Vol. 54, No. 2 JOURNALOF VIROLOGY, May1985, p.625-629
0022-538X/85/050625-05$02.00/0
CopyrightC 1985, AmericanSociety forMicrobiology
The pX
Region of the Bovine Leukemia Virus Is Transcribed
as a
2.1-Kilobase mRNA
R. Z.
MAMOUN,'
T.ASTIER-GIN,'
R. KETTMANN,2* J. DESCHAMPS,2 N.REBEYROTTE,'
AND B. J. GUILLEMAIN'
InstitutNational de la Sante etde la RechercheMedicale, Unite 117, 33076 Bordeaux Cedex, France,1 and Universite Libre deBruxelles, Departement de Chimie Biologique, 1640Rhode-Ste-Genese, Belgium2
Received 26 October1984/Accepted 31 January 1985
Thebovineleukemia virus mRNAs expressedinculturedbovine cells of variousorigins are a9.0-kilobase genomic RNA, a 5.1-kilobase env RNA, and a newly detected 2.1-kilobase RNA corresponding to the transcription ofpX sequenceslocated in betweentheenvgeneand the3' end of the provirus.
The bovine leukemia virus (BLV), anexogenous
compe-tent retrovirus, is known to be the etiological agent ofthe
enzootic formofthe B-type cell bovine leukemia (7-9, 13,
19, 21).Theprovirus is foundintegratedin the tumorcellsin
which itremains unexpressed (1, 9, 24).
The modelsknowntobe
applicable
for virusleukemoge-nesiscannotexplain cell transformation afterBLVinfection
for the following reasons. (i) Because BLV sequences are
entirelyexogenous, the minkcellfocus-forming mechanism
isexcluded,forsuchvirusarise at leastpartially via
recom-bination between endogenous proviral sequences. (ii) A
virus-coded oncogene is not implicated because proviral
sequences encountered in tumor cells have no homology with normal cellsequences (3);furthermore, noevidence of
onc or onc-bearing viral proteins specific forthe tumorous statecould be detected by immunoprecipitation techniques
(15). (iii)The downstreampromotion (20) ofan onccellular
geneinthetumordoesnot seem tobelikely, forBLVdoes
notintegrateatspecific chromosomal sitesorregions (5, 12),
and no cellularmRNAcontaining viral long terminalrepeat
(LTR) is synthetized (11).
5' ss
u3 R i5
P P B Bg
I II I
6 2 A
could play animportant role in tumor genesis.Moreover,an indication of a major role of this region resides in its constant conservation in tumors in which the BLVprovirusis deleted (10).
Toinvestigate the BLV products, we established a number of cell linesexpressing BLV (i.e., normalcells, normal cells experimentally infected with BLV, and cells free of BLV or expressing BLV originating from a tumor). The way in which these cell lines arereferredtoonly gives anindication
of their origin but does not specify their actual nature, neoplastic or not (14, 15). The analysis of BLV mRNAs in these cells shows that, in addition to the genomic
(9.0-kilobase [kb]) and env (5.1-kb) RNAs, a 2.1-kb mRNA is alsotranscribedfromthe pXregion.
PolyadenylatedRNAs werepreparedfromanoncultured
tumor (T3010), three BLV-infected cells from cultured
tu-mors(LB59Ly,3010gg, and3894gg), non-BLV-infectedcells
from cultured tumor (LB44K), normal cells (3894c and
3010c), in vitro BLV-infected normal cells (FLK-BLV and
BESP-LB59), and non-BLV-infected cells cultured from a
sporadic tumor case (2412Ly). After size separation and
B X
I I
X B E SS 3' Proviral
I
I-
D N A4C 3
4 B
4A 1B
1A
Probes
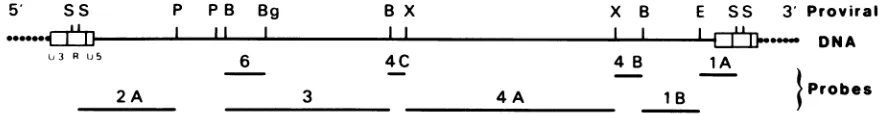
FIG. 1. Localization of the clonedBLVfragment probesversusthemapof BLVprovirus (3) obtained with restrictionenzymesSacl(S),
PstI(P),BamHI(B),BglII(Bg),XbaI(X),andEcoRI(E). Theseprobesweresubcloned with the pBR322 and pAT153plasmids.Restriction of the cloned BLV DNAs were generated by the specific restriction endonuclease digestions. Each fragment was further purified by preparativeagarosegelelectrophoresis.DNAs werelabeled by nick translation inthe presenceof[32P]dCTP(600Ci/mmol), with 5 U ofDNA polymerase asdescribed by Maniatis (17). Probes were isolated fromtriphosphates by centrifugation through Sephadex G-50. The final specific activitywas ca. 108cpm/,ug.
All of these findings also apply to the human T-cell
leukemia virus (HTLV) as described in studies by both
Burny
and Gallo (International Conference on Tumor Virus in RNA, in press); thus, the bovine system is quiteappro-priateas anexperimental model. Interestingly, the analysis
of the sequence at the 3' endofthe envgene of both HTLV and BLVrevealed theexistenceofopenreadingframes,the
putative product of which was tentatively called pX and
* Correspondingauthor.
Northern blotting, the RNAs were hybridized with the
probes as detailed in Fig. 1. These were subclones ofthe
8.3-kb SacI clone containing the BLV
information,
except for 104 basesmissingbecause the LTR of this virus contains twoSacIsites (3).The 1A probe (Fig. 1) represents the 3' end sequence
locatedbetween the EcoRI and Sacl sites andincludingthe entire U3 and the first 45 bases of the R region. It thus
contains the 3' part of the so-called pX region ofthe BLV
genome.Figure2showsarepresentativepattern
indicating
a625
on November 10, 2019 by guest
http://jvi.asm.org/
[image:1.612.86.528.489.546.2]m
-s s c.
-CT It
X e C" u)
c to 0 It co 0 m
- m in in
i:13 J
co 02
-fl
o C:
a
0
Cl)
0
0 C),
9
FIG. 2. Northernblot ofpolyadenylated RNAs hybridized with probes 1Aand1B. RNAwasextracted in TNEbuffer(0.01M Tris[pH 7.9], 0.1 MNaCl, 0.001 M EDTA) containing 0.5% sodium dodecyl sulfate and0.5%Macaloid three times withphenolchloroformsatured in
TNE. Polyadenylated RNA was selected onoligodeoxythymidylate-cellulose and denatured with either glyoxal (18) or methylmercuric hydroxide (2). Size separations (2to5 ,ug of RNA)werecarriedoutby electrophoresis on1.2%agarosegels.The RNAs weretransferred (Northern blotting) on diazobenzyloxymethyl paperand then hybridized with 32P-labeled DNAprobes. Conditions ofhybridization and washing wereperformedasdescribed by Maniatis (17), but thetemperature ofhybridizationwas 37°C. The origins of theRNAs were as
follows: culturednormal cells(3010c and 3894c,normal corneal cells from enzooticlymphosarcomacells[3010and3894]);cultured normal cells in vitro infected withBLV(FLK-BLV, reference fetallambkidneycells(25); BESP-LB59,bovineembryo spleencells infected with the BLV produced by LB59Ly cells); cells from acultured tumorwith no BLV expression (2412Ly, leukemic leukocytesfrom a sporadic lymphosarcomacase [2412]; LB44K,lymphomatous kidney fromanenzootic lymphosarcomacase [LB44]);BLVexpressingcultured cells from enzooticlymphosarcomacasesLB59,3010,and 3894(Ly, leukemic leukocytes;gg,leukemiclymphnodes[3894gghasaverylow BLV expression]);andT3010, noncultured 3010 leukocytes.
'r
.Ai \ r n 1, s,b .46tAtt
.60
5 Kt
,NM fl.iPf
ua fi 2 1 k b
Ai yr
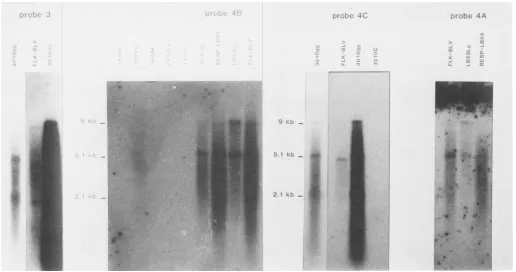
FIG. 3. Northernblot ofthe
polyadenylated
RNAshybridized
withprobes
3, 4A, 413,or4C. See thelegend
toFig.
2.f-n
I... .:. ." a_
-1 a)
I ", .:L
..'T
-., -n
0:
i.
2.it k1.)
- -.j
'Z. -j il,
LI' M Lji
I.,
Alp-A!
tall
I
626
on November 10, 2019 by guest
http://jvi.asm.org/
[image:2.612.73.544.41.320.2] [image:2.612.54.570.440.712.2]NOTES 627
9.0 kbmRNA
5.1 kb mRNA
_mON .AAAA
.- -.
?0.-.0lo" -.
P P
P P B Bg
Proviral DNA
x B E SS
I,l0,
I I IBx
gag / Po/ /"
I U3 R U5
env I pX ?
I
I
I
2.1 kbmRNA
_ one. AAAAA
Bam Hi
XbaI Splice Splice HTLV
I
N"IK
I
a
aw-1I:I 'U:::i:.:q : D
l1
-8 0 52 124 261
Proviral DNA
Eco RI TGA Sacl
I
I
I
IlI
Il
723 967 1242 1513
I?
2.1 Kb m-RNA
pX M.W. > 34 Kd. Putative protein
FIG. 4. (A) Theupperpartof thefigurerepresentstheorigin of the mRNAsversusthe BLVprovirus; the restrictionenzymesitesare
shownasdescribed in thelegendtoFig. 1. (B) In the lowerpart,thepX region of the provirus is detailed. The BamHI site is consideredas
pointzeroof the sequence;HTLV, commonsequencesbetweenBLV, HTLV-1,andHTLV-2; splice, possibleacceptorsplice sites;TGA (-877), termination codon of the envgene;TGA(+967), possible end translation codon for the pX. The dotted lines of the 2.1-kb mRNA
meansthat itsorigin is located in the centralpartof theBLVgenome.
clearhybridization in infected cells ofnotonlythe 9.0- and 5.1-kb mRNAs but also a 2.1-kb mRNA species. Such a resultis evocative ofthe avian lymphoma model, in which
cellular mRNAs often contain viral sequences originating
from the3' endofthe viralgenome. However, thisdoesnot fit the BLV system, for although BLV has no specific
integration site, the size of the small mRNA observed in these experiments remains constant (2.1 kb) regardless of thecellline in which itwasdetected. Furthermore,the sole
contribution of the 45 bases of the Rregion cannotexplain
the clear hybridization observed at this size. We therefore undertook the same investigations, but with a fragment probe (1B) 1 kb distal from the R region (Fig. 1) and
includingthe 5' partof thepX region. Again, all BLV-pro-ducingcellspresentedthesamepattern, indicatingthatsuch a transcript (2.1 kb) does not originate from a LTR down-stream promotionof cellular sequences (Fig. 2).
Hybridization was then performed with 4A, 4B, and 4C Ss
LiR Ub
LTR
TGA
-877 -382
M-12=.-
II I I I I II II I 0 MEE I I %Fw " %w-I
I I if I -t I I I I I
VOL.54,1985
I
on November 10, 2019 by guest
http://jvi.asm.org/
[image:3.612.119.489.71.567.2]probes (Fig. 1). 4A contains all of the open reading frameof the env gene (gp5l and gp35); 4B is located in between the open reading frames of env and pX; 4C is on the 5' side of 4A. With the same transfer (Fig. 3), one can essentially see the putative env mRNA (5.1 kb) in addition to the genomic 9.0-kb mRNA. Noteworthyis the fact that no 2.1-kb mRNA was visualized, indicating that such an mRNA is coded by sequences lying in between the LTRSacl site and the next upstreamBamHI site.
By use of cloned probe (no. 3) containing essentially pol sequences (Fig. 1), a 9.0-kb mRNA and two faint bands corresponding to 5.1 and 2.1 kb were obtained (Fig. 3). Because the 5.1-kb mRNA is dimly recognized, this suggests that only a small portion located at the 3' end of the probe contains sequences of the5'end of the env gene. As a 2.1-kb mRNA is also dimly recognized, this RNA should contain sequences of the 3' end of thepol representative probe. With the gag-specific probe 2A or 5'pol-specific probe 6 (Fig. 1), only the 9.0-kb mRNA is recognized (data not shown), confirming that this RNA is the gag-pol messenger.
All of these hybridizations, as shown in Fig. 2 and 3, were also performed with mRNAs originating from normal, non-BLV-infected cells (3010c and 3894c). As no hybridization could be observed, this confirms the viral nature of the mRNAs described above. The same findings and the same conclusions hold true for cells from a sporadic lymphosar-cornatous cow (2412Ly) or for non-BLV-infected cells de-rived from an enzootic lymphosarcomatous cow (LB44K). The known absence of BLV expression in noncultured
tumor cells was also confirmed with T3010 cells. These
results were extended for the 2.1-kb mRNA.
In all experiments performed with mRNAs of cells in-fected with BLVs of lymphosarcomatous animals, the hy-bridizations corresponded to the three same sizes. It is interesting to notice that the three mRNAs detected in FLK-BLV cells,which are known to contain a BLV isolated from a nontumorous cow, were of a smaller size than that reported above.
The viral mRNAs synthesized byBLV-infected producing cells are mapped in Fig. 4A. Briefly, three species were observed: a 9.0-kb mRNA, a 5.1-kb mRNA, and a new species, i.e., a2.1-kb mRNA. The 9.0-kb mRNA transcribed from the entire BLV provirus may be compared with that of other retroviruses (23) and thus is responsible for gag-pol gene translation (4, 16). The 5.1-kb mRNA is the transcript of the env part of the BLV provirus, extending from a few nucleotides on theleft-hand side of the centralBamHI site to the 3' end of the provirus.
The 2.1-kb mRNA was found to be specific for BLV-in-fected cells. Like the two other mRNAs, the 2.1-kb mRNA was observed only in cellsexpressing the viral proteins and was also notdetectable in noncultured tumor cells. Neither is it specific for the tumoral state, as cultured cells from a sporadic case of lymphosarcoma do not express such a 2.1-kb mRNA. Thehybridization pattern indicates that such an mRNA is representative of two regions because of a splicing event. One region is located in sequences corre-sponding to probe no. 3, and the other is located in the pX sequences. The pX regions of BLV, HTLV-1, and HTLV-2 werepreviously sequenced (6, 21, 22; Burny, in press); the compiled results (Fig. 4B) support previous findings that the splicing occurs at one of the two splicing acceptor sites at positions -8 and +52. Under these conditions, the four common sequences to the three viruses (between positions +124 and +261) ate present in the mRNA. Thus, it is likely that the transduction of the pX region starts before the
splicing
sites with an openreading
frame extending to theendtranslationTGA codon (no. +967). With aframe
iden-ticaltothatproposed forHTLVs(21a),onecan assumethat
1,000 nucleotides is the minimalcoding capacity ofthe pX
gene, thusallowingthe synthesis ofaproteinwitha
molec-ular weight ofat least 34,000.
Although the protein(s) coded by the pX sequences
re-mains as yetunidentified, its role in leukemogenesis can be
envisaged because both BLV and HTLV are competent
viruses able to code for proteins ofthe pX sequences in
addition
toallof their structural proteins. However, thepX
cannot be considered as strictly homologous for a viral oncogene, because ithas nocellularproto-oncogene
coun-terpart. In this case, the pX would have to be modified
(for
example, by mutation), and its role would reside in the
initiation of the lymphoproliferative process. Argumentsfor
suchamodification of the pX include the following. (i)
Only
afew of the BLV-infected animals develop persistent
lym-phocytosis and lymphosarcoma. (ii) As evidenced in our
epidemiological surveys, in herds in which a BLV-induced
lymphosarcoma occurs, the leukemia risk ofthe other
ani-mals drastically increases (this situation mimics the
emer-gence of an "acute BLV" in tumor cells which then is
horizontally transmissible within the herd). (iii) Finally, the
role ofthe pX as the "initiator" ofcell multiplication by a
"hit-and-run" mechanism is supported by the complete
absence ofBLV
expression
in tumor cells.We are indebted to J. M. Miller and M. J. Van der Maaten for
supplyingthe FLK-BLV cell line. We thank J. Quesnel foraccess to alarge variety of tumor-bearing cattle. We also acknowledge the
technicalassistance of C. Bourget and thecareful preparationofthe manuscript by L. Couillaud.
LITERATURECITED
1. Burny, A., C. Bruck, H.Chantrenne, Y. Cleuter, D.Dekegel, J. Ghysdael, R. Kettmann, M. Leclercq, J. Leunen, M. Mam-merickx, and D. Portetelle. 1980. Bovine leukemia virus: mole-cularbiology and epidemiology, p. 231-289. In G. Klein (ed.), Viral oncology. Raven Press, NewYork.
2. Chandler, P. M., D. Rimkus, and N. Davidson. 1979. Gel electrophoretic fractionation of RNAs by partial denaturation with methylmercurichydroxide. Anal. Biochem. 99:200-206. 3. Deschamps, J., R. Kettmann, and A. Burny. 1981. Experiments
with cloned complete tumor-derived bovine leukemia virus information prove thatthe virus is totallyexogenoustoitstarget animals species. J. Virol.40:605-609.
4. Ghysdael, J., R. Kettmann, and A. Burny. 1979. Translation of bovine leukemia virus virionRNAsinheterologous protein-syn-thesizing systems. J. Virol. 29:1087-1098.
5. Gregoire, D., D. Couez, J. Deschamps, S. Heuertz, M.-C. Hors-Cayla, J. Szpirer, C. Szpirer, A. Burny, G. Huez, and R. Kettmann. 1984. Different bovine leukemia virus-induced tu-mors harbor the provirus in different chromosomes. J. Virol. 50:275-279.
6. Haseltine, W. A., J. Sodroski, R.Patarca, D.Briggs,D.Perkins, andF.Wong-Staal.1984.Structuralof 3' terminal region of type II humanTlymphotropic virus: evidence fornewcoding region. Science 225:419-421.
7. Kawakami, T. G., A.L.Moore, G.H.Theilen,and R. J. Munn. 1970. Comparisons ofvirus-likeparticles from leukoticcattle to feline leukosis virus. Bibl. Haemat. 36:471-475.
8. Kenyon, S. J., and C. E. Piper. 1977. Properties of density gradient-fractionated peripheral blood leukocytes from cattle infected with bovine leukemiavirus. Infect. Immun. 16:898-903. 9. Kettmann, R., A. Burny, Y. Cleuter, J. Ghysdael, and M. Manmerickx. 1978. Distribution of bovine leukemia virus pro-viral DNAsequences in tissues of animals with enzootic bovine
on November 10, 2019 by guest
http://jvi.asm.org/
NOTES 629 leukosis. Leuk.Res.2:23-32.
10. Kettmann, R., D. Couez, and A. Burny. 1981. Restriction endonuclease mapping of linear unintegrated proviral DNA of bovine leukemiavirus.J. Virol.38:27-33.
11. Kettmann,R., J. Deschamps, Y.Cleuter, D. Couez, A. Burny, and G. Marbaix. 1982. Leukemogenesis by bovine leukemia virus: proviral DNA integration and lack of RNA expression of viral longterminalrepeatand3'proximatecellular sequences. Proc. Natl. Acad. Sci. U.S.A. 79:2465-2469.
12. Kettmann, R., J. Deschamps, D. Couez, J.-J. Claustriaux, R. Palm,and A. Burny. 1983.Chromosomeintegration domain for bovineleukemia provirus intumors.J. Virol. 47:146-150. 13. Kettmann, R., D. Portetelle, M. Mammerickx, Y. Cleuter, D.
Dekegel, M. Galoux, J. Ghysdael, A. Burny, and H. Chantrenne. 1976.-Bovine leukemia virus: an exogenous RNA oncogenic virus. Proc. Natl. Acad. Sci. U.S.A. 73:1014-1018.
14. Mamoun, R. Z., T. Astier, and B. Guillemain. 1981. Establish-ment and propagation ofabovine leukaemia virus-producing cell lines derived from the leukocytes ofaleukaemic cow. J. Gen.Virol. 54:357-365.
15. Mamoun, R. Z., T. Astier, B. Guillemain, and J. F. Duplan. 1983. Bovinelymphosarcoma: expression of BLV-related pro-teins in culturedcells. J. Gen. Virol. 64:1895-1905.
16. Mamoun, R. Z., T. Astier, B. Guillemain, and J. F. Duplan. 1983. Bovinelymphosarcoma: processing of bovine leukaemia viruscodedproteins. J. Gen.Virol. 64:2791-2795.
17. Maniatis, T., E. F. Fritsch, andJ.Sambrook. 1982.Molecular cloning,alaboratory manual. Cold Spring HarborLaboratory, ColdSpring Harbor,N.Y.
18. McAster,G.K., andG. G. Garmichael. 1977.Analysis ofsingle anddouble stranded nucleic acidsonpolyacrylamide andagarose
gels byusingglyoxal andacridineorange. Proc.Natl. Acad.Sci. U.S.A. 74:4835-4838.
19. Miller, J. M., L. D. Miller, C. Olson, and K. G. Gillette. 1969. Virus-like particles in phytohemagglutinin-stimulated lympho-cytecultures withreferencetobovinelymphosarcoma.J. Natl. CancerInst.43:1297-1305.
20. Neel, B. G., W. S. Hayward, H. L. Robinson,J. Fang, and S. M. Astrin. 1981. Avian leukosis virus-induced tumors have com-mon proviral integration sites and synthesize discrete new RNAs: oncogenesis bypromoterinsertion. Cell 23:323-334. 21. Paul, P.S.,K. A.Pomeroy, A. E.Castro, D. W. Johnson, C. C.
Muscoplat, and D. K. Sorensen. 1977. Detection of bovine leukemia virus in B-lymphocytes by the syncytia induction assay.J. Natl.CancerInst.59:1269-1272.
21a.Rice, N. R., R. M. Stephens, D. Couez, J. Deschamps, R. Kettmann, A. Burny, and R. V. Gilden. 1984. The nucleotide sequence ofthe env gene post-env region of bovine leukemia virus. Virology 138:82-93.
22. Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide se-quenceof theprovirusgenomeintegratedinleukemia cellDNA. Proc. Natl. Acad. Sci. U.S.A.80:3618-3622.
23. Stephenson, J. R. (ed.). 1980. Type C structural and transfor-mation-specific proteins, p. 245-297. In Molecular biology of RNA tumorviruses. Academic Press, London.
24. Stock,N.D., andJ. F. Ferrer. 1972.ReplicatingC-type virus in phytohemagglutinin-treated buffy-coat cultures of bovine origin. J. Natl. CancerInst.48:985-996.
25. Van derMaaten, M.J., andJ.M. Miller. 1976. Replication of bovineleukemia virusin monolayer cell cultures. Bibl.Haemat. 43:360-362.
VOL. 54, 1985