JOURNAL OF VIROLOGY, Apr. 1980, p.108-118 Vol. 34, No. 1 0022-538X/80/04-0108/11$02.00/0
Short-Lived Minus-Strand
Polymerase
for Semliki Forest
Virus
DOROTHEA L.SAWICKI* AND STANLEY G. SAWICKI Departmentof Microbiology, MedicalCollege of Ohio, Toledo, Ohio 43699
Semliki Forest virus (SFV)-infected BHK-21, Vero,and HeLa cells
incorpo-rated[3H]uridineinto42S and 26Splus-strandRNA and into viral minus-strand
RNA (complementary to the 42S virion RNA) early in the infectious cycle.
Between3 and 4 hpostinfection, the synthesisof minus-strand RNA ceased in
thesecultures,
although
thesynthesis
ofplus-strand
RNA continuedat amaximalrate. At thetime ofcessation of minus-strandRNAsynthesis,two
changes
inthepatternofviralproteinsynthesisweredetected: adecrease in the translation of
nonstructural proteins and anincrease inthe translation ofthe viral structural
proteins. Additionofcycloheximide andpuromycintocultures of SFV-infected
BHKcellsactivelysynthesizing both viral
plus-
and minus-strand RNAresultedwithin 15 to 30 min in the selective shutoff of minus-strand RNA synthesis.
Removalof thecycloheximide-containingmedium ledtotheresumptionof
minus-strandsynthesis and toan increased rate of viral RNAsynthesis. We conclude
that the minus-strand polymerase regulates the rate of SFV plus-strand RNA
synthesis by determining the number ofminus-strand
templates
and that thesynthesis of the minus-strand
templates
isregulated
atthe level of translationby
amechanismwhichutilizes one or moreshort-livedpolymerase proteins.
Semliki Forest virus
(SFV)
is analphavirus
and contains as its genome a single strand of RNA of
plus
polarity having
amolecularweight
of 4 x 106 to 4.5 x 106 and a sedimentationcoefficient of 42S (8, 12). The
replication
ofalphaviruseshas
recently
been reviewed (9, 18,29). In orderfor SFV toreplicate, the parental
42S plus-strandRNAmustbe transcribed into
a complementary minus strand that in turn
serves as a
template
for thesynthesis
ofbothadditional 42S
plus-strand
RNA andsubge-nomic 26S mRNA. The 42S plus-strand RNA
serves as a
template
for the synthesis ofnon-structural viral proteins and is packaged into
virions.Thesubgenomic26S mRNA serves as a
template for the translation of the structural
viral proteinsand is notfoundinvirions.
In animal viruses that contain a plus-strand
RNA genome, it is generally assumedthat the
RNApolymerasesresponsible for the replication
ofthe viral genome are composed of nonstruc-turalpolypeptides becausetranslation must
oc-curbeforeviral RNAsynthesis commences and
purifiedvirions lack detectable
RNA-polymer-izing activity.Thenonstructural proteins (ns) of
SFV that are repeatedly detected in infected
cells are three in number, having molecular
weights ofapproximately 70,000 (ns7O), 86,000
(ns86), and 72,000 (ns72), all of which are
ini-tially
translated fromthe42S plus-strand RNAas onepolyprotein (9, 29). The products of the
alphavirus RNA polymerase are three distinct
RNA species: the 42S minus-strand RNA and
the plus-strand 42S and 26S RNA. The SFV
minus-strand RNA, unlike the plus-strand 42S
or 26S RNA, does not accumulate as
single-strandedRNA; rather,it isdetectedonlyaspart of the replicative intermediates (RIs) (26, 27).
Apparently, immediately afteraminus strand is
synthesized, it is used as a template for
plus-strandsynthesis.
Recently, thenatureof thealphavirus
polym-eraseresponsible forthesynthesis of SFVRNA 4 to 8hpostinfection (p.i.) waspartially
eluci-dated. Three laboratories have data which
indi-catethat the viralns7Ois themajor polypeptide
associated with membrane-bound replication
complexes transcribing42S and 26S RNA (4, 19;
P. J. Gomatos,unpublisheddata). Two of these
laboratories also consistently found smaller amountsof ns86 and ns72insuch complexes (19;
Gomatos,unpublisheddata). Theoverallaim of
our studies istocharacterizethe SFV polymer-aseresponsible for thesynthesisof minus-strand RNA. The resultspresented in this paper indi-cate that atrelatively early times in the infec-tious cycle, the synthesis of SFV minus-strand RNA ceases while the synthesis of plus strand 42S and 26S RNAcontinues at a maximal rate; this wasfound tobe true for SFV replicating in three different cell types: BHK-21, Vero, and HeLa. Whereas Brutonand Kennedy (2)
previ-108
on November 10, 2019 by guest
http://jvi.asm.org/
SFV MINUS-STRAND RNA 109
ously reported a similar observation for
SFV-infectedBHK-21 cells,wealso foundthat
cyclo-heximide andpuromycin treatment of
SFV-in-fected cells which were in the process of
tran-scription of both plus- and minus-strand RNA also resulted in the selective shutoff of
minus-strandtranscription, leaving untouched the
on-going plus-strandRNA synthesis. Although
pre-vious investigators have considered the alpha-virus RNA polymerase to be one species of en-zyme, ourresultssuggest that theremay actually exist two forms of the SFV RNA polymerase: one form responsible for the synthesis of
minus-strandRNA which is detected only early in the
infectiouscycle and whichrequires continuous
protein synthesisfor activity; and a second, more
stable polymerase which is responsible for the
synthesis of 42S and 26S plus-strand RNA
throughout the infectious cycle.
MATERIALS AND METHODS
Cell culture. BHK-21cells, acontinuouscell line derived frombabyhamsterkidneycells, weregrown in plastic petri dishes in Dulbecco-modified Eagle minimal essential medium (DMEM) supplemented with5%fetal bovineserum.HeLa (CCL-2)and Vero (CCL-81) cellswereobtainedfrom the AmericanType Culture Collection andweregrowninDMEM supple-mented with 5% fetal bovineserum.
Virus. Wild-type SFV, obtained from the second
passage ofSFV-infected mouse brain suspension in BHK cells, wasused in these experiments. Growth andpurification of virus and determination of its in-fectivity by plaque assaywere described previously (22).
Preparation of infected-cell extracts. BHK monolayers were infected with wild-type SFV at a multiplicity of infection (MOI) of100 as previously described (22), exceptthatactinomycinD (AMD) at aconcentration of1 ug/mlwaspresent.Thetime of addition of virus is designated zerotime. HeLa and Vero monolayers were similarly infected with wild-typeSFV except that AMDwasadded30minbefore labelingof the infected HeLamonolayersbecause of the sensitivity of HeLa cells to the cytotoxicity of AMD(24).
SFV RNA was labeled with [5,6-3H]uridine at a final concentration of 250
ILCi/ml.
For the protein synthesis inhibition studies, cycloheximide or puro-mycinat afinal concentration of50ug/mlwasaddedto DMEM containing 0.2% bovine serum albumin (BSA)and1
iLg
of AMD per ml.[3H]uridinewasaddedto a freshportionof this medium foranalysisof RNA
synthesizedinthe presence ofeachdrug. Cyclohexi-mideinhibitionwasreversedbyremoval of the drug-containing medium followedbyfoursuccessive washes with37°C DMEM and further incubation in DMEM containing BSA and AMD.
Atthe timesindicated forharvest, the cellswere
washed with ice-cold phosphate-buffered saline, a
small volume of ET buffer (0.01 M EDTA-0.01 M
Tris-hydrochloride,pH 7.4)containing2% sodium
do-decyl sulfate (SDS) was added, the lysed cells were scraped from the dish, and the cellular DNA was sheared by passing the extracts four times through a 27-gauge needle. For analysis of SFV RNA, the total cell extract waslayered directly onto 15 to 30% sucrose gradients in NET buffer (0.1 M NaCl-0.01 M EDTA-0.01 M Tris-hydrochloride, pH 7.4) containing 0.2% SDS and sedimented in an SW27 rotor at 95,400 x g for14.5hat20°C. Fractions of 1 ml werecollected by using a peristaltic pump, and the radioactivity in each was determined by counting a portion in a toluene-based scintillation fluid containing Triton X-100.
Labeling with [3S]methionine was carried out in methionine-free DMEM, using 50 ,uCi/ml per 60-mm petri plate. At thetimes indicated in individual exper-iments, infected BHK cells were exposed for 30 min to 335 mM NaCl (16, 21) in methionine-free DMEM containing0.2% BSA and 1 tgof AMD per ml. This medium was removed, and new medium containing
[3S]methionineand0.1M sucrose was added for the
timesindicated, followed by the addition of DMEM containing 20-fold the normal concentration of methi-onine.Aftera90-min chase period, the cell monolayers werewashed several timeswith ice-cold phosphate-buffered saline and harvested as described above.
Isolation of RF RNA, the RNase-resistant
coresof SFV RIs. The intracellular double-stranded SFV RNA (RIs) was obtained from material sedi-mentingatthe35 to20Sregion ofsucrosegradients of SDS-treated whole-cellextracts.After ethanol precip-itation at -20°C for 18h, the 35 to 20S RNA was dissolved in0.5ml ofdigestionbuffer(0.15 M NaCl-0.02MTris-hydrochloride, pH7.4); 25 ngof pancreatic RNasewasadded; and the samplewasincubatedat
room temperature for 15min. SDS was added to a final concentration of 2%. The digested sample was overlaid onto a 15 to 30%sucrose gradient in NET buffer containing 0.2% SDS. Centrifugation was at
154,000xgfor16h inanSW40rotor.Fractions of0.5
ml were collected; the fractions containing the RF RNA, the RNase-resistant cores of the RIs which sedimented at 20 to15S, werepooled,and the RNA was ethanolprecipitated in the presence of25 tLg of carrierratliver RNA.
Hybridizationof RF RNA with unlabeled 42S viron RNA. The procedures followed were as de-scribedpreviously (23).Briefly,theRF RNA thatwas obtained fromone100-mmpetriplate (approximately
7 x 106 cells)wasdissolved in0.2mlof1mMEDTA, pH 7.4. One-quarter of the dissolved RF RNA was dilutedto3.0ml with1mMEDTAat100°C,heated
at100°Cfor2min,quicklycooledto0°C,and divided into threeequalportions.Oneportionof the RF RNA served as a control for the fraction of the RNA
re-maining double stranded afterheatingandquick cool-ing.Thesecondand thirdportionsweresubjectedto hybridization conditions(0.4MNaClat68to70°Cfor
30minfollowedby30minat roomtemperature),one
inthe absence andthe otherinthe presence of10yg of unlabeled virion RNA.Toone-half of eachsample
wasaddedanequalvolume ofasolutioncontaining
0.3 M NaCl, 0.03 M sodium citrate, and pancreatic RNase(10
i&g/ml),
andthemixturewasincubatedat370Cfor 30min. Theuntreated and RNase-treated portions were precipitated with trichloroacetic acid VOL. 34, 1980
on November 10, 2019 by guest
http://jvi.asm.org/
110 SAWICKI AND SAWICKI
and theacid-insoluble radioactivity wasdetermined. Theheated andquick-cooledportion and the portion hybridized in the absence of anyunlabeled virion RNA gaveessentially equalRNase resistance of1 to 12%. Thisvaluewassubtracted from fractions of RNase-resistantradioactivity obtained from the correspond-ingportionhybridized in the presence ofan excessof unlabeled virion RNA. This RNase-resistant radioac-tivity was taken as the relative amount oflabeled minus-strand RNA in the RF RNA, since use of RNase-derived RF RNA results in the loss ofsome
nascentminus-strand RNA.
SDS-polyacrylamide gel electrophoresis. Poly-acrylamide slab gels with 8% separation gel and 5% spacergelwereprepared accordingtothe method of Laemmli (11). The gelswerefixed in 20% trichloroace-tic acid for60 min before treatmentwith dimethyl sulfoxide and2,5-diphenyloxazole (PPO) (1).
Materials. [5,6-3H]uridine (41Ci/mmol) was pur-chased from Amersham Radiochemicals (Arlington Heights,Ill.) and New England Nuclear Corp. (Boston, Mass.). [3S]methionine (600Ci/mmol) wasobtained from NewEngland Nuclear Corp. Cycloheximide was purchased from Calbiochem (SanDiego, Calif.) and puromycin was purchased from Boehringer Mann-heim Corp. (Indianapolis, Ind.). All other materials
wereobtainedfrompreviously described sources (22, 23).
RESULTS
Time course of SFV minus-strand RNA
synthesis. Functionalminus-strand SFV RNA
polymerase was detected by assaying for the
synthesis of SFV minus-strand RNA in BHK
cells infected with SFV
(100
PFU/cell)
in thepresence of 1
,tg
of AMD per ml.Replicate
cultures were labeled with [3H]uridine for 30
min atintervals
starting
1.5hp.i.
At the end ofthepulse period, the cellsweresolubilizedwith
2% SDS in ET bufferand
centrifuged
on 15 to30%sucrose
gradients;
the RIssedimenting
from 35 to20S werecollected, treated withpancreatic
RNase, and again
subjected
to rate zonalcen-trifugation on 15 to30%sucrose
gradients.
Theresulting double-strandedcores (RF RNA)
sed-imentingat 20 to15Swereused in theannealing
reactions instead of the intact RIstoeliminate
any
possible
interferenceby
the vast excess ofnascent, labeled plus-strand RNA chains in the
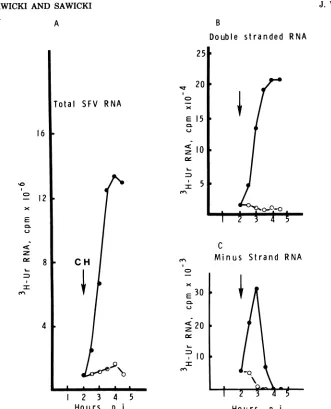
RIs. Figure 1 shows the amount of
[3H]uridine
incorporated intoRF RNA obtained from a
por-tion (one-seventh) of eachsample. LabeledRF
RNA wasreadilydetected 1.5 to 2 h p.i. Between 1.5and 3 hthere was a threefold increase in RF
RNA synthesized in each 30-min period. At
about 3 to 3.5 hp.i., the synthesis of RF RNA became constant. The synthesis of single-stranded SFV RNAalsoincreased between 1.5 and 3 h p.i. and became constant at 3.5 h p.i.
(datanotshown).
The fractionof
[3H]uridine
incorporated intoJ. VIROL.
200
100
x
E
50
2C
I0
5
2
total ds
/ minus strand
I'
IJ
I '
I
\
I I
0
0
2 3 4 5
Hours p. i.
FIG. 1. Time course of SFV minus-strand RNA synthesis. BHK-21 cells in100-mmpetri plateswere
infected with SFV(MOI of100) in the presence of AMD.Startingat1.5hp.i.,[3H]uridine(250,uCi/ml)
wasaddedforsequential30-minperiods, followedby preparationofinfected-cellextracts asdescribed in thetext. In allfigures, the amountofRNA synthe-sizedduringapulse is indicatedatthe endof the pulseperiod. [3H]uridine-labeledSFV RNA which sedimentedin the35 to20Sregionof15 to30%sucrose
gradients waspooled and treated with pancreatic RNase, and the RF RNA was resedimented on a
secondsucrosegradient. Anequalportion (one-sev-enth) of the labeled RF RNA (ds RNA)wasanalyzed forits[3H]uridine incorporation (0),and,afterheat denaturation and hybridization in the presenceof
excessunlabeled 42S virionRNA,fortheamountof
[3H]uridine incorporated into minus-strand RNA
(0). The[3H]uridine-labeledRF RNA which
specif-ically hybridizedtounlabeled virion RNA was con-sidered minus-strand RNA (see text).
minus strands of the RF RNA wasdetermined
by hybridization of denatured RF RNA with
unlabeled 42S virion RNA. Incorporation of
[3H]uridine into minus-strand RNA reached a
maximum at 3 hp.i.,afterwhich timeit declined
(Fig. 1).After3.5h, essentiallynominus-strand
synthesiswas detectable, although plus-strand
RNAcontinued to besynthesizedat amaximal rate.
Weinvestigated the time course of SFV
mi-nus-strandRNA transcription in two othercell
types. Monolayers of Verocellsand HeLacells
were infectedwith SFV at an MOI of 100, the
viral RNAwas labeled with
[3H]uridine
in the presence of AMD for sequential 30-min periods, andthe RF RNA was isolated and analyzed asdescribedabove for the BHKcellextracts
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.508.284.430.67.284.2]SFV MINUS-STRAND RNA 111
ble 1). Both cell types produced viral RNA,
although HeLa cells incorporated only about
one-fourthasmuch
[3H]uridine
into viral RNAasdid eitherVero or BHKcells.Thesynthesis
of SFV minus-strandRNAdeclined3 to 4 h p.i.
in eachofthese celltypes: Vero cellsexhibited
asimilar ifnotidenticaltimecourse as the
BHK-21cell cultures; inHeLacells,theinhibition was
not asabrupt.
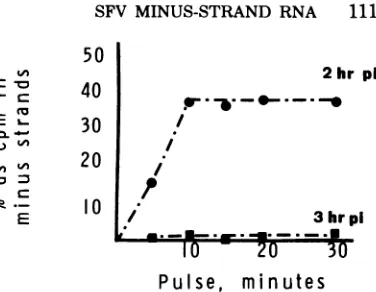
IfSFV minus-strand RNA were synthesized
butrapidly degraded after3hp.i., we might not
have detected synthesis of new minus-strand
RNA after 3 h, using 30-min labeling periods.
Therefore,wedetermined theamountof
incor-poration of [3H]uridine into nascent
minus-strand RNA, using shorter pulse-labeling times.
A 5-min pulse was sufficient to label
minus-strand RNA when thepulsewasgiven at 2 h p.i.
(Fig. 2). This resulted in thevastmajority of the
labeledRNAsedimentingasRIsand only very
smallamounts oflabeled RNA sedimenting as
completed 42S and 26S species (datanotshown).
Pulse times of10to 30min at 2hp.i. resultedin
maximallevels of labeled minus strands in the
RF RNA.However,duringa 5- to30-minpulse
of
[3H]uridine
given toinfected BHK cellcul-tures at 3hp.i., little or nosynthesis of
minus-strand RNAwasdetected (Fig.2). We conclude
from these results that transcription of
minus-strandRNA, and thusthe activity of the
polym-erase synthesizing the minus strands, ceases
TABLE 1. TimecourseofSFV minus-strand synthesis in threedifferentcelltypesa
[3H]uridine
time % of RF RNA inminus-strand RNA of addition BHK-21(h p.i.) cells Vero cels HeLacells
2-2.5 46 44 45
2.5-3 24 28 41
3-3.5 4 6 38
3.5-4 0 5 26
4.5-5 0 1 13
5.5-6 0 8
6.5-7 6
7.5-8 6
8.5-9 4
aLabeled viral RNA in total
cell
extractsharvestedatthe end of thepulseperiodwascentrifugedon15to 30%sucrosegradients;RNAsedimentinginthe35to
20Sregionwascollectedby ethanolprecipitation;and the double-stranded RF RNA was prepared as de-scribed in the text. Between 1,000 and 50,000 cpm (approximately0.03
pg)
ofRF RNAwasusedineach annealingreaction with10jugof unlabeled42S virion RNA. Hybridization of SFV RF RNA isolated from infectedBHK-21 cell extracts, which hadbeenlabeledwith[3H]uridinefrom0to4hp.i.,gave valuesranging from38 to47% ofthe total labeled RNA in
minus-strand RNA.
qA
C
-E
50
40
30
20
10
2 hr pi
i
i
L. M. _
_6-3hr pi
* _.n _. _.
-IU LU0 U
Pulse,
minutes
FIG. 2. Presence of nascent SFV minus-strand RNA inRFRNA. SFV-infected BHK-21 cell cultures
wereexposed at2or 3h p.i. to[3H]uridine (250
ACil
ml)for periods of 5 to 30 min, after which time they wereharvestedasdescribed in the text. The RFRNAwas obtained and, after heat denaturation, the
amount of labeled RF RNA in minus-strand RNA
was determinedby hybridization with excess unla-beled virion RNA (see Fig. 1). Symbols: 0, Minus-strand RNA in RF RNA obtained from cultures pulsed at2hp.i.; , minus-strand RNA in RF RNA obtainedfiomculturespulsed at 3 h p.i.
after3h.Furthermore, since the rate of
[3H]-uridine
incorporation
intonewly
synthesizedplus-strand RNA remainedconstantfor1to 2 h
after the cessation of minus-strand synthesis,
previously synthesized minus-strand RNAmust
continuetofunctionastemplate for plus-strand
synthesis andnotbesubjecttodegradation after
3h.
Patternof
virus-specific
proteins.We nextdeterminedwhether there was a changein the
pattern
of viralprotein synthesis
atthetime ofminus-strand RNA inhibition. At different
times
after infection with
SFV,
BHKcellswerelabeledwith
[3S]methionine
aftertreatmentof thecul-tures for 30 min with hypertonic medium to
reduce the
background
of hostprotein synthesis.Figure 3 represents a
fluorogram
of the viralproteinssynthesized during 30-min intervals
be-tween1and4.5hafter infection.The
synthesis
ofns70 andns86wasdetectable
beginning
at1.5handwas
maximnal
between 2.5and3h;
there-after the
synthesis
of ns7O and ns86 declinedprecipitously.
On the otherhand,
thesynthesis
of the structural
proteins,
capsid
and the twoenvelope
proteins
(El
andE2),
was firstob-servedat2h and
only
became maximalat3to3.5 h p.i. The structural
proteins
became thepredominant
proteins
synthesized by
infectedcells at 3.5 h.
Thus,
the time atwhichminus-strand
synthesis
ceased wasnearly
the sametime that the infected cell
began
tosynthesize
almost
exclusively
structuralproteins.
Effect of
protein
synthesis
inhibition onIf%
VOL.34,198
on November 10, 2019 by guest
http://jvi.asm.org/
[image:4.508.264.452.54.200.2] [image:4.508.54.250.417.542.2]112
SAWICKI AND SAWICKI1 1.5 2 2.5 3 3.5 4 X
...g._ ...
* m
__
beginningat 1.5hp.i.,
cycloheximide
wasaddedto a final concentration of 50
,ug/ml.
Thirty
minutes after the
cycloheximide
wasadded, halfof the cultures were labeled for 30 min with
[3H]uridine.
Theother halfwerelabeled for30min with
[3H]uridine
at 4.5 h after infection.Replicate cultures of SFV-infected cells that
re-ceivedno
cycloheximide
alsowerelabeled for30minwith
[3H]uridine
at0.5-hintervalsbeginning
1.5hp.i. Between1.5and2.5h
p.i.,
therewas an86
exponential
riseintherateof RNAsynthesis
inSFV-infected cultures (Fig. 4). The addition of
cycloheximide
blocked the increase in the rateof SFV RNA
synthesis.
Addition ofcyclohexi-nz7O mide to infected cultures after 3 h p.i. had no
effecton the rate of RNA
synthesis.
RF RNAwasisolated from cellsthateitherweretreated
with
cycloheximide
at 1 or 1.5 hp.i.
or wereE2
E1
FIG. 3. SFV viralproteins synthesized at various timesafterinfection. BHK-21 cell cultures in 60-mm petri plateswereinfectedwithSFV at an MOI of 100. The timesindicated represent the time of addition of medium containing 0.335 M NaCl (16, 21) to the individual cultures. Startingat 1 hp.i., the cultures
wereexposed to the hypertonic medium for 30 min, followed by a 30-min pulse of[35S]methionine in mediumcontaining0.1Msucrose and a90-min chase period. Whole cells were collected into ET buffer containing 2% SDS, and portions containing equal
amountsof total cell protein were electrophoresed on
an8%polyacrylamide slab gel as described in the
text.At3hp.i.,aduplicate culture was pulsed with
[365]methionine for 7min after synchronization of
translation by hypertonic treatment and chased for
90minin order to label specifically theN-terminal
twopolypeptides translatedfromthe 42Splus-strand RNA, ns7Oandns86(lane X). The portions applied
tothe gelcontained approximately the following cpm: (1 h) 200,000; (1.5 h) 160,000; (2 h) 120,000; (2.5 h) 130,000; (3 h) 90,000; (3.5 h) 90,000; (4 h)120,000. The exposure time was 20 h.
minus-strand synthesis. We then asked
whether the cessation of synthesis of minus-strand RNAwouldoccurwhen protein synthesis wasinhibited before the
time
that the infectedcells began to synthesize predominantly viral
structuralproteins.
Replicate
cultures of BHKcellswereinfected withSFV. At 0.5-h intervals
20
10
'U,
II55
U a
I.2
I
a
s
1 2 3 4 S
Moore p.1.
FIG. 4. Effectofcycloheximidetreatment onSFV RNAsynthesis.BHK-21 cell cultures in35-mmpetri plates were infected with SFV (MOIof 100). Un-treated cultureswerelabeled in the presenceofAMD for30 minwith [3H]uridine (50 uCi/ml) at 30-min intervals between 1 and5 hp.i. At 0.5-h intervals beginning 1.5 hp.i., replicate cultureswere treated withcycloheximide (50 pg/ml). The arrowsindicate the timeofcycloheximide addition. Half ofthe
cul-tures were labeledfor a30-minperiod with
[3H]-uridinebeginning30minafteradditionofthedrug. The otherhalfwaspulsed with[3H]uridine
at 4.5 to 5hp.i. Theinfectedcellswereharvestedatthe end ofthepulseperiod by solubilization in ET-SDSbufferasdescribed in thetext. Portions induplicatewere
assayedforacid-insolubleradioactivity andfor pro-tein by the methodof Lowryetal. (13).Symbols: 0,
[3H]uridine
SFV RNA in untreated cultures; 0,[3H]uridine
SFV RNA incycloheximide-treatedcul-tures.
I
J. VIROL.
.: oo:.kfw
"Ie.
O' I
I
0
--l.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:5.508.69.245.60.372.2] [image:5.508.269.455.276.485.2]VOL. 34, 1980
untreated and thatwerelabeledat6h
p.i.
for1h with [3H]uridine. At 1 to 1.5h
p.i.,
therewasnodetectable synthesis of 26S SFV RNA (data
notshown) or ofviralstructural proteins (Fig.
3). The presence oflabeled minus-strand RNA
intheRFRNAwasdeterminedbyhybridization
to an excessof unlabeledvirionRNA.Nolabeled
minus-strand RNAwasdetected when the label
was added 5 h after cycloheximide
(data
notshown). We
conclude, therefore,
that thesyn-thesisofminus-strand, butnot
plus-strand,
RNAisprevented by
long-term cycloheximide
treat-ment,and that the accumulation of viral
struc-tural proteins is not
required
to shut off thesynthesis
of minus-strand RNA.A
possible
requirement for continuedtransla-tion of the viral nonstructuralproteinsforSFV
minus-strand
transcription
wassuggested fromthese results and from those ofFig. 3 andwas
investigated
further. The effect ofcycloheximideon minus-strand synthesis was monitored at
shortintervals after its addition totheinfected
monolayers. At 2 h p.i., when transcription of
minus-strand RNA was at a maximum,
cyclo-heximide was added to a final concentration
of 50 ug/ml. The cultures were pulsed with
[3H]uridine
for 30-min periods between 2 and4.5hp.i. The addition of cycloheximide at 2 h
p.i. stoppedthe increase in viral RNA synthesis
(Fig.5A) and the increase in RF RNA (Fig.5B).
Compared with the increasing amounts of
la-beled RF RNAisolated from untreated cultures
between2 and 3.5 h p.i., a constant amount of
labeled RF RNA was obtained from infected
cultures thatwere treated withcycloheximide at
2hp.i. andmaintained in the presence of
cyclo-heximide.
Hybridization ofthe labeled RFRNA with an
excess ofunlabeled virion RNA indicated that
minus-strand RNAsynthesis rapidly ceased in
thepresence of cycloheximide (Fig.
50).
Wheninfected cells were labeled at 2 h p.i. with
[3H]uridine
for30mininthepresence ofcyclo-heximide, the amount of labeled minus-strand
RNAdetected in theRF RNA wasthe same as
that foundininfected cellsthatwere labeled at
1.5hp.i. for30mininthe absenceof
cyclohexi-mide. When infected
cells
were treated withcycloheximide
at 2h p.i. and labeledfor 30minwith
[3H]uridine
at2.5 to 3or 3 to3.5h,essen-tially
no labeled minus-strand RNA wasde-tected inthe RFRNA.Sucrosegradient analysis
demonstratedthat 42S and 26SRNA was
syn-thesized during cycloheximide treatment (data
not shown). Therefore, minus-strand, but not
plus-strand,RNAsynthesis rapidly declined in
thepresenceofcycloheximide.
An identicalexperimentwascarriedout, using
SFV MINUS-STRAND RNA 113
puromycin to inhibitprotein synthesis. At 2 h
p.i., medium containing puromycin (50 ug/ml) wasadded toSFV-infectedcultures whichwere
subsequently labeled with [3H]uridine for
30-min intervals over the next 2 to 3h (Fig. 6A).
Compared with the untreated cultures,
puro-mycin addition resultedin a50% decreaseinthe
amountof SFV RNAsynthesized in the first30
min of
drug
addition. This was followed by aconstant or even decreased level of RNA
syn-thesis for thenext 2h.Similartocycloheximide
treatment, the addition of puromycin stopped
the increase in total RF RNA, and the total
amountoflabeled RF RNApresentin
puromy-cin-treated cultures remained relatively
con-stant.Also similartotheeffect of
cycloheximide,
puromycin addition ledtotheselective and
rapid
shutoff of minus-strand RNAtranscription (Fig.
6B).
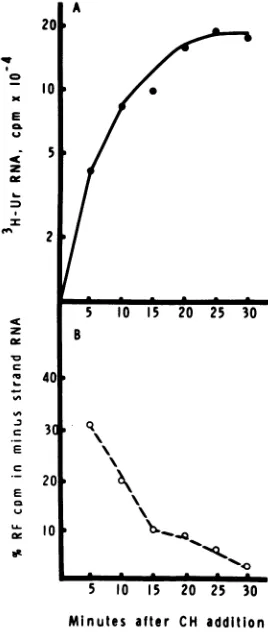
A closer examination of minus-strand
tran-scription was undertaken
by
labeling
with[3H]uridine the SFVRNAsynthesized in5-min
intervalsduring the first30minof
cycloheximide
treatment.Cycloheximidewasaddedat2h
p.i.,
and the cells were given a 5-min
pulse
labelbetween2 and2.5h p. i.
(Fig.
7A). TherateofSFV
transcription
increasedduring
thefirst 15min of
cycloheximide
treatment and becameconstant by 20 min. The amount of labeled minus-strand RNA in isolated RF RNA
de-creasedata
rapid
ratefor thefirst 15minafterdrug
addition, continuing
to decreaseslowly
thereafter
(Fig.
7B).Therefore,
labeled minusstrands detected after
cycloheximide
additioninFig.5Cwere
synthesized mainly
during
the first15 minof the30-min
labeling period.
Since thesynthesis
of minus-strand RNAessentially
ceased 20 min after the addition of
cyclohexi-mide but the
synthesis
ofplus-strand
RNAcon-tinued at a
given
rate, the number ofminus-strand RNA
templates
mostprobably
deter-mines the rate of
plus-strand
RNAsynthesis.
Theseresults demonstratethattheSFV
polym-erase
responsible
for thesynthesis
of minusstrands is
short-lived,
whereas the SFVpolym-erase
responsible
for thesynthesis
ofplus
strands is
long-lived.
Reversal of inhibition of minus-strand
RNA
synthesis.
Since our resultsimplicated
continued
protein
synthesis
as arequirement
forminus-strand
transcription,
theresumption
oftranslation afterremoval of the
cycloheximide-containingmediumand
thorough
washing
of thecell
monolayers
shouldinturnbefollowed
by
aresumption of minus-strand
transcription.
Whencycloheximide
wasaddedat2hp.i.andremoved30 or 60 min later and then the
newly
synthe-sized RNA was labeled with[3H]uridine,
theon November 10, 2019 by guest
http://jvi.asm.org/
114 SAWICKI AND SAWICKI A
16
2
x
E
0L.
(z
I
12
8
4
J. VIROL.
B
Double stranded RNA
v 20
1
x
E 15
< 10
M,
I 5E30
0..
< 20
10
I
2 3 4
Ho urs, p. i.
C
Minus Strand RNA
Hours, p. i.
FIG. 5. Effectof cycloheximidetreatment onSFVminus-strand RNAsynthesis.Starting 1.5 h p.i.,duplicate culturesofSFV-infectedBHK-21 cells(100-mm petri plates)werelabeled with[3H]uridine (250pLCi/ml)for sequential30-minperiods. Onesetofcultureswasleftuntreated(0);tothe otherset, mediumcontaining50 pgofcycloheximide(CH)permlwasaddedbeginningat2 hp.i.(0). The cultureswereharvestedatthe end ofthepulseperiodasdescribed in the text. (A) Total acid-insoluble[3H]uridine incorporation into SFV RNA.(B)Total[3HJuridine incorporationinto RF RNA obtainedfromuntreated andcycloheximide-treated
cultures. The RF RNAwasobtainedasdescribed in thetext.(C) Theamountoflabeled RF RNA in minus-strand RNA.TheRFRNAwasdenatured andhybridizedin the presenceofexcessunlabeled virion RNAas described inFig.1.
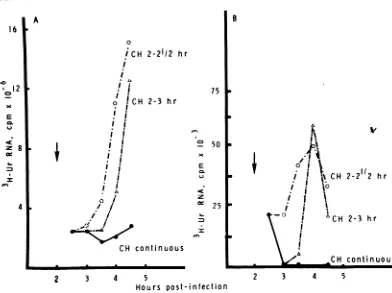
rate ofsynthesis of total SFV RNA increased afteralag period ofapproximately30min (Fig. 8A). The amount of labeled double-stranded
RNA also increased, and was proportional to
thatobserved for single-stranded RNA. Analysis of thepolarityofthelabeled RNA in the isolated RF RNAconfirmedthe presence of newly
syn-thesized minus strands in these RIs (Fig. 8B).
Transcription of minus-strand RNA began
within the first 30 min of drug removal and
increased substantially during the next 30 min
(drugremovedat3h) to 60 min (drug removed
at2.5h). Interestingly, the transcription of mi-nus strands declined between 4 and 4.5 h in
on November 10, 2019 by guest
http://jvi.asm.org/
[image:7.508.99.430.55.464.2]SFV MINUS-STRAND RNA
115
thesis ofplus-strand RNA, is temporally
regu-lated and that the polymerase
activity
respon-sible for minus-strand RNA
synthesis
isshort-lived and
disappears quickly
afterinhibition ofprotein synthesis with cycloheximide or
puro-mycin.
Our resultsstrengthen
and extendthoseof Bruton and
Kennedy (2),
who alsodemon-strated the
temporal regulation
of minus-strandRNAsynthesis in SFV-infected BHK cells and
suggested that therateof
plus-strand
RNAsyn-thesismight be correlated with theamount of
minus-strand RNA. We observed that therate
of
plus-strand
RNAsynthesis
becomesconstantwhenminus-strand RNA
synthesis
ceases, andthat blockage of
protein
synthesis,
whichin-hibits
rapidly
thesynthesis
of minus-strand201
a40 ">
C.
30
E 20 .9,
20~~~~~~
%A 10. \
1 2 3 4 S
HOURS, POST-INFECTION
FIG. 6. Effect ofpuromycinadditiononSFVRNA
synthesis. (A) Total [3H]uridine incorporation into
SFVsingle- and double-stranded RNA during 30-min pulse periods from1.5to4.5hp.i. BHK-21 cell
cultures in 100-mmpetri plates were infected with
SFVatanMOIof100,labeled with [3H]uridine(250
.Ci/ml),
andharvested asdescribed inFig. 5.Me-diumcontaining puromycin (50pg/ml)wasaddedto
culturesat2 hp.i. (arrow). (B) Percentageof total
[3HJuridine incorporatedintoRF RNA thatwasin
minus-strand RNA. The RF RNA was isolated as
described in thetext, denatured by heating,and hy-bridized in thepresenceofexcess unlabeled virion
RNA. Symbols: 0, [3H]uridine-labeled SFV RNA
obtainedfromuntreatedcultures;0,
[3HJuridine-la-beled SFV RNA obtained from puromycin-treated
cultures.
these cultures, although only 40 to 75% ofthe amountoftotal RF RNApresentin untreated cultures had beenfornedby this time.
DISCUSSION
Ourresults demonstratethat thesynthesisof
SFVminus-strandRNA,in contrast tothe
syn-C
c E
z
cc
to
Sc
l0o
5
2
40 3a 201
10I
A0
5 10 15 20 25 30 B
N\
[image:8.508.65.247.52.430.2]5 10 1 5 20 25 30 Minutes after CH addition
FIG. 7. Kineticsofminus-strand inhibitionby
cy-cloheximide. SFV-infected BHK-21 cell cultures in
100-mmpetri platesweretreated withcycloheximide (50 pg/ml) at2 hp.i. andpulsedwith [3HJuridine
(250,Ci/ml) for sequential5-minperiods between2
and 2.5 hp.i. (A)Total[3H]uridine incorporatedinto
acid-insoluble SFV single- and double-stranded
RNA.(B) Percentage ofthe total[3HJuridine
incor-poratedintoRF RNA in minus-strandRNA,
deter-minedasdescribedinFig.1.
VOL. 34, 1980
< 16
E
C)
12
z
,2 8
r-=4 ?.
c0
on November 10, 2019 by guest
http://jvi.asm.org/
[image:8.508.293.427.248.567.2]116 SAWICKI AND SAWICKI
0
ICH 2-2112
hri Ii i
I!
i
Ii
I
1i
,c
CH continuous
75
° 50
x
E
Ca.
z
I25
B
4
CH
2-2I/2
hrCH 2-3 hr
CH continuous
2 3
4
5
Hours post-infect ion
2 3 4 5
FIG. 8. Resumption ofSFV minus-strand RNAsynthesis afterremovalofcycloheximide.Medium
contain-ing cycloheximide (50 pg/ml)wasadded at 2 hp.i.toall cultures(arrow);in certaincultures,itwasremoved
30min (0) or60min (A) later (see text).[3H]uridine(250 g.Ci/ml) wasaddedfor30minat0.5-hintervals either in thepresence (cultures continuously treated with cycloheximide) orin the absence(cultures from
which the drug hadpreviously been removed) of cycloheximide. The cpm shown represents the total
incorporationobtainedfrom each 100-mmpetri plate. Thecultureswereharvested atthe endofthepulse period. (A)[3H]uridineincorporation into SFVsingle- anddouble-stranded RNA. (B)[3H]uridine-labeled
RF RNA in minus-strand RNA. Between 2 and 4.5 hp.i.,theamountoflabeledminus-strand RNAdetected
in these cultureswasasfollows: (cycloheximide,2to2.5h) 625,000cpm;(cycloheximide,2to3h)356,000cpm;
(untreated)820,000cpm;(cycloheximidecontinuousfrom2h)100,000cpm.
RNA but not ofplus-strand RNA,also results in
a constant rate of RNAsynthesis. Polymerase
proteinspresent in SFV-infected cells that have had theirprotein synthesisarrested continue to function in thesynthesisofplus-strandRNAby transcribing the template minus-strand RNA which was formed before protein synthesis
in-hibition; thus,the number ofminus-strand tem-platesappearstocontrol therateofplus-strand synthesis. Only when minus-strand synthesis
wasallowedtoresumebyremoval ofthe cyclo-heximide didweobserveanincreaseintherate
ofplus-strand synthesis.
Weconclude fromthese results that transcrip-tion of SFV minus-strand RNA is regulatedat
the level of translation bya mechanism which
utilizesone or moreshort-lived polymerase
pro-teins that either are not involved in the
tran-scriptionofplus-strandRNAor aremodified by
cleavage or other posttranslational modifica-tions to form the stableplus-strand polymerase. Thus, there may exist in SFV-infected cells
either two forms of theviralpolymeraseortwo distinctpolymerases.
Nothingis known ofthe location within the SFV-infected cell ofminus-strandpolymeraseor
ofthe replication complex transcribing minus-strandRNA. The recentreportof Dasguptaet
al. (5) indicates that thepresumptive poliovirus minus-strand polymerase (replicase) was
de-tected inasoluble ratherthanmembrane-bound
fraction of theinfected cell.The SFV plus-strand polymerase is associated with a
membrane-boundreplication complex (4,7, 9, 15, 18, 28, 29). Recently, the SFV nonstructural protein ns70 has been shown to be the major viral poly-peptideassociated with thereplication complex (4, 19; Gomatos,unpublished data). In addition, A
16
No
1 12
2 I.x
Z 8
4L
4i
J. VIROL.
on November 10, 2019 by guest
http://jvi.asm.org/
[image:9.508.60.452.63.356.2]VOL. 34,1980
smalleramountsof ns72 andns86werefoundin
this complex (19; Gomatos, unpublished data). Themigration of ns70 in polyacrylamidegels is always as a broad band, suggesting that this
polypeptide chain may have undergone
post-translational modification. Suchamodification
could account for temporal alteration of
tem-plate preference of the SFV polymerase. Also uncleavedorincompletelycleavedprecursorsof
thenonstructuralproteins could function in al-phavirus minus-strandsynthesis. Insupport of these possibilitiesisthe finding that continued proteinsynthesisisnecessaryfor the replication
ofvesicular stomatitis virion RNA (17, 31) and that there is an apparent correlation between
the cleavage of certain poliovirus proteins and replicase instability in poliovirus-infected cells
(10).
It has been reported previously that the amount but notthe ability of the viral
polym-erase to synthesizeplus-strand RNA in alpha-virus-infected cellscould be varied bythe
addi-tionofpuromycinorcycloheximideatdifferent
timesearly in infection (30). However, by 3to4 h p.i., essentially all of the normal amount of viralpolymeraseresponsible for the synthesis of
plus-strandRNA had been formed and
inhibi-tion ofprotein synthesis after this time didnot affect the amount ofRNA synthesized (6, 25, 30). We found,asdid others (6, 25, 30), that the
SFVplus-strandpolymerase formed before the
addition of cycloheximide or puromycin
re-mained active formanyhours in theabsence of
continuedprotein synthesis. Our results indicate that this is not the case for the synthesis of
minus-strand RNA. The addition of cyclohexi-mide orpuromycintoculturesactively
synthe-sizing both plus- and minus-strand SFV RNA
led veryquicklytothe selective loss of minus-strand transcription. The fact thattwoprotein synthesisinhibitors whose mechanisms of inhi-bitionare differentresult in thesame phenom-enonleadsustoconcludethat this isduetothe inhibition of translation.Thus,wearguethatit is thesynthesisoftheminus-strandpolymerase thatregulatestherate ofplus-strandRNA
syn-thesis by determining the number of minus-strandtemplatesformed in the infected cell.
Our characterization of the synthesis of mi-nus-strand RNAsuggestedthat its cessationwas
correlated with two changes in the pattem of viralprotein synthesis: adecrease in the
trans-lation of nonstructuralproteinsand the
appear-anceoflargeamounts ofstructuralproteins.The
nonstructural proteins are translated from the
42Splus-strandRNAas apolyproteinfollowed
by cleavageinto the individualproteins,whereas
the structural proteins are read from the 26S
mRNA, also initiallyas a polyprotein (2, 9, 18,
SFV MINUS-STRAND RNA
117
29). Therefore, the
ability
toregulate thepro-ductionof the
nonstructural
proteinsindepend-ently of the structural proteins exists for the
alphavirusesbut notfor otherplus-strandRNA
virusessuchaspoliovirus,whichproducesallof
itsviralproteins from thesamepolyprotein(20).
The observed
instability
of the minus-strandpolymerasecoupled with the decreased
transla-tionof
all
nonstructural proteins could be oneexplanation
for theselectivecessation ofdetect-able minus-strand transcriptionatabout 3 to 4
hp.i.
Weproposethe
following
scheme for the early events Mi SFV replication. The SFV parental genomeisfirsttranslated into the viralnonstruc-tural
proteins,
someof whichfunction
totran-scribe thissameRNA molecule into
complemen-taryminus strands. For thistooccur,the
minus-strand
polymerase
mustinitiate RNAsynthesisat the 3' end of the 42S parental plus strand
(22). Because ribosomes bind to the 5' end of
42Splus-strand RNA and failtoproceedpast a
termination signal located approximately
two-thirdsof the distance from the 5' end(9, 18, 29),
the3'terminus of the
parental
plus strand mightbe
readily
accessible for recognition byminus-strand
polymerase.
We have been unable todetect the
synthesis
ofonly minus-strand
RNA;RIs synthesizing
bothplus
and minus strandswere always present
early
in infection, inap-proximately
a5:1 ratio(see
Fig.
2). Atapproxi-mately 3.5 h after
infection,
minus-strandsyn-thesis ceased
although
plus-strand
RNAcontin-ued toaccumulate: 26S on polysomes and42S
in
nucleocapsids.
Apossible
advantage conferredby thecessation of minus-strand RNA
synthesis
is that of allowing "free" 42S
plus
strands toaccumulate for the fonnation of nucleocapsids;
otherwise, 42S
plus
strandsmight continuetobeconverted into RIs at the expense of progeny
virion
production.
ACKNONLEDGMENTS
Wethank ThomasSawyerforexcellenttechnical asistance and Peter J.GomatosandPaul Lehmann forhelpful discus-sions.
This investigation was supported by funds from Public HealthService Research grant AI-15123 from the National Institute forAllergyand Infectious Diseases and from BRS grant5S07-RR-05700-09awardedbytheBiomedical Research
Support Resources,National Institutes ofHealth.
ITMRATURE CITED
1.Bonner,M.W.,andR.Laskey.1974. Afilm detection method for tritium-labeledproteins and nucleic acidsin
polyacrylamide gels.Eur. J. Biochem.46:83488.
2. Bruton,C. J., and S. L.T. Kennedy. 1975. Semliki Forestvirusintracellular RNA:propertiesof the multi-stranded RNA species and kinetics of positive and negativestrandsynthesis.J. Gen.Virol.28:111-127. 3. Clegg,J.C.S.,H.Brzeski,and S.L.T.Kennedy.1976.
RNApolymerasecomponentsinSemliki Forest
on November 10, 2019 by guest
http://jvi.asm.org/
118 SAWICKI AND SAWICKI
induced cells: synthesis from largeprecursors.J.Gen. Virol.32:413-430.
4. Clewley, J.P., and S. L.T.Kennedy. 1976.Purification and polypeptide composition of Semliki Forest virus RNApolymerase.J. Gen. Virol. 32:395-411.
5. Dasgupta, A.,M. H.Baron, andD.Baltimore. 1979. Poliovirusreplicase:asolubleenzymeabletoinitiate copying of poliovirus RNA. Proc. Natl. Acad. Sci. U.S.A.76:2679-2683.
6. Friedman,R.M.,and P. M.Grimley. 1969.Inhibition ofarbovirusasserhbly by cycloheximide. J. Virol. 4: 292-299.
7. Friedman, R.M.,J. G.Levin,P. M.Grimley, and I. K.Berezesky.1972.Membrane associatedreplication complex in arbovirus infection. J.Virol.10:504-515. 8. Friedman,R.M.,H. B.Levy,and W. B. Carter. 1966.
Replication of Semliki Forest virus: three forms of viral RNAproduced duringinfection. Proc.Natl. Acad. Sci. U.S.A.56:440-446.
9. Kaariainen, L., and H. Soderlund.1978.Structureand replication of alpha viruses. Curr. Top. Microbiol. Im-munol.82:15-69.
10.Korant, B. D.1975.Regulation of animal virus replication byprotein cleavages, p.621-644. In E. Reich, D. B. Rifkin, and E. Shaw (ed.), Proteasesand biological control, Cold Spring Harbor ConferenceonCell Prolif-eration, vol.2. ColdSpring Harbor Laboratory, Cold Spring Harbor, N.Y.
11.Laemmli, U. K. 1970. Cleavage of structural proteins during theassemblyofthe head ofbacteriophage T4. Nature(London)227:680-685.
12. Levin,J. G., andR. M.Friedman. 1971. Analysis of arbovirusribonucleic acid formsbypolyacrylamide gel electrophoresis.J. Virol.7:504-514.
13.Lowry,0.H.,N. J.Rosebrough,A.L.Farr, andR.J.
Randall. 1951. Protein measurement with the Folin phenolreagent.J.Biol.Chem. 193:265-275.
14. Martin,B.A.,and D. C.Burke. 1974. Thereplicationof SemlikiForest virus. J. Gen. Virol.24:45-66. 15. Martin,E. M. 1969. Studiesonthe RNApolymerase of
sometemperature-sensitivemutantsofSemliki Forest virus.Virology 39:107-117.
16.Nuss,D.L.,H.Opperman,andG. Koch.1975.Selective blockage of initiation of hostcellprotein synthesis in RNA-virusinfected cells.Proc. Natl.Acad.Sci. U.S.A. 72:1258-1262.
17. Perlman, S. M., and A. S. Huang.1973.RNA synthesis ofvesicular stomatitisvirus. V.Interactions between transcription and replication.J.Virol. 12:1395-1400.
18. Pfefferkorn,E.R.,and D.Shapiro.1974.Reproduction
oftogaviruses, p. 171-230.InH. Fraenkel-Conrat and R. R. Wagner(ed.), Comprehensive virology, vol. 2. Plenum Press, New York.
19. Ranki, M., and L. Kaariainen.1979.Solubilized
repli-cationcomplex from Semliki Forest virus-infectedcells.
Virology98:298-307.
20.Rekosh,D. M. K.1977.Themolecularbiologyof
picor-naviruses, p.63-103.In D. P.Nayak(ed.),The molec-ular biology ofanimal viruses. Marcel Dekker, New York.
21. Saborio, J.L., S. S. Pong, andG.Koch. 1974. Selective and reversible inhibition of initiation of protein synthe-sis inmammalian cells. J. Mol. Biol. 85:195-212. 22. Sawicki,D.L.,and P. J.Gomatos. 1976.Replication of
Semliki Forest virus:polyadenylateinplus-strand RNA andpolyuridylateinminus-strand RNA. J. Virol. 20:
446-464.
23. Sawicki,D.L.,L.Kaariainen,C.Lambek,and P. J.
Gomatos.1978.Mechanism for control ofsynthesis of Semliki Forest virus 26S and 42S RNA. J. Virol. 25:19-27.
24. Sawicki,S.G.,andG. C. Godman.1971.On the differ-entialcytotoxicity of actinomycinD.J.CellBiol.50: 746-761.
25. Scheele,C.M.,and E. R. Pfefferkorn.1969.Inhibition ofinterjacent (26S) ribonucleic acid synthesis in cells infected with Sindbis virus. J. Virol. 4:117-122. 26. Segal, S.,and T.Sreevalsan.1974.Sindbis virus
repli-cativeintermediates:purification and characterization. Virology59:428-442.
27. Simmons, D.T., and J. H. Strauss. 1972. Replication of Sindbis virus. II. Multiple forms of double-stranded RNAisolated from infectedcells.J. Mol. Biol. 71:615-631.
28. Sreevalsan,T., and F. H. Yin. 1969. Sindbis virus-in-duced viral ribonucleic acidpolymerase.J. Virol. 3:599-604.
29. Strauss, J. H., and E. G. Strauss. 1977. Togaviruses, p. 111-166. InD. P.Nayak (ed.), The molecular biology of animal viruses. MarcelDekker, New York.
30. Wengler, G., and G. Wengler. 1975. Studies on the synthesis of viral RNA polymerase-template complexes inBHK-21 cells infected with Semliki Forest virus. Virology 66:322-326.
31.Wertz, G., and M. Levine. 1973. RNA synthesis by vesicular stomatitis virus and asmallplaquemutant: effects ofcycloheximide. J. Virol. 12:253-264.
J. VIROL.
![FIG. 1.preparationpulseAMD.gradientsforsizedRNase,[3H]uridinesynthesis.denaturationsideredwasthesedimentedsecondexcessenth)infectedically(0)](https://thumb-us.123doks.com/thumbv2/123dok_us/1497162.102411/3.508.284.430.67.284/fig-preparationpulseamd-gradientsforsizedrnase-h-uridinesynthesis-denaturationsideredwasthesedimentedsecondexcessenth-infectedically.webp)
![FIG. 3.petriperiod.followedRNA,[365]methioninemediumexposuremedium90timestranslationtwotext.tocontainingamountsindividualwerean(1130,000;The h) the min SFV viral proteins synthesized at various after infection](https://thumb-us.123doks.com/thumbv2/123dok_us/1497162.102411/5.508.69.245.60.372/petriperiod-followedrna-methioninemediumexposuremedium-timestranslationtwotext-tocontainingamountsindividualwerean-proteins-synthesized-infection.webp)