increase much compared with the rate (62%) of recombination between the two identi-cal sequences on the same RNA molecule.
MATERIALS AND METHODS
Nomenclature.Plasmids are designated as, for example, pJZ442; viruses made from these plasmids are designated as, for example, JZ442. Some infectious Moloney murine leukemia virus (MLV) vectors contained a 290-bp sequence (39 hyg). When the 290-bp sequence was inserted at the 59end of theneogene, the number of nucleotides inserted is on the left of the “N” (N stands forneo) (for example, pL290N). When the 290-bp sequence was inserted at the 39end of the
neogene, the number of nucleotides inserted is on the right of the “N” (for example, pLN290).
Vector constructions.All recombinant techniques were carried out by conven-tional procedures (14). All vector sequences are available upon request.
(i) Construction of pJZ442 and pJZ44213*Hyg (Fig. 1A and B).The pJZ442 construct, from 59 to 39, was assembled as follows. The 5.4-kbNdeI-BamHI fragment (from positions 2990 to 1630) was isolated from pLN (12) and contains the neogene and the two MLV long terminal repeats (LTRs). The 0.7-kb
BamHI-SalI fragment (from positions 1631 to 2387) was isolated from pEGFP-1 (Clontech, Palo Alto, Calif.). The 0.6-kbSalI-NdeI fragment (from positions 2388 to 2989) was isolated from pCITE-1 (Novagen, Madison, Wis.) and contains the internal ribosome entry segment (IRES) sequence. In pJZ442139Hyg, the
SacII-HindIII fragment (290 bp long) of the 39hygsequence was inserted at the
NotI andClaI sites of pJZ442, which are located downstream of the open reading frame of thegfpgene.
(ii) Construction of pJZ211, pLN290, pL290N, and pL290N290 (Fig. 2).The pJZ211 and pLN290 constructs were described previously (17, 18). The 290-bp * Corresponding author. Mailing address: 206 Combs Research
Bldg., University of Kentucky, 800 Rose St., Lexington, KY 40536-0096. Phone: (606) 257-4456. Fax: (606) 257-8940. E-mail: jzhan1 @pop.uky.edu.
5912
on November 9, 2019 by guest
http://jvi.asm.org/
hygsequence in pL290N and pL290N290 was cloned as theSacII-HindIII frag-ment from pJZ211.
Cells, transfection, and infection.The processing of D17 cells (a dog osteo-sarcoma cell line; ATCC CRL-8468), PA317 helper cells (ATCC CRL-9078),
and PG13 helper cells (ATCC CRL-10686), DNA transfections, virus harvesting, and virus infections were as previously described (17).
Introduction of JZ44213* Hyg (and JZ442) into helper cell line PG13. Plasmid DNA of pJZ442139Hyg (and pJZ442) was transfected into an MLV amphotropic helper cell line, PA317 (10). The supernatant media containing the viruses were collected and designated STEP 1 virus stock. The STEP 1 viruses were used to infect an MLV xenotropic helper cell line, PG13 (11). The viruses released from infected PG13 cells were unable to infect NIH 3T3 derivatives, including PG13 (11). This procedure ensured that the infection of D17 cells with viruses collected from PG13 cells represented only a single round of infection. Infected cells were selected for hygromycin resistance (Hygr). Visible colonies
appeared about 10 days after selection. The cells of well-separated green colo-nies were isolated and designated STEP 2 cells.
Fluorescence microscopy.A fluorescence inverted microscope (Zeiss Axiovert 25) with a mercury arc lamp (100 W) and a fluorescence filter set (CZ909) consisting of a 470- to 40-nm exciter, a 515-nm emitter, and a 500-nm beam splitter was used to detect green fluorescent protein in living cells.
RESULTS
[image:2.612.56.293.74.198.2]Determination of the recombination rate within one retro-viral RNA molecule.A bicistronic MLV-based vector (pJZ442) that carries a drug resistance gene,hyg, and an unselected color reporter gene,gfp(3), along with an IRES sequence between the two genes, has been constructed (Fig. 1A). The IRES sequence of the encephalomyocarditis virus origin allows the ribosome to bind to the internal AUG that initiates the trans-lation of the second gene independently of the upstream gene (1, 2). To measure the recombination rate between two iden-tical sequences within the same RNA molecule, another vector (pJZ442139Hyg) that also contains thehygandgfpgenes but also includes the insertion of a sequence homologous to 290 bp
FIG. 1. Structures of retrovirus vectors used for determination of the recom-bination rate between two identical sequences within the same RNA molecule. (A) Structure of the retrovirus vector containing thehyggene and thegfpgene.
Thehyggene is expressed from the 59MLV LTR, and thegfpgene is expressed
from an encephalomyocarditis virus IRES. (B) Structure of the retrovirus con-taining two identical sequences. JZ442139Hyg is similar to JZ442, except that JZ442139Hyg also contains 290 bp of the 39hyggene sequence downstream of
thegfpgene. (C) Structure of the recombinant provirus. After one round of
replication, the downstream 39hyggene sequence will recombine with the
iden-tical upstreamhyggene sequence and result in the deletion of thegfpgene.
Recombinants, therefore, contain only thehyggene. The broken lines between
JZ442, JZ442139Hyg, and the recombinant provirus indicate the identical 39
hyggene sequences.
FIG. 2. Chimeric RNA vector, infectious virus vectors, and resulting recombinants. JZ211 contains only the 59MLV LTR, while the infectious vectors LN, LN290,
L290N, and L290N290 contain two MLV LTRs. The recombinant proviruses containing thehyggene form only when recombination occurs between JZ211 and an
infectious vector such that thehyggene is flanked by two LTRs. Recombination between JZ211 and LN is nonhomologous (17). Most recombinations between JZ211
and LN290, L290N, and L290N290 occurred between the 290-bp identical sequences. The broken lines between the chimeric RNA vector and the infectious vectors indicate the identical 290-bp 39hygsequences in the two vectors. The resulting recombinants correspond to individual pairs of chimeric RNA and infectious vectors. The lengths of theBamHI-HindIII fragments which hybridized to ahygprobe are shown for the recombinants. Two recombinants resulted from recombination between JZ211 and L290N290: one, utilizing the upstreamhygsequence, gives a 2.4-kbBamHI-HindIII fragment, while the other, utilizing the downstreamhygsequence, gives a 1.1-kbBamHI-HindIII fragment. Rates of recombination are shown on the right.
VOL. 73, 1999 RETROVIRUS RECOMBINATION 5913
on November 9, 2019 by guest
http://jvi.asm.org/
[image:2.612.57.537.412.660.2]of the 39 hyggene into the 39untranslated portion of thegfp gene (downstream of thegfpgene or after the stop codon of thegfpgene) has been constructed (Fig. 1B).
pJZ442 1 39 Hyg was used to transfect PG13 cells, and transfected cells were selected for Hygr. Hygrcells were ana-lyzed under a fluorescence microscope. Green cells contained parental JZ442139Hyg, and clear cells contained agfpgene deletion (or mutation) in the transfected provirus. Approxi-mately 48.8%610.2% of the transfected HygrPG13 cells were clear. Therefore, transfection alone caused a high frequency of deletion (or mutation) between the two identical sequences in the same plasmid DNA. To avoid a high frequency of deletion during transfection, JZ442 and JZ442 139 Hyg were intro-duced by infection into the helper cell line PG13 described in Materials and Methods. The viruses released from each PG13 clone, which contained JZ442 or JZ442 1 39 Hyg provirus, were used to infect D17 cells; the infected D17 cells were selected for Hygr. After about 12 days of selection, visible Hygr colonies appeared, and these were designated STEP 3 cells. Because D17 cells do not contain viralgag-poland envgene products for retroviral replication, no progeny viruses were released from these cells (17). Therefore, each colony of Hygr cells represented a single viral infection.
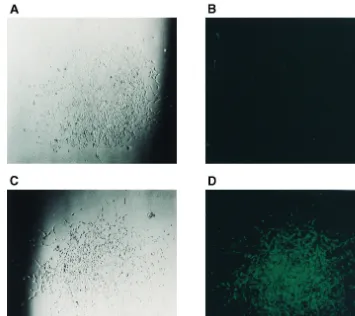
Individual Hygr colonies were examined under a fluores-cence microscope. The clear colonies (Fig. 3A and B) repre-sented cells containing the proviruses with the gfp deletion. The green colonies (Fig. 3C and D) and represented cells containing the parental proviruses. Results from counting
green and clear colonies (Table 1) indicated that the rate of deletion between the two identical sequences in the same RNA molecule was very high (62%69% per replication cycle). In comparison, only 1% of the colonies infected with JZ442 were clear. This result indicates that the rate of mutation of thegfp gene in the JZ442 vector during a single round of retroviral replication is about the same as previously reported (13). To determine the nature of the recombinants, genomic DNAs from clear and green STEP 3 cells were digested withEcoRV. EcoRV digested within the LTRs of the vectors; the parental provirus produced a 4.2-kb fragment, while the recombinant provirus with the deletion of thegfpgene produced a 2.5-kb fragment (Fig. 4). The recombinant formed a distinct 2.5-kb band, indicating that most deletions of the gfpgene resulted from recombination between the two identical sequences.
[image:3.612.126.481.72.388.2]Intermolecular recombination between a noninfectious RNA and an infectious RNA containing two identical se-quences.The retroviral vectors and protocol used to measure rates of recombination have been described previously (17). In order to study recombination between chimeric and infectious MLV RNAs, a chimeric RNA, JZ211 (Fig. 2), was prepared. JZ211, derived from spleen necrosis virus (SNV), contained a deletion in the U3 region of the 39SNV LTR as well as anXhoI restriction site linker in the deletion site (Fig. 5). This vector also contained a truncated MLV vector between the two SNV LTRs, in a transcriptional orientation opposite that of the SNV LTRs. In this truncated MLV vector, thehyg gene was ex-pressed from the 59 MLV LTR, and a herpes simplex virus
FIG. 3. Microscopic analyses of D17 cells infected with viral vector JZ442139Hyg containing thegfpgene. (A) Visible-light microscopy of a hygromycin-resistant colony containing recombinant JZ442139Hyg provirus. (B) Fluorescence microscopy of a hygromycin-resistant colony (same colony as in panel A) containing recombinant JZ442139Hyg provirus. (C) Visible-light microscopy of a hygromycin-resistant colony containing parental JZ442139Hyg provirus. (D) Fluorescence microscopy of a hygromycin-resistant colony (same colony as in panel C) containing parental JZ442139Hyg provirus.
on November 9, 2019 by guest
http://jvi.asm.org/
thymidine kinase poly(A) addition signal replaced the com-pletely deleted 39 MLV LTR. The infectious MLV vectors contained aneo gene between the two MLV LTRs (17, 18) (Fig. 2 and 5). pJZ211 DNA was transfected into the SNV C3A2 helper cell line (containing the SNV gag-pol and env genes) (16) (Fig. 5). The cells were selected for Hygr, and resistant cells were pooled and designated STEP 1 cells (Fig. 5, STEP 1). Virus from STEP 1 cells was used to infect the MLV helper cell line PG13. Infected cells were selected for Hygr, and individual clones were isolated and designated STEP 2 cells. The structure of the proviruses formed from the SNV U3-negative vector in the PG13 cells was monitored by South-ern (DNA) analysis. TheXhoI linker in JZ211 was duplicated in the 59LTR during the formation of the STEP 2 provirus
(Fig. 5, STEP 2) (7). The STEP 2 clones that contained the expectedXhoI fragment, which hybridized to ahygprobe, were used for further analysis (Fig. 5, STEP 2). To test whether any virus capable of forming Hygrcolonies was produced by STEP 2 cells, the supernatant medium (3 ml) from each STEP 2 cell clone was used to infect D17 cells, and the infected cells were selected for Hygr. No HygrD17 colonies were detected. This was because the deletion of the U3 region (promoter and enhancer) in the SNV 59 LTR prevented transcription from the SNV vector (6).
[image:4.612.54.552.83.200.2]The infectious MLV vectors LN, LN290, L290N, and L290N290 (Fig. 2) were transfected individually into the helper cell line PA317 (Fig. 5). Cells exhibiting the Neorphenotype were pooled, viruses from the PA317 cells were used to super-infect STEP 2 cells containing JZ211, and the super-infected cells were selected for Neor. Individual Neorclones were isolated and designated STEP 3 cells (Fig. 5, STEP 3). Each STEP 3 cell clone contained a single JZ211 integration and a single integration of LN, LN290, L290N, or L290N290. Viruses from each STEP 3 clone were used to infect D17 cells, and the infected cells were selected separately for Hygrand for Neor. The resulting cells were designated STEP 4 cells (Fig. 5, STEP 4). With this approach, Hygrcolonies form only when recom-bination between JZ211 and any one of the three vectors occurs at the shared 290-bp homologous sequences, so that the hyggene is flanked by two LTRs. If a nonhomologous recom-bination event occurs between JZ211 and LN, it results in a Hygrcolony. The rate of recombination is much lower than the rates for vectors containing the 290-bp sequence when there are shared homologous sequences (Fig. 2A) (17, 18). The tar-get cells do not contain viralgag-polandenvgene products for retrovirus replication; therefore, no progeny virus can be re-leased from them (17). Consequently, these vector viruses had undergone only one cycle of replication. LN290 and L290N contained the same 290-bp 39 hyg sequence, except that the 290-bp sequence of LN290 was inserted at the 39end ofneo and served as the 39 untranslated sequence, whereas the 290-bp sequence of L290N was inserted at the 59end ofneo and served as the 59untranslated sequence (Fig. 2B and C). This 290-bp sequence does not contain an ATG motif; there-fore,neotranslation should not be affected by the 59insertion (Fig. 2A and B). The ratios of HygrCFU to NeorCFU pro-duced were 8310246631024and 17310246531024, respectively, for LN290 and L290N. L290N290 contained two copies of sequences identical to that in JZ211. Specifically, the 290-bp 39hygsequence was inserted both upstream and down-stream of theneogene within this vector. The rate of recom-bination between JZ211 and L290N290 was 60310246133
FIG. 4. Southern analysis of chromosomal DNA of Hygrcells. Cellular DNA
isolated from STEP 3 cells was digested withEcoRV and hybridized with ahyg
gene probe. Hygrcells had a deletion between the two identical sequences in one
RNA molecule of vector JZ442139Hyg. Pooled DNA was isolated from more than 500 HygrSTEP 3 colonies. The clear clone is an individual Hygrcolony
resulting from deletion of thegfpgene between the two 39hyggene segments. The green clone is an individual Hygrcolony. Molecular sizes are shown on the
[image:4.612.116.229.410.657.2]left.
TABLE 1. Microscopic analysis of D17 cells infected with JZ442 and JZ442139Hyga
JZ442 clone No. of colonies that were JZ442Hyg clone139 No. of colonies that were
Green Clear Mixture Green Clear Mixture
1 124 1 1 1 27 55 1
2 124 1 0 2 14 70 3
3 105 1 0 3 33 53 5
4 112 2 0 4 13 19 0
5 72 0 1 5 19 25 0
6 132 2 0 6 33 46 0
7 28 32 0
Total 669 7 2 Total 167 300 9
aEach clone was an individual clone of PG13 cells containing the JZ442 or JZ442139Hyg provirus. The colonies were analyzed under a fluorescence microscope.
The rates ofgfpdeletion were 1% for JZ442 clones and 62%69% for JZ442139Hyg clones.
VOL. 73, 1999 RETROVIRUS RECOMBINATION 5915
on November 9, 2019 by guest
http://jvi.asm.org/
1024, and the rate of recombination between JZ211 and LN was only 0.33102460.231024.
To determine whether recombination had occurred at the 59 or at the 39end of thehygsequence, the DNA of pooled STEP 4 cells was digested withBamHI andHindIII and hybridized with ahygprobe. The proviruses resulting from recombination between JZ211 and L290N290 utilizing upstream and
down-stream hyg sequences produced different sizes of
BamHI-HindIII fragments that hybridized with a hyg probe (Fig. 2). The recombinants utilizing the upstreamhygsequence produced a 2.4-kb BamHI-HindIII hyg fragment, and those utilizing the downstream hyg sequence produced a 1.1-kb
BamHI-HindIIIhygfragment. The pooled recombinants from JZ211 and L290N290 yielded a 1.1-kb fragment and a 2.4-kb fragment, indicating that both identical sequences were used to form the recombinants between the chimeric RNA and the infectious vector (Fig. 6).
DISCUSSION
[image:5.612.130.478.74.538.2]Because they have two genomic RNA molecules in their virions, retroviruses undergo recombination at a high rate. Our data indicate that the high rate of recombination between two identical sequences within the same RNA molecule was not
FIG. 5. Outline of an experimental approach for the determination of the rate of recombination during a single cycle of retroviral replication between a chimeric RNA vector, JZ211, and infectious vectors. No plasmid backbone sequences are shown. The directions of transcription in SNV and MLV are shown by the long thin arrows. Transfections are indicated by test tube shapes. Infections are indicated by virion shapes. The different backgrounds represent the indicated cell lines. SV, late
polyadenylation signal of simian virus 40;Cand E, encapsidation sequences of MLV and SNV, respectively; tk, thymidine kinase. The lines in the LTR separate the
U3, R, and U5 regions.
on November 9, 2019 by guest
http://jvi.asm.org/
dependent on the packaging signal. The rate of intermolecular nonhomologous deletion in a single RNA molecule (1025bp per replication cycle) is about 1,000 times higher than that of intermolecular nonhomologous recombination (1028 bp per replication cycle) (13).
Recombination between sequences within the same RNA molecule can be intermolecular or intramolecular. Recombi-nation between a chimeric RNA vector and infectious vectors containing one or two copies of the identical sequences was examined. The results indicated that the presence of two cop-ies of identical sequences on the infectious RNA increased the rate of recombination very little (631023) compared with a 60% increase in the rate of recombination (631021) between the same 290-bp identical sequences in the same RNA mole-cule. In addition, the actual titers of the infectious viruses, as measured by Neor, were even higher. This result would be due to intrastrand recombination within the two identical se-quences on either side of the neogene. From the data pre-sented in this study, the deletion rate should be nearly 50%, which would account for the discrepancy observed. However, it is interesting to observe that most intermolecular recombina-tion events either have involved the downstream stretch of identical sequence or have already involved an intramolecular recombination event between the two copies of the 290-bp
stretch. This finding suggests the phenomenon of negative in-terference, which indicates that intramolecular recombination increases the chance of intermolecular recombination or vice versa. However, the individual rate of inter- versus intramo-lecular recombination between two identical sequences within the same RNA molecule remains unresolved.
ACKNOWLEDGMENTS
We thank William Bargmann, Chih-Li Hsu, Alan Kaplan, Alan Simmons, and Ting Li for helpful comments on the manuscript.
This research was supported by Public Health Service research grant CA70407 from the National Institutes of Health.
REFERENCES
1.Adam, M. A., N. Ramesh, A. D. Miller, and W. R. Osborne.1991. Internal initiation of translation in retroviral vectors carrying picornavirus 59 non-translated regions. J. Virol.65:4985–4990.
2.Boris-Lawrie, K. A., and H. M. Temin.1993. Recent advances in retrovirus vector technology. Curr. Opin. Genet. Dev.3:102–109.
3.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher.1994. Green fluorescent protein as a marker for gene expression. Science263:802– 805.
4.Coffin, J. M., S. H. Hughes, and H. Varmus.1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
5.Delviks, K. A., W.-S. Hu, and V. K. Pathak. 1997.C2vectors: murine leukemia virus-based self-inactivating and self-activating retroviral vectors. J. Virol.71:6218–6224.
6.Dornburg, R., and H. M. Temin.1988. Retroviral vector system for the study of cDNA gene formation. Mol. Cell. Biol.8:2328–2334.
7.Dougherty, J. P., and H. M. Temin.1988. Determination of the rate of base-pair substitution and insertion mutations in retrovirus replication. J. Vi-rol.62:2817–2822.
8.Hu, W. S., and H. M. Temin.1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA87:1556–1560. 9.Julias, J. G., D. Hash, and V. K. Pathak.1995. E2vectors: development of
novel self-inactivating and self-activating retroviral vectors for safer gene therapy. J. Virol.69:6839–6846.
10. Miller, A. D., and C. Buttimore.1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol.6:2895–2902.
11. Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden.1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol.65:2220–2224.
12. Miller, A. D., and G. J. Rosman.1989. Improved retroviral vectors for gene transfer and expression. BioTechniques7:980–982, 984–986, 989–990. 13. Pulsinelli, G. A., and H. M. Temin.1991. Characterization of large deletions
occurring during a single round of retrovirus vector replication: novel dele-tion mechanism involving errors in strand transfer. J. Virol.65:4786–4797. 14. Sambrook, J., E. F. Fritsch, and T. Maniatis.1989. Molecular cloning: a
laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
15. Skalka, A. M., and S. Goff.1993. Reverse transcriptase. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
16. Watanabe, S., and H. M. Temin.1983. Construction of a helper cell line for avian reticuloendotheliosis virus cloning vectors. Mol. Cell. Biol.3:2241– 2249.
17. Zhang, J., and H. M. Temin.1993. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science259: 234–238.
18. Zhang, J., and H. M. Temin.1994. Retrovirus recombination depends on the length of sequence identity and is not error prone. J. Virol.68:2409–2414. FIG. 6. Southern analysis of recombinants between JZ211 and L290N290.
Chromosomal DNA of HygrSTEP 4 cells was used. Cellular DNAs isolated from
STEP 4 cells were pooled from more than 100 Hygrcolonies, digested with BamHI andHindIII, and hybridized with ahyggene probe (Fig. 2). LN290, L290N, and L290N290 represent Hygrcells resulting from recombination
be-tween JZ211 and LN290, L290N, and L290N290, respectively. Molecular sizes are shown on the left.
VOL. 73, 1999 RETROVIRUS RECOMBINATION 5917