Copyright © 2000, American Society for Microbiology. All Rights Reserved.
Use of Inhibitors To Evaluate Coreceptor Usage by Simian and
Simian/Human Immunodeficiency Viruses and Human
Immunodeficiency Virus Type 2 in Primary Cells
YI-JUN ZHANG,1BERNARD LOU,1RENU B. LAL,2AGEGNEHU GETTIE,1
PRESTON A. MARX,1,3ANDJOHN P. MOORE1*
Aaron Diamond AIDS Research Center, The Rockefeller University, New York, New York 100161; National Center
for Infectious Diseases, DASTLR, Centers for Disease Control and Prevention, Atlanta, Georgia 303332;
and Tulane University Medical Center, New Orleans, Louisiana 704333
Received 21 January 2000/Accepted 2 May 2000
We have used coreceptor-targeted inhibitors to investigate which coreceptors are used by human immuno-deficiency virus type 1 (HIV-1), simian immunoimmuno-deficiency viruses (SIV), and human immunoimmuno-deficiency virus type 2 (HIV-2) to enter peripheral blood mononuclear cells (PBMC). The inhibitors are TAK-779, which is specific for CCR5 and CCR2, aminooxypentane-RANTES, which blocks entry via CCR5 and CCR3, and AMD3100, which targets CXCR4. We found that for all the HIV-1 isolates and all but one of the HIV-2 isolates tested, the only relevant coreceptors were CCR5 and CXCR4. However, one HIV-2 isolate replicated in human PBMC even in the presence of TAK-779 and AMD3100, suggesting that it might use an undefined, alternative coreceptor that is expressed in the cells of some individuals. SIVmac239 and SIVmac251 (from macaques) were also able to use an alternative coreceptor to enter PBMC from some, but not all, human and macaque donors. The replication in human PBMC of SIVrcm(from a red-capped mangabey), a virus which uses CCR2 but not CCR5 for entry, was blocked by TAK-779, suggesting that CCR2 is indeed the paramount coreceptor for this virus in primary cells.
Like human immunodeficiency virus type 1 (HIV-1), simian immunodeficiency viruses (SIV) and human immunodeficiency virus type 2 (HIV-2) use seven-transmembrane receptors as coreceptors during the process of virus-cell fusion (reviewed in references 6, 8, 9, 25, 74, and 87). Usually, this process requires an initial interaction of the viral envelope glycoproteins with CD4 (48, 107, 114). However, rare examples of CD4-indepen-dent HIV-1 isolates have been described (29, 47), and several HIV-2 and SIV strains can interact with coreceptors quite efficiently in the absence of CD4 (14, 30, 33, 35, 65, 89, 97). The first coreceptor described for HIV-1 was CXCR4, which serves to mediate the entry of the so-called T-cell line-tropic or syn-cytium-inducing (SI) viruses (39), now designated X4 isolates (7). The major coreceptor for the macrophage-tropic, non-syncytium-inducing (NSI) viruses, now designated R5 isolates, was found to be CCR5 (2, 20, 22, 27, 28). A plethora of other coreceptors has also been described to function for HIV-1 entry, especially for SI viruses, at least in the context of core-ceptor-transfected cells in vitro (5, 19, 20, 22, 27, 31, 32, 38, 40, 50, 60, 62, 84, 85, 92, 93, 96, 102, 104, 105, 116).
HIV-2 isolates can also use CCR5 and CXCR4 for entry into coreceptor-transfected cells in vitro. In general, coreceptor usage by HIV-2 is broader than that for HIV-1, in that many seven-transmembrane receptors have been reported to support HIV-2 entry when transfected into cell lines (10, 14, 22, 35, 45, 48, 52, 68, 77, 80, 89, 106).
The first coreceptor to be identified as supporting SIV entry was CCR5, which was shown to function with SIVmacisolates
soon after it was found to be an HIV-1 coreceptor (12, 18, 38).
The use of CCR5 by several other SIV strains, including pri-mary isolates from the natural host, has since been docu-mented (14, 22, 30, 31, 33, 48, 53, 54, 64, 65, 93). CXCR4 usage by SIV strains is, however, very rare, although an example of SIV entry via CXCR4 is known, albeit for an isolate obtained from mandrills (SIVmnd) (98). However, the initial reports of
SIVmacentry via CCR5 concluded that additional coreceptors
used by SIV may be substitutes for CXCR4 (12, 18, 38). Since then, several seven-transmembrane receptors have been re-ported to support SIV entry in vitro, often with an efficiency comparable to that of CCR5 (3, 22, 31, 32, 38, 93, 96). Argu-ably, the most efficient among these SIV coreceptors are the ones variously designated BOB/GPR15 and Bonzo/STRL33/ TYMSTR (3, 22, 31, 62).
The question arises, however, as to whether these other coreceptors are as important as CCR5 and (for HIV-1 and HIV-2) CXCR4 for viral replication in vivo. There is mounting evidence that many, if not all, of the other coreceptors have only limited, if any, relevance to viral replication in primary cells and hence in vivo, except perhaps in specialized tissues and cell types (13, 31, 43, 70, 81, 86, 101, 102, 117). Care must always be taken when evaluating viral entry mediated via trans-fected seven-transmembrane receptors (66). We have contin-ued to address this issue here using inhibitors targeted at CCR5 and CXCR4 (117). Our conclusion is that CCR5 is the most important SIVmac coreceptor in primary CD4⫹T cells
but that an alternative coreceptor(s) may indeed be relevant, at least in cells from some macaques. SIVrcm, however, uses
CCR2 and not CCR5. These observations may be useful in studies of SIV-infected nonhuman primates as model systems for the development of HIV-1 vaccines (23, 57, 73). For the same reason, we have also studied the coreceptor usage of selected simian/human immunodeficiency viruses (SHIV).
* Corresponding author. Present address: Weill Medical College of Cornell University, 1300 York Ave., New York, NY 10021. Phone: (212) 746-4462. Fax: (212) 746-8340. E-mail: jpm2003@med.cornell .edu.
6893
on November 9, 2019 by guest
http://jvi.asm.org/
MATERIALS AND METHODS
Coreceptor inhibitors.The bicyclam AMD3100, a small-molecule inhibitor of HIV-1 entry via CXCR4 (26, 37, 56, 100), and TAK-779, a small-molecule inhibitor of HIV-1 entry via CCR5 (4), were both gifts from Annette Bauer, Michael Miller, Susan Vice, Bahige Baroudy, and Stuart McCombie (Schering Plough Research Institute, Bloomfield, N.J.). Aminooxypentane-RANTES (AOP-RANTES), a derivatized CC-chemokine that interacts with CCR5, was provided by Amanda Proudfoot, Serono Pharmaceutical Research Institute, Geneva, Switzerland (34, 63, 103, 113). The human chemokines monocyte che-motactic peptide (MCP) 1 (MCP-1), MCP-3, and stromal cell-derived factor 1␣
(SDF-1␣) were purchased from Peprotech Inc. (Norwood, Mass.).
Viral isolates and preparation of virus stocks.The HIV-1 primary isolates 5160 and 5073, derived from individuals with AIDS, have been described previ-ously (115), as have two other primary isolates, M6-v3 and P6-v3, obtained from an HIV-1-infected mother-child transmission pair (116, 117). All these viruses have the SI phenotype, except for P6-v3. Six HIV-2 primary isolates have also been described elsewhere (41, 80). Three of these (7924A, 77618, and GB122) were isolated from individuals with AIDS, one (7312A) was isolated from an individual with lymphadenopathy, and two (310340 and 310342) were isolated from blood donors whose clinical conditions were unrecorded (80). The origins of HIV-1 SF162, DH123, and NL4-3 have been described elsewhere, as have their coreceptor usage profiles (116, 117). All HIV-1 and HIV-2 isolates were propagated and titrated in phytohemagglutinin-activated human peripheral blood mononuclear cells (PBMC) before use.
The SIV strains SIVmac251, SIVmac239, SIVmac251/1390, SIVmac239/5501,
SIVsm(variant SIVsmpbj), and SIVrcmwere all provided by Preston Marx and
Zhiwei Chen (14, 15). SIVmac251/1390 and SIVmac239/5501 were isolated from
macaques which progressed to AIDS after infection with SIVmac251 and
SIVmac239, respectively (14, 67). SIVrcmwas originally isolated from a
red-capped mangabey by cocultivation with human PBMC (15). All SIV strains were propagated and titrated in rhesus macaque PBMC, except for SIVrcm, for which
human PBMC were used (15).
SHIV strains 89.6, 89.6P, and 89.6PD were obtained from David Montefiori (90, 91). SHIV strain SF33A was obtained from Cecilia Cheng-Mayer (46), and SHIV strain KU-2 was obtained from Opendra Narayan (51). All SHIV stocks were prepared in macaque PBMC, except for a second stock of 89.6PD, which was grown in human PBMC for comparison (89.6PD-hu).
Virus replication in PBMC.Human PBMC were isolated from various healthy blood donors by Ficoll-Hypaque separation and stimulated for 3 days with phytohemagglutinin (5g/ml) and interleukin-2 (IL-2; 100 U/ml) (a gift from Hofmann-La Roche, Inc., Nutley, N.J.). These donors were all homozygous for the CCR5 wild-type allele. PBMC from three individuals known to be homozy-gous for the CCR5⌬32 allele (⌬32-CCR5) were also used. Activated PBMC (2⫻
105/well) were cultured in 96-well plates with 150l of RPMI 1640 medium
containing 10% fetal calf serum and IL-2. Virus inocula (100 or 1,000 50% tissue culture infective doses [TCID50] in 75l) were added to duplicate or triplicate
wells. Three wells lacked cells to provide a control for the viral antigen input. Rhesus macaque PBMC were prepared by similar procedures, except that they were stimulated for 3 days with staphylococcal enterotoxin B (Sigma Chemical Co., St. Louis, Mo.) at 5g/ml in RPMI 1640 growth medium containing IL-2 (46).
CEMx174 cells in RPMI 1640 growth medium were used at concentrations of 4⫻104/well. Culture supernatants were harvested on days 7 and 11
postinfec-tion, and fresh medium was added to replenish the cultures.
Viral antigen detection.Virus production was measured using a Gag antigen capture enzyme-linked immunosorbent assay. A commercial diagnostic kit (Cel-lular Products Inc., Buffalo, N.Y.) was used, with modifications, to quantitate HIV-2 and SIV p27 antigen. Briefly, p27 antigen in a 100-l volume was captured onto wells of a 96-well plate by the adsorbed anti-p27 monoclonal antibody provided with the kit. The captured p27 antigen was then detected using the biotin-labeled anti-SIV Gag polyclonal antibodies provided with the kit. To increase the sensitivity of antigen detection, we used a modified protocol that involved streptavidin-conjugated alkaline phosphatase (DAKO, Carpinteria, Calif.) and a chemiluminescent alkaline phosphatase substrate (ELISA-Light; Tropix Inc., Bedford, Mass.). The plates were read with a microtiter plate luminometer (Dynex Technologies Inc.), and the amount of antigen detected was calculated using a standard antigen curve prepared in each assay. The use of the chemiluminescent detection system increased the sensitivity of HIV-2 or SIV p27 detection by more than 100-fold. HIV-1 p24 antigen was detected as described previously (109, 111), except that the chemiluminescent detection system was used.
Determination of coreceptor usage by viral isolates using GHOST cells ex-pressing CD4 and coreceptors.Coreceptor usage was determined essentially as described previously (109, 116, 117). Human osteosarcoma (GHOST) cells ex-pressing CD4 and one of the following coreceptors were obtained from Dan Littman and Vineet KewalRamani (Skirball Institute, New York University School of Medicine, New York, N.Y.): CCR1, CCR2, CCR3, CCR4, CCR5, CCR8, CXCR4, BOB, Bonzo, GPR1, APJ, V28, and US28. These cells were cultured in complete Dulbecco’s minimal essential medium containing G418 (5
g/ml), hygromycin (1g/ml), and puromycin (1g/ml). GHOST cells
express-ing only CD4 (GHOST-CD4 cells) served as controls; they were cultured in the same medium, except that puromycin was omitted.
GHOST cells (105/ml; 500l per well) were maintained in 24-well plates for
24 h. The medium was then removed, and 200l of fresh medium was added, along with a viral inoculum of 1,000 TCID50. On the next day, residual virus was
removed and the cells were washed once with 1 ml of medium. A 750-l aliquot of fresh complete medium containing the selection antibiotics was then added. At approximately day 5 postinfection, Gag antigen production in 100l of harvested culture supernatant was measured. For a few slowly replicating SIV isolates, it was necessary to replenish the cultures and repeat the antigen assay on day 7 or 10 postinfection. In all cases, the amount of antigen produced in control GHOST-CD4 cells was subtracted from the amount produced in coreceptor-transfected GHOST-CD4 cells. Whether this is a sufficient correction for use by some isolates of the low level of endogenous CXCR4 in GHOST-CD4 cells is discussed in Results. Attempts were made to quantify CXCR4 expression on the various coreceptor-transfected GHOST-CD4 cell lines. All the lines do express CXCR4, but at very low levels that are difficult to quantify accurately by fluo-rescence-activated cell sorting (FACS). Thus, we could not accurately quantitate the extent to which CXCR4 expression varied among the various lines. This situation is consistent with the experience of others (Dan Littman, personal communication).
Effect of coreceptor-targeted inhibitors on viral replication.Human PBMC were used with HIV-1, HIV-2, and SIVrcm, rhesus macaque PBMC were used
with other SIV isolates, and both human and macaque PBMC were used with SHIV. Stimulated PBMC (75l) were cultured in 96-well plates at 2⫻105per
well for human cells and 1⫻105per well for macaque cells. A range of
concentrations of inhibitors (75l) was incubated with the cells, in duplicate or triplicate wells, for 1 h at 37°C before addition of the viral inoculum (100 TCID50
in 75l). The final inhibitor concentrations used, unless otherwise specified, were as follows: AMD3100, 400, 40, and 4 nM; AOP-RANTES, 40, 4, and 0.4 nM; TAK-779, 3.3M, 330 nM, and 33 nM; and MCP-1 and MCP-3, 400, 40, and 4 nM. For each virus tested, five wells without drugs and five wells containing only virus served as positive and negative controls for virus production, respec-tively. Culture supernatants (200l) were harvested for measurement of Gag antigen content (in 100l) by an enzyme-linked immunosorbent assay on days 4, 7, and 10. Inhibitors were added back each time. Only when sufficient antigen had been produced was the effect of the inhibitors on virus production calcu-lated.
To determine the specificity of the inhibitors, GHOST-CD4 cells and a core-ceptor were used. The cells were cultured as described above. Briefly, 24 h after the cells were plated, inhibitors in a total volume of 200l were added to each well of a 24-well plate. AMD3100 was used at 1.2M, AOP-RANTES was used at 120 nM, and TAK-779 was used at 10M. After incubation for 1 h at 37°C, a viral inoculum of 1,000 TCID50was added for overnight incubation. The cells
were then washed, and 750l of fresh medium was added. The production of p24 antigen and the effect of the inhibitors were determined as for the PBMC cultures, except that the supernatants were harvested on days 3, 6, and 10.
RESULTS
Coreceptor usage by HIV-1, HIV-2, SHIV, and SIV in trans-fected cells.We assembled a panel of HIV-1, HIV-2, SHIV, and SIV isolates to study their replication in primary cells. We first determined which coreceptors these viruses could use, at least under artificial conditions, by measuring their replication in human GHOST-CD4 cell lines stably transfected with one of several seven-transmembrane receptors (Table 1).
CCR5 and CXCR4 were clearly the coreceptors most widely and efficiently used by HIV-1, HIV-2, and SHIV isolates. None of the SIV used CXCR4, a feature that distinguishes SIV from HIV-1 and HIV-2 (3, 10, 22, 31, 32, 34, 38, 45, 48, 68, 77, 80, 89, 93, 96, 106), but all the SIV except for SIVrcmused CCR5
(Table 1). SIVrcm was originally isolated from a red-capped
mangabey, a monkey species with a high frequency of a mu-tated, inactive CCR5 gene, the⌬24-CCR5 allele (15). SIVrcm
has a unique pattern of coreceptor usage in that it uses CCR2 and not CCR5 as its major coreceptor (15). We confirmed this fact and found that SIVrcmcan also use Bonzo/STRL33, V28,
and US28 efficiently (Table 1).
Consistent with previous reports, some HIV-2 and SIV iso-lates were able to enter cells expressing several other corecep-tors (3, 10, 22, 31, 32, 34, 38, 45, 48, 68, 77, 80, 89, 93, 96, 106). For instance, some SIV isolates were able to use BOB/GPR15 and Bonzo/STRL33 efficiently—notably, SIVmac239/5501
(Ta-ble 1). HIV-1 and SHIV isolates of the SI phenotype, i.e.,
on November 9, 2019 by guest
http://jvi.asm.org/
viruses that could use CXCR4 efficiently, were usually able to replicate in GHOST-CD4 cells expressing various coreceptors (Table 1). Any differences in coreceptor usage patterns be-tween this and previous reports (80, 115) probably arises from the use of different GHOST-CD4 cell clones and/or isolates with a different passage history.
The broad tropism of SI viruses in coreceptor-transfected cell lines is well known (5, 19, 20, 22, 27, 31, 32, 38, 40, 50, 60, 62, 84, 85, 92, 93, 96, 104, 105, 116). However, the growth of HIV-1, HIV-2, and SHIV isolates in GHOST-CD4 cells
[image:3.612.56.551.92.354.2]trans-fected with other coreceptors was only rarely comparable to the replication of the same viruses in CCR5- or CXCR4-ex-pressing cells. Examples of relatively efficient replication in-clude that of HIV-1 P6-v3 and M6-v3 in Bonzo-transfected cells, HIV-2 7924A in APJ- or US28-transfected cells, and SHIV 89.6PD in V28-transfected cells (Table 1). Whether
TABLE 1. Coreceptor usage by HIV-1, SHIV, HIV-2, and SIV isolates in GHOST-CD4 cells expressing a transfected seven-transmembrane, G-protein-coupled receptor
Viral isolate Inoculum(TCID
50)
Replication in the presence of the following receptora:
CCR1 CCR2 CCR3 CCR4 CCR5 CCR8 CXCR4 BOB Bonzo GPR1 V28 APJ US28
HIV-1 P6-v3 1,000 ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫺ ⫺
HIV-1 P6-v3 1,000 ⫺ ⫺ ⫺ ⫹⫹⫹ ⫹ ⫹⫹⫹ ⫺ ⫹⫹⫹ ⫺ ⫹ ⫹ ⫺
HIV-1 5073 1,000 ⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹ ⫺ ⫺ ⫺ ⫹ ⫹ ⫹
HIV-1 5160 1,000 ⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹ ⫺ ⫺ ⫺ ⫹ ⫹ ⫹
HIV-1 NL4-3 1,000 ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺
SHIV 89.6PD 500 ⫺ ⫺ ⫹⫹⫹ ⫺ ⫹⫹⫹⫹⫹ ⫹⫹⫹ ⫹⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫹⫹⫹⫹ ⫹⫹ ⫹⫹⫹
SHIV 89.6PD-hu 500 ⫺ ⫺ ⫹⫹ ⫺ ⫹⫹⫹⫹⫹ ⫹⫹ ⫹⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫹⫹ ⫹⫹
SHIV KU-2 500 ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫹⫹ ⫹⫹
SHIV SF33A 500 ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫺ ⫹⫹ ⫹ ⫺
HIV-2 310340 1,000 ⫺ ⫺ ⫺ ⫹⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺
HIV-2 310342 1,000 ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺
HIV-2 7312A 1,000 ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫹ ⫹ ⫺ ⫺ ⫺ ⫺
HIV-2 GB122 1,000 ⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫹⫹ ⫹⫹ ⫹⫹
HIV-2 77618 1,000 ⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫺ ⫹⫹ ⫹⫹ ⫹
HIV-2 7924A 1,000 ⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫹⫹ ⫹⫹⫹⫹ ⫹⫹⫹
SIVrcm 100 ⫺ ⫹⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫹⫹⫹⫹ ⫺ ⫹⫹⫹⫹⫹
SIVrcm 500 ⫺ ⫹⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹⫹⫹ ⫺ ⫹⫹⫹⫹⫹ ⫺ ⫹⫹⫹⫹⫹
SIVmac239 500 ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹⫹ ⫺ ⫺ ⫹⫹ ⫹⫹⫹ ⫹ ⫺ ⫹⫹ ⫺
SIVmac251 500 ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫹ ⫹ ⫺ ⫺ ⫺ ⫺
SIVmac239/5501 500 ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹⫹ ⫺ ⫺ ⫹⫹⫹⫹ ⫹⫹⫹⫹ ⫹⫹⫹ ⫺ ⫹⫹⫹ ⫺
SIVmac251/1390 500 ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹ ⫺ ⫺ ⫹ ⫹ ⫺ ⫺ ⫺ ⫺
SIVsmpbj 500 ⫺ ⫺ ⫺ ⫺ ⫹⫹⫹⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺
aAbility to replicate in GHOST-CD4 cells expressing the seven-transmembrane, G-protein-coupled receptor indicated. The extent of replication (Gag antigen
production) is recorded as follows:⫺,⬍0.1 ng/ml;⫹, 0.1 to 1 ng/ml;⫹⫹, 1 to 5 ng/ml;⫹⫹⫹, 5 to 20 ng/ml;⫹⫹⫹⫹, 20 to 100 ng/ml; and⫹⫹⫹⫹⫹,⬎100 ng/ml. For each CXCR4-utilizing virus, the amount of p24 antigen produced in the parental GHOST-CD4 cells was subtracted from the amount produced in the coreceptor-expressing GHOST-CD4 cells. Whether this is always a sufficient correction for the use of the CXCR4 that is endogenous to GHOST-CD4 cells is discussed in the text.
TABLE 2. Replication of HIV-1 and SHIV isolates in PBMC from wild-type and⌬32-CCR5 donors and in CEMx174 cells
HIV-1 or SHIV isolate
Virus (ng of p24 or p27 antigen/ml) produced in the following cells on the indicated day postinfectiona:
Wild-type
PBMC ⌬32-CCR5PBMC CEMx174
7 11 7 11 7 11
SF162 15.2 13.4 0 0 0 0
P6-v3 10.2 14.4 0 0 0 0
NL4-3 2.6 10.1 5.7 14.3 8 17.3
DH123 14.1 11.6 13.2 6.2 7.3 15
M6-v3 12.6 13.9 6.5 15.5 4.1 18.2
5073 7 13.2 5.6 13.8 2.9 17.3
89.6PD 85.2 134.2 53.3 154.3 41.5 181.9
KU-2 37 102 15.7 70 94 134
SF33A 73 131 72 107 51.4 155
aThe inoculum was 1,000 TCID
50, but an identical pattern of data was found
at 100 TCID50(data not shown). PBMC were from donors A (wild type) and 1
[image:3.612.310.549.529.719.2](⌬32-CCR5).
TABLE 3. Replication of HIV-2 isolates in PBMC from wild-type and⌬32-CCR5 donors and in CEMx174 cells
HIV-2
isolate TCID50
Virus (ng of p27 antigen/ml) produced in the following cells on the indicated day postinfectiona:
Wild-type
PBMC ⌬32-CCR5PBMC CEMx174 7 11 7 11 7 11
310340 100 1,250 1,509 0 0 0 0
1,000 1,329 1,717 0 0 0 0
7312A 100 2.3 10.3 0 0.7 0.4 0.8
1,000 15 69.8 0.2 5.5 1.5 8.9
GB122 100 58 87 85 130 121 114
1,000 82.9 105 108 133 116 135
77618 100 37.7 58.2 42.6 159 167 186
1,000 125 182 113 195 168 237
7924A 100 88 141 78 141 29.9 89
1,000 84 68 91 99 59 161
aPBMC were from donors B (wild type) and 1 (⌬32-CCR5).
on November 9, 2019 by guest
http://jvi.asm.org/
[image:3.612.54.294.556.702.2]virus entry into the various GHOST-CD4 cell lines actually occurs via the transfected coreceptor is discussed below.
Replication of HIV-1, HIV-2, SHIV, and SIV isolates in PBMC from donors expressing or not expressing CCR5 and in CEMx174 cells.The above experiments showed that many of the test isolates can apparently use multiple coreceptors to en-ter transfected human cell lines. To gain insights into the impor-tance of CCR5 for viral replication in primary cells, we compared the abilities of the isolates to replicate in human PBMC from either donors who had wild-type CCR5 alleles or donors who were homozygous for the⌬32-CCR5 mutation and so did not express functional CCR5 proteins (21, 61, 95). We also used the CEMx174 human B/T-hybrid line because these cells can support high-level SIV replication. CEMx174 cells are CXCR4⫹
but CCR5⫺(18, 58, 110) and strongly express the SIV
mac251
and SIVmac239 coreceptor BOB/GPR15 (22, 31, 86).
Among the six HIV-1 isolates tested, the R5, NSI viruses SF162 and P6-v3 were unable to replicate in the ⌬32-CCR5 PBMC from donor 1 (Table 2). Similar results were obtained with PBMC from two other ⌬32-CCR5 donors (data not shown; see also Table 5). These observations are consistent with the known dependence of these viruses on CCR5, so they
validate the use of the⌬32-CCR5 cells for subsequent studies of HIV-2 and SIV replication. HIV-1 SF162 and P6-v3 also failed to replicate in CEMx174 cells (Table 2). In contrast, the X4 HIV-1 clone NL4-3 and the multitropic HIV-1 isolates DH123, M6-v3, and 5073 all replicated in both wild-type and
⌬32-CCR5 PBMC (donor 1) as well as in CEMx174 cells. This finding was also true of the three SHIV tested, 89.6PD, KU-2, and SF33A (Table 2). Hence, all seven of these HIV-1 and SHIV isolates can use a coreceptor other than CCR5 to enter PBMC and CEMx174 cells, consistent with their replication patterns in the various GHOST-CD4 cell lines (Table 1).
One of the five HIV-2 isolates tested, 310340, failed to replicate in⌬32-CCR5 PBMC from donor 1 and in CEMx174 cells (Table 3). This virus was also unable to use any coreceptor other than CCR5 to enter GHOST-CD4 cells (Table 1). An-other HIV-2 isolate, 7312A, grew very poorly, but detectably, in⌬32-CCR5 PBMC and CEMx174 cells; the extent of 7312A production in⌬32-CCR5 PBMC was 5 to 10% that in wild-type PBMC (Table 3). Of note is that HIV-2 7312A could use BOB/GPR15 and Bonzo/STRL33 inefficiently; the amount of p24 produced from GHOST-CD4 cells expressing BOB or Bonzo was approximately 5% that derived from GHOST-CD4 cells expressing CCR5 (Table 1; also data not shown). The remaining three HIV-2 isolates, GB122, 77618, and 7924A, all replicated to comparable extents in the wild-type and ⌬ 32-CCR5 PBMC and replicated efficiently in CEMx174 cells (Ta-ble 3). These results are consistent with the ability of these three isolates to use CXCR4 and other coreceptors (Table 1). The replication of SIVmac239 and SIVmac251 in human
PBMC from an individual homozygous for the⌬32-CCR5 al-lele has been taken as strong evidence that these viruses can use a coreceptor other than CCR5 to enter primary, CD4⫹
cells (18). We sought to confirm this. In the first experiment, the extent of SIVmac239, SIVmac251, SIVmac239/5501, and
SIVmac251/1390 replication in⌬32-CCR5 PBMC from donor 1
was never more than 5% and usually was less than 1% the replication of the same viruses in wild-type PBMC (Table 4). A second experiment also included⌬32-CCR5 PBMC from two more donors, 2 and 3. There was, again, little or no production of SIVmac251 and SIVmac239 in⌬32-CCR5 PBMC from donor
1 (Table 5). However, both isolates replicated well in PBMC from⌬32-CCR5 donors 2 and 3, although antigen production from SIVmac251 in cells from donor 2 was lower than that from
[image:4.612.52.295.91.275.2]typical CCR5 wild-type donors (Table 5). Thus, PBMC from some, but not all, human donors must express a coreceptor
TABLE 4. Replication of SIV isolates in PBMC from wild-type and
⌬32-CCR5 donors and in CEMx174 cells
SIV
isolate TCID50
Virus (ng of p24 or p27 antigen/ml) produced in the following cells on the indicated day
postinfectiona:
Wild-type
PBMC ⌬32-CCR5PBMC CEMx174 7 11 7 11 7 11
SIVmac239 100 69.9 414.8 0.4 4.1 146 444
500 373 289 5.2 1.9 209 555
1,000 303 381 8 5.6 755 470
SIVmac239/5501 100 394 366 4.5 7 923 455
SIVmac251 100 121 606 6.8 6.8 47 402
SIVmac251/1390 100 22 299 0.9 0.5 20.9 442
SIVrcm 100 2,536 2,413 266 678 12 4
1,000 7,550 2,975 5,016 2,707 79 46
aPMBC were from donors C (wild type) and 1 (⌬32-CCR5).
TABLE 5. Replication of SIV and HIV-1 isolates in PBMC from wild-type and⌬32-CCR5 donors and in CEMx174 cells in two different experiments
SIV or HIV-1
isolate TCID50
Virus (ng of p24 or p27 antigen/ml) produced in the following cells on the indicated day postinfectiona:
Wild-type
PBMC (D) ⌬PBMC (1)32-CCR5 ⌬PBMC (2)32-CCR5 PBMC (E)Wild-type ⌬PBMC (3)32-CCR5
7 11 7 11 7 11 7 11 7 11
SIVmac239 100 10.9 87.7 0.2 0.5 3 44.2 34 383 8.7 201
500 27.4 112 0.5 0.8 7.7 53.4 134 460 112 298
1000 37.6 105 1.6 3.4 7.6 52 ND ND ND ND
SIVmac251 100 4.4 146 0.5 1.4 0.6 9.9 85 481 46 309
SIVrcm 100 154 707 65 710 55 637 ND ND 294 358
SF162 100 18.1 16.8 0 0 0 0 0 0 0 0
aTwo experiments are recorded: one that compared donors D, 1, and 2 and the other that compared donors E and 3 (donor designations in parentheses after cell
types). ND, not done.
on November 9, 2019 by guest
http://jvi.asm.org/
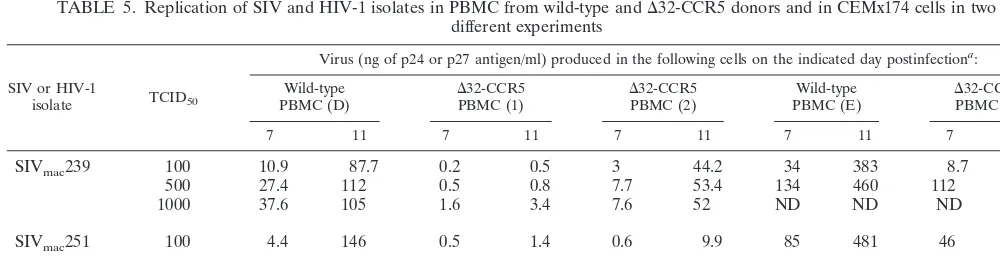
[image:4.612.52.552.575.711.2]FIG. 1. Testing of the specificity of coreceptor-targeted inhibitors. The replication of the test viruses in GHOST-CD4 cells expressing the coreceptor indicated in the presence and absence of AMD3100 (1.2M), AOP-RANTES (AOP-R) (120 nM), TAK-779 (10M), or SDF-1␣(500 nM) was evaluated. The extent to which replication was inhibited by each agent was recorded.
on November 9, 2019 by guest
http://jvi.asm.org/
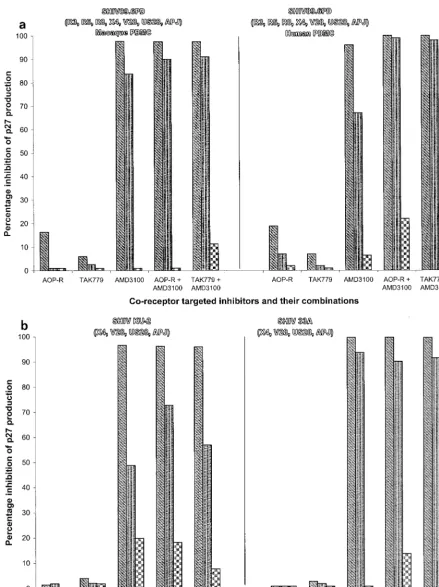
FIG. 2. Effects of coreceptor-targeted inhibitors on HIV-1 replication in human PBMC. The replication of the HIV-1 isolates P6-v3 and M6-v3 (a) and 5073 and 5160 (b) in human PBMC in the presence and absence of AOP-RANTES (AOP-R) (40 nM [left bar], 4 nM [middle bar], and 0.4 nM [right bar]), TAK-779 (3.3M, 330 nM, and 33 nM), or AMD3100 (400 nM, 40 nM, and 4 nM) or with combinations of AMD3100 and either AOP-RANTES or TAK-779 was evaluated. When combinations were used, the concentration of each agent was the same as when the agents were used alone. The extent to which replication was inhibited by each agent or combination was recorded. The coreceptors that can be used by each isolate in GHOST-CD4 cells are indicated below the isolate designation in parentheses.
on November 9, 2019 by guest
http://jvi.asm.org/
other than CCR5 that can be used with reasonable efficiency by members of the SIVmacgroup of viruses.
SIVrcm replicated efficiently in wild-type and ⌬32-CCR5
PBMC (Tables 4 and 5), consistent with its lack of dependence on CCR5 for entry into PBMC (15). SIVmac239 and SIVmac251
also replicated efficiently in CEMx174 cells, as found previ-ously (18), but SIVrcmreplication was inefficient in these cells
(Table 4). Thus, neither the major coreceptor for SIVrcm,
CCR2, nor the minor ones Bonzo, US28, and V28 are ex-pressed in CEMx174 cells.
Evaluation of the specificity of coreceptor-targeted inhibi-tors.Coreceptor-targeted inhibitors are useful for evaluating which coreceptors are relevant for viral entry into PBMC. One suitable inhibitor of entry via CXCR4 is the bicyclam AMD3100 (26, 37, 56, 100). Inhibitors of entry via CCR5 are the TAK-779 molecule (4) or the CC-chemokine derivative AOP-RANTES (62, 103, 113). The specificity of these agents is an important issue. Previous studies have found that AMD3100 is specific for CXCR4 (26, 37, 56, 100) and that TAK-779 can interact with both CCR5 and CCR2 (4). Although RANTES fully activates all of its receptors, AOP-RANTES is able to do this only for CCR5; it has half the activity of RANTES for CCR3 and is very inefficient at acti-vating CCR1 (79, 88). AOP-RANTES is therefore a moderate inhibitor of CCR3-mediated HIV-1 infection, compared to its effect on entry mediated by CCR5 (34).
To confirm these specificities, we determined whether AMD3100, TAK-779, and AOP-RANTES could inhibit viral entry into GHOST-CD4 cells transfected with other corecep-tors by using viruses that were broadly tropic in these cells. For each test virus, AMD3100 was used at 1.2M, AOP-RANTES was used at 120 nM, and TAK-779 was used at 10M (Fig. 1). SHIV 89.6PD replication in GHOST-CD4 cells expressing CCR5 was sensitive to both TAK-779 and AOP-RANTES but not to AMD3100, as expected (Fig. 1a). We also found that AOP-RANTES, but not TAK-779, inhibited SHIV 89.6PD entry into GHOST-CD4 cells expressing CCR3, consistent with an interaction between AOP-RANTES and CCR3, a known RANTES receptor (data not shown). However, neither TAK-779 nor AOP-RANTES had any significant effect on SHIV 89.6PD replication in GHOST-CD4 cells expressing CXCR4, CCR8, V28, US28, or APJ (Fig. 1a; also data not shown). Both TAK-779 and AOP-RANTES inhibited
SIV-mac239 entry into GHOST-CD4 cells expressing CCR5, but the
entry of this virus into GHOST-CD4 cells expressing either BOB, Bonzo, GPR1, or APJ was unaffected by TAK-779 or AOP-RANTES (Fig. 1a). The entry of SIVrcminto
GHOST-CD4 cells expressing CCR2 was completely inhibited by TAK-779, whereas AOP-RANTES had only a marginal effect on entry via CCR2 (Fig. 1a). SIVrcmreplication in GHOST-CD4
cells expressing Bonzo, V28, or US28 was, however, insensitive to TAK-779 or AOP-RANTES (Fig. 1a), as was HIV-2 7924A replication in cells expressing V28, APJ, or US28 (data not shown). Neither TAK-779 nor AOP-RANTES inhibited the replication of HIV-1 P6-v3 in GHOST-CD4 cells expressing Bonzo (data not shown).
Taken together, these data suggest that AOP-RANTES can block viral entry via CCR5 and CCR3 and that TAK-779 in-hibits entry via CCR5 and CCR2. The latter result is consistent with the report that TAK-779 binds to both CCR2 and CCR5 but not to CCR1, CCR3, or CCR4 (4). TAK-779 and AOP-RANTES have no effect on viral replication in GHOST-CD4 cells expressing any one of the eight coreceptors that we were able to evaluate: CXCR4, CCR8, V28, US28, APJ, BOB, Bonzo, or GPR1.
A less clear-cut pattern of inhibition was observed with
AMD3100. As expected, the efficient replication of SHIV 89.6PD in GHOST-CD4 cells expressing CXCR4 was blocked by AMD3100 (Fig. 1a). However, AMD3100 also prevented the inefficient replication of SHIV 89.6PD in GHOST-CD4 cells expressing either CCR3, V28, APJ, US28, or CCR8 and significantly inhibited the limited replication of HIV-2 7924A in GHOST-CD4 cells expressing V28, APJ, or US28 (Fig. 1b; also data not shown). However, AMD3100 had no detectable effect on SIVmac239 entry into GHOST-CD4 cells expressing
BOB, Bonzo, GPR1, or APJ or on SIVrcmentry into
GHOST-CD4 cells expressing CCR2, Bonzo, V28, or US28 (Fig. 1a). The replication of HIV-1 P6-v3 in GHOST-CD4 cells express-ing Bonzo was also unaffected by AMD3100 (data not shown). Thus, entry via V28, US28, and APJ in GHOST-CD4 cell lines can apparently be either sensitive or insensitive to AMD3100, depending upon the test virus.
There are two possible explanations for the unusual pattern of inhibition shown by AMD3100. One is that AMD3100 is broadly reactive with multiple coreceptors but that certain viruses, particularly SIV, can still interact with some of these coreceptors even in the presence of AMD3100. The other is that the apparent cross-reactivity of AMD3100 is an artifact of the presence of low levels of endogenous CXCR4 in corecep-tor-transfected GHOST-CD4 cells (109, 110). To address this possibility, we tested the sensitivity of SHIV 89.6PD and HIV-2 79HIV-24A replication in several GHOST-CD4 cell lines to SDF-1␣. In all cases, whenever AMD3100 inhibited the replication of the test viruses, so did SDF-1␣(Fig. 1b; also data not shown). Since SDF-1␣ is specific for CXCR4 (6, 8, 9, 71, 84), these findings strongly suggest that the entry of SHIV 89.6PD and HIV-2 7924A into several coreceptor-transfected GHOST-CD4 cell lines occurs via endogenous CXCR4. This coreceptor may well be expressed to different levels in different individual GHOST-CD4 cell lines, although we were unable to accurately quantitate this expression by FACS.
The inhibitory effect of AMD3100 in coreceptor-transfected GHOST-CD4 cell lines is, therefore, most probably explained by its antagonism of viral entry via endogenous CXCR4. The coreceptor usage information presented in Table 1 should be interpreted with this caveat in mind. Overall, we can find no evi-dence that AMD3100 is anything other than specific for CXCR4. Effect of coreceptor-targeted inhibitors on HIV-1, SHIV, and HIV-2 replication in PBMC.The replication of each test virus in mitogen-stimulated PBMC in the presence and absence of AMD3100, TAK-779, or AOP-RANTES was evaluated. Com-binations of AMD3100 with TAK-779 and AMD3100 with AOP-RANTES were also tested. Each inhibitor, alone and in combination, was used at three different concentrations: 400, 40, and 4 nM for AMD3100; 3.3M, 330 nM, and 33 nM for TAK-779; and 40, 4, and 0.4 nM for AOP-RANTES. Prelim-inary experiments had indicated that the effects of the inhibi-tors usually titrated out over these ranges. Human PBMC from CCR5 wild-type donors were used in experiments with HIV-1 and HIV-2 isolates and SIVrcm; rhesus macaque PBMC were
used with other SIV; and both human and macaque PBMC were used with SHIV.
Four HIV-1 primary isolates that could use multiple core-ceptors, as determined by the GHOST-CD4 cell assays (Table 1), were evaluated with human PBMC (Fig. 2). P6-v3, a virus able to use CCR5 and Bonzo, was completely inhibited by both TAK-779 and AOP-RANTES but not by AMD3100 (Fig. 2a). The more broadly tropic virus M6-v3 was partially sensitive to each of the three inhibitors, but its replication was fully blocked by combinations of either TAK-779 or AOP-RANTES with AMD3100 (Fig. 2a). Isolates 5073 and 5060 were able to replicate in several different coreceptor-expressing
on November 9, 2019 by guest
http://jvi.asm.org/
FIG. 3. Effects of coreceptor-targeted inhibitors on SHIV replication in PBMC. The experimental design was like that described in the legend to Fig. 2. The SHIV isolates evaluated were 89.6PD in macaque and human PBMC (a) and KU-2 and SF33A in human PBMC (b).
on November 9, 2019 by guest
http://jvi.asm.org/
CD4 cell lines, including GHOST-CD4 cells expressing CXCR4, but their replication was completely inhibited in PBMC by AMD3100 (Fig. 2b). Thus, none of the tested HIV-1 isolates appeared to enter PBMC from the donors included in these studies via a coreceptor other than CCR5 or CXCR4.
Results similar to those obtained with the broadly tropic HIV-1 isolates were found when SHIV were evaluated (Fig. 3). Thus, SHIV 89.6PD replication in either macaque or human PBMC was fully inhibited by AMD3100, while TAK-779 and AOP-RANTES had no effect (Fig. 3a). The same was true of SHIV KU-2 and SHIV SF33A in human PBMC (Fig. 3b) and also of SHIV 89.6 and SHIV 89.6P (data not shown). The paramount, and most probably exclusive, coreceptor for all of these SHIV in PBMC therefore appears to be CXCR4. This finding was unexpected for SHIV 89.6, 89.6P, and 89.6PD, con-sidering that these viruses efficiently use CCR5 in transfected GHOST-CD4 cells (Table 1 and Fig. 1a; also data not shown).
Among the HIV-2 isolates tested, 310342 and 7312A were both completely inhibited by TAK-779 and AOP-RANTES but were insensitive to AMD3100 (Fig. 4a). Although HIV-2 7312A can use BOB and Bonzo, to a limited extent, in GHOST-CD4 cells (Table 1), this property does not allow the virus to evade CCR5-directed inhibitors in PBMC (Fig. 4a). HIV-2 77618 and GB122 were almost completely (⬎95%) blocked by AMD3100, whereas TAK-779 and AOP-RANTES had no effect on these viruses (Fig. 4b; also data not shown). All of these HIV-2 isolates probably use only CCR5 or CXCR4 to enter PBMC. An exception was, however, noted with HIV-2 7924A. This virus was partially sensitive to AMD3100, but the extent of inhibition did not exceed 30% even at the highest AMD3100 concentration, 400 nM (Fig. 4b). HIV-2 7924A was completely insensitive to TAK-779 or AOP-RANTES, and combining these agents with AMD3100 did not increase the extent of inhibition caused by AMD3100 alone (Fig. 4b).
HIV-2 isolate 7924A has an unusual pattern of sensitivity to coreceptor-targeted inhibitors. The insensitivity of HIV-2 7924A to AMD3100 is unusual, since this virus can use CXCR4, and perhaps only CXCR4, to enter GHOST-CD4 cells (Table 1 and Fig. 1b). Usually, 50% inhibitory concentra-tions (IC50s) of AMD3100 against viruses that use CXCR4 in
PBMC are 4 to 40 nM (Fig. 2b, 3a and b, and 4b; also data not shown). To evaluate whether the insensitivity of HIV-2 7924A to AMD3100 in PBMC was donor dependent, we tested much higher AMD3100 concentrations in cells from four CCR5 wild-type donors (Fig. 5a). Donor-to-donor variation in the potency of AMD3100 was significant, with IC50s ranging from 2.1M
(donor 1) to 34M (donor 2). However, if sufficient AMD3100 (40M) was used, inhibition of HIV-2 7924A was complete in cells from three of the four donors. Whether at a concentration as high as 40M AMD3100 remains specific for CXCR4 is not known, although no overt toxicity was observed.
We also tested AMD3100 (4M) against HIV-2 7924A in PBMC from a⌬32-CCR5 homozygous donor (donor 1). For the first 4 days of culturing, AMD3100 at 4M completely suppressed HIV-2 7924A replication; however, by day 7, the virus had broken through, and the extent of inhibition was negligible thereafter. In contrast, HIV-1 5160 was completely inhibited by 4M AMD3100 throughout the duration of cul-turing (data not shown).
To gain more insight into whether HIV-2 7924A could use CXCR4 for entry into PBMC, we determined its sensitivity to SDF-1␣in cells from the same four CCR5 wild-type donors as those used in the AMD3100 experiment. Even at the highest concentration tested (400 nM), SDF-1␣did not inhibit HIV-2 7924A replication in PBMC from any of the four donors, whereas HIV-1 NL4-3 replication was efficiently blocked (Fig.
5b). Taken together with the insensitivity of HIV-2 7924A to TAK-779 and AOP-RANTES (Fig. 4b), the limited or nonex-istent effect of AMD3100 and SDF-1␣on HIV-2 7924A rep-lication suggests that this virus uses an undefined coreceptor other than CXCR4 to enter PBMC. An alternative explanation is that HIV-2 7924A uses CXCR4 in a highly unusual, inhibi-tor-insensitive manner. If this is so, how this virus uses CXCR4 must be cell type dependent, since we determined that the IC50
of AMD3100 for this virus in GHOST-CD4 cells was 0.47M. This value contrasts markedly with the IC50s of 2.1 to 34M
for the same virus in PBMC.
Effect of coreceptor inhibitors on SIV replication in ma-caque PBMC. To evaluate the inhibitor sensitivities of SIV isolates, we used macaque PBMC. In cells from the first donor macaque tested, SIVmac251, SIVmac239, SIVmac251/1390, and
SIVmac239/5501 were all inhibited by both TAK-779 and
AOP-RANTES to an extent that was complete, or virtually so (⬎95%), whereas AMD3100 had no effect (Fig. 6a and b; also data not shown). Thus, these SIV isolates all use CCR5, and only CCR5, to enter PBMC from this macaque donor. However, there are issues of donor cell dependency to consider (see below).
Because SIVrcmuses CCR2 but neither CCR5 nor CXCR4
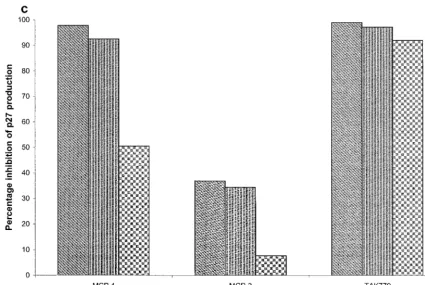
for entry (Table 1), we tested chemokine ligands of CCR2 for their abilities to inhibit SIVrcmreplication in human PBMC. Of
these, MCP-1 almost completely inhibited SIVrcmreplication,
whereas MCP-3 had only a limited effect (Fig. 6c). TAK-779 was also an effective inhibitor of SIVrcmreplication in human
PBMC (Fig. 6c), just as it was in GHOST-CD4 cells expressing CCR2 (Fig. 1a). However, AOP-RANTES had no effect on SIVrcm replication in human PBMC (data not shown). This
virus appears to make truly exclusive use of CCR2 as a core-ceptor in primary human PBMC.
The effect of TAK-779 on SIVmac239 replication in macaque PBMC is donor dependent.We showed above that there is a donor dependency in the ability of SIVmac239 and SIVmac251
to replicate in human PBMC from⌬32-CCR5 homozygous individuals (Table 4). There is also a donor dependency in the potency with which CCR5-targeted inhibitors inhibit
SIV-mac239 replication in macaque PBMC. Thus, the extent to
which TAK-779, at 3.3 M, inhibited SIVmac239 replication
varied from ⬎99% to ⬍50% in PBMC from four different macaques (Fig. 7a). The IC50s of TAK-779 ranged from 240
nM (macaque 3) to 12.6M (macaque 1), a 60-fold variation. However, at the very high concentration of 33M, TAK-779 completely inhibited SIVmac239 replication in all four donors
(Fig. 7a). Similar results were obtained with SIVmac251 in the
two donors tested; the IC50s were 0.18M (donor 3) and 20 M (donor 1) (Fig. 7a).
There was less variation in the potency of TAK-779 against HIV-1 replication in human PBMC. For instance, HIV-1 P6-v3 was inhibited by TAK-779 in PBMC from four donors at IC50s
ranging from 15 nM to 24 nM (Fig. 7b). This result suggests that major variations in inhibition potency are not an inherent feature of TAK-779.
When the inhibitor sensitivities of SIVmac239 and SIVmac251
were evaluated with CEMx174 cells, both viruses were insen-sitive (⬍5% inhibition) to AMD3100 (400 nM), TAK-779 (3.3
M), or AOP-RANTES (40 nM), alone or in combination (data not shown). In contrast, HIV-1 NL4-3 replication in these cells was completely blocked by AMD3100 but not by TAK-779 or AOP-RANTES (data not shown). Thus, what-ever coreceptor(s) SIVmac239 and SIVmac251 use to enter
CEMx174 cells, it is not CCR2, CCR3, CCR5, or CXCR4. Whether this is the same coreceptor that these viruses can use to enter human or macaque PBMC from some donors is not yet known.
on November 9, 2019 by guest
http://jvi.asm.org/
FIG. 4. Effects of coreceptor-targeted inhibitors on HIV-2 replication in human PBMC. The experimental design was like that described in the legend to Fig. 2. The HIV-2 isolates evaluated were 310342 and 7312A (a) and 77618 and 7924A (b).
6902
on November 9, 2019 by guest
FIG. 5. Effects of coreceptor-targeted inhibitors on HIV-2 isolate 7924A in PBMC from different donors. The replication of HIV-2 7924A in PBMC from four different human donors in the presence of AMD3100 at 40M, 4M, 400 nM, and 40 nM (a) and SDF-1␣at 400 nM, 40 nM, 4 nM, and 0.4 nM (b) was evaluated. HIV-1 NL4-3 was also tested with SDF-1␣. In each case, replication was measured after 7 and 10 days. IC50s of AMD3100 were calculated and are shown in panel a. The data shown were obtained on day 10, but values from day 7 were similar.
on November 9, 2019 by guest
http://jvi.asm.org/
FIG. 6. Effects of coreceptor-targeted inhibitors on SIV replication in PBMC. The experimental design was like that described in the legend to Fig. 2. The SIV isolates evaluated were SIVmac251 and SIVmac239 in macaque PBMC (a), SIVmac251/1390 and SIVmac239/5501 in macaque PBMC (b), and SIVrcmin human PBMC (c). MCP-1 and MCP-3 were used at 400, 40, and 4 nM (left to right); TAK-779 was used at 3.3M, 330 nM, and 33 nM.
on November 9, 2019 by guest
http://jvi.asm.org/
DISCUSSION
Primate lentiviruses can use about 12 different seven-trans-membrane receptors as coreceptors in transfected cell lines. However, questions have been raised as to whether corecep-tors other than CCR5 and CXCR4 are relevant for viral entry into primary cells and, hence, for viral replication in vivo (31, 43, 70, 86, 101, 116, 117). This issue affects the development of antiviral drugs aimed at coreceptors. Must multiple corecep-tors be targeted, or just CCR5 and CXCR4 (117)? Does the ability of SIV and SHIV to use multiple coreceptors in vitro influence the interpretation of vaccine experiments with pri-mates (73)?
We addressed these issues by using coreceptor-targeted in-hibitors to block viral replication in primary PBMC, focusing here on HIV-2 and SIV isolates. As inhibitors, we used AMD3100 for CXCR4 and TAK-779 and AOP-RANTES for CCR5. These agents are not completely specific: TAK-779 and AOP-RANTES also inhibit viral entry via CCR2 and via CCR3, respectively. However, we could find no evidence that AMD3100 is anything other than specific for CXCR4, as found previously with other assay systems and test viruses (26, 37, 56, 100).
The low-level entry of viruses such as SHIV 89.6PD and HIV-2 7924A into GHOST-CD4 cells expressing V28, US28, APJ, and others actually occurs via endogenous CXCR4 and not via the transfected coreceptor, since it is inhibited by both SDF-1␣ and AMD3100. Of note is that SHIV 89.6PD and HIV-2 7924A use CXCR4 very efficiently, so they may be able to enter coreceptor-transfected GHOST-CD4 cells that ex-press very low levels of CXCR4; the levels of exex-pression of this coreceptor may also vary slightly among different GHOST-CD4 clones, making some transfected cell lines particularly susceptible to viruses that use CXCR4, although we could not
accurately quantitate such variation by FACS. Whatever the explanation, ambiguities can arise when the coreceptor usage of CXCR4-tropic viruses is determined with transfected GHOST-CD4 cell lines. Many, if not all, of the “positives” for use of coreceptors other than CCR5 and CXCR4 by CXCR4-tropic viruses in GHOST-CD4 cell lines (Table 1) may simply reflect entry via endogenous CXCR4 and not the transfected coreceptor. This caveat may also apply to other studies that have used these cell lines. A similar conclusion was recently reached by others (59).
We previously concluded from an inhibitor-based study that coreceptors other than CCR5 and CXCR4 made, at most, only a limited contribution to HIV-1 replication in PBMC (117). Our inhibitor studies are now strengthened by the recent avail-ability of TAK-779 (4). This CCR5-targeted inhibitor does not inhibit viral entry via CCR3, whereas AOP-RANTES can do so, albeit inefficiently compared to its effect on CCR5-medi-ated entry (34). Since TAK-779, by itself, is able to block the replication in PBMC of all of the R5, NSI HIV-1 and HIV-2 isolates that we tested, CCR3 is not relevant to their entry. Any possible use of CCR2 that might be masked by TAK-779 is not supported by the complete inhibition of the same isolates by AOP-RANTES. This chemokine derivative does not block vi-ral entry via CCR2, at least for SIVrcm, which is the only truly
[image:13.612.89.514.72.356.2]CCR2-tropic virus yet identified (15). Another advantage of TAK-779 is that it avoids the potential complications of AOP-RANTES-induced enhancement of attachment and entry of X4 HIV-1 isolates (44, 108). However, we did not observe infectivity enhancement with human or macaque PBMC at the AOP-RANTES concentrations tested in this study. Overall, the use of TAK-779 reinforces our previous conclusion about the paramount role of CCR5 and CXCR4 in HIV-1 replication in PBMC (116). This is not to say that other coreceptors are FIG. 6—Continued.
on November 9, 2019 by guest
http://jvi.asm.org/
FIG. 7. Donor-dependent variation in the effects of coreceptor-targeted inhibitors in PBMC. (a) SIVmac239 replication in PBMC from four different macaques was evaluated in the presence of TAK-779 at 33M, 3.3M, 330 nM, and 33 nM. SIVmac251 was similarly evaluated with cells from two donors. (b) HIV-1 P6-v3 replication in PBMC from four different human donors was evaluated in the presence of TAK-779 at 3.3M, 330 nM, 33 nM, and 3 nM. In each case, replication was measured after 7 and 10 days, and IC50s of the inhibitor were calculated. The data shown were obtained on day 10, but values from day 7 were similar.
on November 9, 2019 by guest
http://jvi.asm.org/
completely irrelevant; Bonzo/STRL33 can be used by rare HIV-1 isolates for entry into a minor subset of PBMC in a donor-dependent manner (102), and CCR3 and CCR8 are po-tential coreceptors expressed on some T-cell subsets (94, 118). The SHIV isolates that we evaluated—89.6, 89.6P, 89.6PD, SF33A, and KU-2—all exclusively used CXCR4 in human and macaque PBMC; AMD3100 was sufficient to completely inhibit their replication, while neither TAK-779 nor AOP-RANTES had any effect. Thus, although HIV-1 89.6 can enter trans-fected cells via several coreceptors, including CCR5 (27), the SHIV derived from it use only CXCR4 to enter PBMC. Of note is that SHIV 89.6, SHIV 89.6P, and SHIV 89.6PD very efficiently enter GHOST-CD4 cells expressing CCR5 (Table 1; also data not shown). Thus, these viruses can use CCR5 for entry, at least in CCR5-transfected cells, but CXCR4 is pre-ferred in primary cells. Why this should be the case and whether it matters for transmission and pathogenesis studies with these viruses in macaques are open questions.
We also conclude that, for most HIV-2 strains, CCR5 and/or CXCR4 are the principal coreceptors relevant to the replica-tion of these strains in PBMC. Thus, TAK-779, AOP-RAN-TES, and AMD3100, alone or in combination, completely or very substantially inhibited the replication of almost all of our test viruses. HIV-2 isolate 7924A is an apparent exception. The replication of this broadly tropic virus in PBMC was inhibited only by very high concentrations of AMD3100 and was com-pletely insensitive to SDF-1␣, TAK-779, or AOP-RANTES. One possibility is that HIV-2 7924A is able to use an alterna-tive coreceptor to enter human PBMC, perhaps the CXCR5 receptor reported recently to function with some HIV-2 iso-lates but not with HIV-1 or SIV isoiso-lates (52). Alternatively, HIV-2 7924A may use CXCR4 in a manner that is relatively insensitive to AMD3100. The latter explanation would be con-sistent with the observation that very high concentrations of AMD3100 do completely inhibit the replication of HIV-2 7924A, although there may be concerns about the specificity of AMD3100 for CXCR4 at such concentrations. Escape mutants of HIV-1 NL4-3 that continue to use CXCR4, but in a drug-insensitive manner, are known to emerge in response to selec-tion pressure from AMD3100 and SDF-1␣(24, 99). It has been suggested that CXCR4 can exist in different isoforms on dif-ferent cell types (69); this property might be one explanation for why AMD3100 is a potent inhibitor of HIV-2 7924A in GHOST-CD4 cells expressing CXCR4 (IC50⫽0.47M) but
can be such a weak one in PBMC (IC50⫽2.1 to 34M,
de-pending upon the donor). Additional studies of HIV-2 7924A are warranted.
Our conclusions for SIVmacisolates are more complicated.
The CCR5 proteins from multiple primate species can function as viral coreceptors (55, 78), and our inhibitor studies are con-sistent with an important role of CCR5 in SIVmac entry into
primary cells. One aspect of coreceptor usage that distinguishes SIV from HIV-2 isolates is the inability of almost all SIV to use CXCR4. This property contrasts with the efficient use of CXCR4 by many HIV-2 isolates. In this sense, HIV-2 more closely resembles HIV-1 than it does SIV, an unexpected finding given the genetic relationships among these virus families and the evolution of HIV-2 from SIVsm(16, 17, 41, 42, 44, 49). The
minimal use of CXCR4 by SIV strains is mirrored by that of HIV-1 isolates from genetic subtype C (1, 11, 82, 83, 112), although SI primary viruses from this subtype are known (111). Although CCR5 is important and CXCR4 is unimportant for SIVmac entry, we found indications that SIVmac239 could
use a coreceptor other than CCR5 to enter PBMC from some human and macaque donors. Thus, in PBMC from one ⌬ 32-CCR5 homozygous human donor, SIVmac239 replication was
negligible. However, in cells from a second such individual, the virus replicated fairly efficiently, as observed previously (18). Furthermore, there was considerable variation in the potency with which TAK-779 inhibited the replication of SIVmac239 in
PBMC from different macaques. This result might be ac-counted for by the use of an additional coreceptor that is expressed in PBMC from only a subset of macaques or that is expressed in cells from all macaques but at different levels that are sometimes below a threshold needed for infection. The expression of both CCR5 and Bonzo/STRL33 varies from do-nor to dodo-nor, in both humans and macaques, to an extent that can affect infection efficiency (102, 107). The ability of
SIV-mac239 to use a coreceptor other than CCR5, perhaps Bonzo/
STRL33, in an animal-dependent manner might influence the highly variable rates at which different infected macaques progress to disease and death (23, 57, 73). However, at least for SIVmne, CCR5 usage is maintained throughout the course of
disease progression in infected macaques (53). This is also true of SIVmac239 and SIVmac251 (14).
One coreceptor used efficiently by SIVmac239 in vitro is
BOB/GPR15 (22, 38). Po¨hlmann et al. have, however, shown that this coreceptor has no relevance to SIVmac239 replication
in vivo, at least in some macaques (86). An unknown, alterna-tive coreceptor(s) also mediates the AMD3100-, TAK-779-, and AOP-RANTES-insensitive entry of SIVmac239 into
CEMx174 cells; this coreceptor cannot, therefore, be CCR2, CCR3, CCR5, or CXCR4. It is not known whether this is the same coreceptor as the one used by SIVmac239 to enter human
or macaque PBMC from some donors.
We could not distinguish SIVmac239 from the closely related
SIVmac251 in terms of their sensitivity to coreceptor inhibitors.
Although SIVmac251 but not SIVmac239 replicates efficiently in
macrophages, there is no correlation between the coreceptor usage profiles of these viruses in transfected cells and their tropism for primary cells (53, 75, 76, 86). There is also no relationship between the in vitro tropisms of SIVmac strains
and their abilities to be transmitted to uninfected animals (36, 71, 72). We have not yet performed coreceptor inhibitor stud-ies with these viruses and purified macrophages and CD4⫹T
cells from macaques, as opposed to unfractionated PBMC. SIVrcmclearly uses CCR2 as its primary coreceptor (15), in
a manner that we have shown is sensitive to TAK-779. The ability of SIVrcmto enter GHOST-CD4 cells expressing US28
and V28 in vitro is likely to be of limited relevance to the replication of this virus in red-capped mangabeys.
Overall, we conclude that there is a greater complexity to coreceptor usage by SIV strains in PBMC than there is for HIV-1 and HIV-2, for which CCR5 and CXCR4 are usually the paramount coreceptors. An unknown coreceptor(s) can perhaps be used by SIVmac239 and HIV-2 7924A to enter
PBMC, at least from some macaque and human donors. In-volvement of the same coreceptor in the entry of both SIVmac239 and HIV-2 7924A might conceivably have
rele-vance to cross-species viral transmission and the evolution of HIV-2 from SIVsm(16, 17, 41, 42, 44, 49).
ACKNOWLEDGMENTS
We thank Annette Bauer, Michael Miller, Susan Vice, Bahige Ba-roudy, and Stuart McCombie for AMD3100 and TAK-779; Amanda Proudfoot, Robin Offord, and Brigitte Dufour for AOP-RANTES; Zhiwei Chen for SIV isolates; David Montefiori, Cecilia Cheng-Mayer, and Opendra Narayan for SHIV isolates; Dan Littman and Vineet KewalRamani for GHOST cells; and James Hoxie and Nelson Michael for preferring hard liquor to blood. We appreciate helpful comments by Amanda Proudfoot and Bob/GPR15 Doms.
on November 9, 2019 by guest
http://jvi.asm.org/
This study was supported by NIH grant RO1 AI41420 and by the Pediatric AIDS Foundation, of which J.P.M. is an Elizabeth Glaser Scientist.
REFERENCES
1.Abebe, A., D. Demissie, J. Goudsmit, M. Brouwer, C. L. Kuiken, G. Pol-lakis, H. Schuitemaker, A. L. Fontanet, and T. F. Rinke deWit.1999. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and core-ceptor usage among Ethiopian patients with AIDS. AIDS13:1305–1311. 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M.
Murphy, and E. A. Berger.1996. CC CKR5: a RANTES, MIP-1␣, MIP-1
receptor as a fusion cofactor for macrophage-tropic HIV-1. Science272:
1955–1958.
3.Alkhatib, G., F. Liao, E. A. Berger, J. M. Farber, and K. W. C. Peden.1997. A new SIV coreceptor, STRL33. Nature388:238.
4.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, and K. Meguro.1999. A small molecule nonpeptide CCR5 antagonist with highly potent and selec-tive anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA96:5698–5703. 5.Bazan, H. A., G. Alkhatib, C. C. Broder, and E. A. Berger.1998. Patterns of
CCR5, CXCR4, and CCR3 usage by envelope glycoproteins from human immunodeficiency virus type 1 primary isolates. J. Virol.72:4485–4491. 6.Berger, E. A.1997. HIV entry and tropism: the chemokine receptor
con-nection. AIDS11(Suppl. A):S3–S16.
7.Berger, E. A., R. W. Doms, E.-M. Fenyo¨, B. T. M. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss.1998. A new classification for HIV-1. Nature391:240.
8.Berger, E. A., P. M. Murphy, and J. M. Farber.1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol.17:657–700.
9.Bieniasz, P. D., and B. R. Cullen.1998. Chemokine receptors and human immunodeficiency virus infection. Front. Biosci.3:44–58.
10.Bron, R., P. J. Klasse, D. Wilkinson, P. R. Clapham, A. Pelchen Matthews, C. Power, T. N. C. Wells, J. Kim, S. C. Peiper, J. A. Hoxie, and M. Marsh.
1997. Promiscuous use of CC and CXC chemokine receptors in cell-to-cell fusion mediated by a human immunodeficiency virus type 2 envelope pro-tein. J. Virol.71:8405–8415.
11.Bjo¨rndal, A., A. Sonnerborg, C. Tscherning, J. Albert, and E. M. Fenyo¨.
1999. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res. Hum. Retrovir.
15:647–653.
12.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh.1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol.71:3932–3939.
13.Chan, S. Y., R. F. Speck, C. Power, S. L. Gaffen, B. Chesebro, and M. A. Goldsmith.1999. V3 recombinants indicate a central role for CCR5 as a coreceptor in tissue infection by human immunodeficiency virus type 1. J. Virol.73:2350–2358.
14.Chen, Z., A. Gettie, D. D. Ho, and P. A. Marx.1998. Primary SIVsmisolates
use the CCR5 co-receptor from sooty mangabeys naturally infected in West Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2 and
SIVmac. Virology245:113–124.
15.Chen, Z., D. Kwon, Z. Jin, S. Monard, P. Telfer, M. S. Jones, C. Y. Lu, R. F. Aguilar, D. D. Ho, and P. A. Marx.1998. Natural infection of a homozygous
⌬24-CCR5 red-capped mangabey with an R2b-tropic simian immunodefi-ciency virus. J. Exp. Med.188:2057–2065.
16.Chen, Z., A. Luckay, D. L. Sodora, P. Telfer, P. Reed, A. Gettie, J. M. Kanu, R. F. Sadek, J. Yee, D. D. Ho, L. Zhang, and P. A. Marx.1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characteriza-tion of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol.71:3953–3960. 17.Chen, Z., P. Telfier, A. Gettie, P. Reed, L. Zhang, D. D. Ho, and P. A. Marx.
1996. Genetic characterization of new West African simian immunodefi-ciency virus SIVsm: geographic clustering of household-derived SIV strains
with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol.70:3617–3627. 18.Chen, Z.-W., P. Zhou, D. D. Ho, N. R. Landau, and P. A. Marx.1997.
Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J. Virol.71:2705–2714.
19.Choe, H., M. Farzan, M. Konkel, K. Martin, Y. Sun, L. Marcon, M. Cayabyab, M. Berman, M. E. Dorf, N. Gerard, G. Gerard, and J. Sodroski.
1998. The orphan seven-transmembrane receptor Apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J. Virol.72:6113–6118.
20.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, G. Gerard, and J. Sodroski.1996. The-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell85:1135–1148.
21.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S.
Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multi-center Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study, and S. J. O’Brien.1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion of the CKR5 structural allele. Science
273:1856–1862.
22.Deng, H., D. Unutmaz, V. N. KewalRamani, and D. R. Littman.1997. Expression cloning of new receptors used by simian and human immuno-deficiency viruses. Nature388:296–300.
23.Desrosiers, R. C.1995. Non-human primate models for AIDS vaccines. AIDS9(Suppl. A):S137–S141.
24.De Vreese, K., V. Kofler-Mongold, C. Leutgeb, V. Weber, K. Vermie`re, S. Schacht, J. Anne´, E. de Clercq, R. Datema, and G. Werner.1996. The molecular target of bicyclams, potent inhibitors of human immunodefi-ciency virus replication. J. Virol.70:689–696.
25.Doms, R. W., and S. C. Peiper.1997. Unwelcome guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology235:179–190. 26.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este´, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore.1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med.4:72–77.
27.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms.1996. A dual-tropic, primary HIV-1 isolate that uses fusin and the-chemokine receptors CKR-5 and CKR-2b as fusion cofactors. Cell85:1149–1159.
28.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Na-gashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton.1996. HIV-1 entry into CD4⫹cells is mediated by the chemokine
receptor CC-CKR-5. Nature381:667–673.
29.Dumonceaux, J., S. Nisole, C. Chanel, L. Quivet, A. Amara, F. Baleux, P. Briand, and U. Hazan.1998. Spontaneous mutations in theenvgene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J. Virol.72:512–519.
30.Edinger, A. L., C. Blanpain, K. J. Kuntsman, S. M. Wolinsky, M. Parmen-tier, and R. W. Doms.1999. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J. Virol.73:4062–4073.
31.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, B. O’Dowd, and R. W. Doms.1998. Use of GPR1, GPR15, and STRL33 as co-receptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology249:367–378.
32.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, Y. Yi, W. Choe, D. C. Kolson, B. Mitrovic, Y. Zhou, D. Faulds, R. G. Collman, J. Hesselgesser, R. Horuk, and R. W. Doms.1998. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol.
72:7934–7940.
33.Edinger, A. L., J. L. Mankowski, B. J. Doranz, B. J. Margulies, B. Lee, J. Rucker, M. Sharron, T. L. Hoffman, J. F. Berson, M. C. Zink, V. M. Hirsch, J. E. Clements, and R. W. Doms.1997. CD4-independent, CCR5-depen-dent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc. Natl. Acad. Sci. USA94:14742–14747. 34.Elsner, J., M. Mack, H. Bruhl, Y. Dulkys, D. Kimmig, G. Simmons, P. R. Clapham, D. Schlo¨ndorff, A. Kapp, T. N. C. Wells, and A. E. I. Proudfoot.
2000. Differential activation of CC chemokine receptors by AOP-RANTES. J. Biol. Chem.275:7787–7794.
35.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. C. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie.1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell87:745–756.
36.Enose, Y., K. Ibuki, K. Tamaru, M. Ui, T. Kuwata, T. Shimada, and M. Hayami.1999. Replication capacity of simian immunodeficiency virus in cultured macaque macrophages and dendritic cells is not prerequisite for in-travaginal transmission of the virus in macaques. J. Gen. Virol.80:847–855. 37.Este´, J. A., C. Cabrera, J. Blanco, A. Gutierrez, G. Bridger, G. Henson, B.
Clotet, D. Schols, and E. DeClerq.1999. Shift of clinical human immuno-deficiency virus type 1 isolates from X4 to X5 and prevention of emergence of the syncytium-inducing phenotype by blockade of CXCR4. J. Virol.
73:5577–5585.
38.Farzan, M., H. Choe, K. Martin, L. Marcon, W. Hofmann, G. Karlsson, Y. Sun, P. Barrett, N. Marchand, N. Sullivan, N. Gerard, C. Gerard, and J. Sodroski. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med.186:405–411.
39.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger.1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein coupled receptor. Science272:872–877.
40.Frade, J. M. R., M. Llorente, M. Mellado, J. Alcami, J. C. Gutierrez Ramos, A. Zaballos, G. del Real, and A. C. Martinez.1997. The amino terminal domain of the CCR2 chemokine receptor acts as a coreceptor for HIV-1 infection. J. Clin. Investig.100:497–502.