0022-538X/94/$04.00+0
Copyright (C 1994, AmericanSociety forMicrobiology
The
tat/rev
Intron
of
Human
Immunodeficiency
Virus
Type 1
Is
Inefficiently
Spliced because of Suboptimal
Signals
in
the 3' Splice Site
ALFREDO STAFFAANDALANCOCHRANE*
Department ofMicrobiology and Immunology, McGill University, Montreal, Quebec, Canada H3A 2B4
Received 24 August 1993/Accepted 16 February 1994
Proportional expression of retroviral genes requires that splicing of the viral primary transcript be an
inefficient process. Much ofourcurrent knowledge aboutretroviral suboptimal splicingcomes from studies
withRoussarcomavirus.Inthisreport,wedescribe theuseof chimeric introns composed of human I-globin
andhumanimmunodeficiency virustype 1 (HIV-1) splice sitestoestablishthe basis forinefficient splicing of the intronwhich comprisesmostof the HIV-1envcodingsequences(referredtoasthetat/revintron). Si RNA analysis of transfected COS-7 cellsrevealed that the3' splice site (3' ss) of this regionwas significantly less
efficientthan the 3' ssofthe first intron of ,B-globin. Deletion ofsequences flanking thetat/revintron3' ss
demonstrated thattherequirements foritsinefficiencyresidewithin the region that is expectedtocomprise the essential signals for splicing (i.e., the branchpoint region,thepolypyrimidinetract,and theAGdinucleotide). Introduction ofanexactcopyof the efficient I-globin branchpointsequencewithinahighly conserved region
rendered thetat/rev intron 3' ss highly efficient.Improvementof the polypyrimidine tractalso increased the splicing efficiency,buttoadegreeslightlyless than thatobtained with the branchpointmutation. Subsequent examination of thetat/rev intron 5' splice siteinaheterologous context revealedthatit isefficiently utilized. These results indicate thatbothapoorbranchpoint region anda poorpolypyrimidinetractareresponsible for
the lowsplicing efficiency of the HIV-1tat/rev intron. It is of fundamental interesttoestablish the basis for inefficient splicing of the HIV-1tat/revintron sinceitmayprovide the keytounderstandingwhynuclearexport ofmRNAs encoding HIV-1 structural proteins is Rev dependent.
Primary transcripts of most higher eukaryotic protein-en-coding genes transcribed by RNA polymerase II are
inter-rupted by introns. These introns are usually selectively and
efficiently removedby splicing,anuclearprocessinwhich the
5' splicesite (5' ss)and 3' splice site (3' ss) arecleaved and
joined prior to export of the mRNA to the cytoplasm. The splicing process involves a complex series of protein-protein
andprotein-RNA interactions between the pre-mRNA, com-ponents of the spliceosome (Ul, U2, U4/U6, and U5 small nuclear ribonucleoproteins [snRNPs]), and other auxiliary factors (18, 30).Given thecomplexity ofthesplicing pathway, there is great potential for elaborate regulation. In fact, splicing of primary transcripts is emerging as an important
level of regulation in gene expression. Species ranging from
Drosophila melanogaster to humans have evolved regulated mechanisms such asalternative splicingand exon skippingto
express multiple protein products from asingle primary
tran-script (26, 39). Some viruses ofhigher eukaryotes have also evolved mechanismstomaximize thecoding potential of their
genomes. While DNA viruses such assimian virus 40 (SV40) utilize mechanisms of regulated splicing similar to those of their cellularhosts(i.e., alternativesplicing) (17),retroviruses have evolved different posttranscriptional regulatory mecha-nisms. In thecaseofretroviruses, onlyafraction of theprimary transcripts are spliced to subgenomic RNAs (8, 46). Propor-tional expression of retroviral genes requires that splicing of the genome-length primary transcript bythe cellular splicing machinerybe aninefficientprocess. Furthermore,unlikemost
*Correspondingauthor. Mailingaddress:Department of
Microbi-ologyandImmunology, McGillUniversity,3775 University St.,
Mon-treal,Quebec, Canada H3A2B4. Phone: (514) 398-6653. Fax:(514)
398-7052. Electronic mail address:cza8@musica.mcgill.ca.
cellular transcripts which arerestricted to the nucleus unless spliced,acertainproportionoffull-length retroviral transcripts mustbe exportedtothecytoplasm toserveas mRNAfor the
gagandpolgeneproductsandasgenomicRNA forpackaging intovirus particles (8, 46).
Althoughthe Lentiviridae and Oncoviridae retroviral classes share similar pathways of replication, they display striking differencesin genome complexityand regulation, particularly
in the metabolism of the viral primary transcript (8, 46). Comparedwithsimpleretroviruses suchasthe Roussarcoma
virus (RSV) of the Oncoviridae class,the human immunodefi-ciencyvirustype1 (HIV-1)hasamorecomplex RNA-splicing pattern (37, 43). In addition to producing the Gag, Pol, and Env proteins common to all replication-competent
retrovi-ruses,the HIV-1 primary transcript alsoservestoproducesix
regulatory proteins (10, 14),twoof which-Tat andRev-are essential for HIV-1 replication (2, 11, 15, 31, 41, 42, 44, 47). Moreover, unlike the RNAs ofsimple retroviruses,which are
constitutively transported from the nucleus to the cytoplasm (43), the unspliced and singly spliced HIV-1 RNAs (which encode structural viralproteins) areretained in the nucleusin the absence of the HIV-1 Revprotein;thatis, onlythedoubly spliced,2-kb class of HIV-1 RNAs isconstitutively transported to the cytoplasm and translated independently of the
expres-sion of any viral proteins (12, 13, 19, 25). Since all HIV-1 RNAs with an intact
tatlrev
intron are not constitutively transported to the cytoplasm (12, 13, 19, 25), it seems that elements within this intron must contribute to the nuclear entrapment of the incompletely spliced RNAs. Therefore, mechanisms uniquetothe lentivirusclass of retrovirusesmustexist which(i)allow themajorityof the RNAto escapesplicing
and(ii) causenuclearretention ofincompletely splicedRNAs
in the absence of viral regulatory proteins. To date, intronic
3071
on November 9, 2019 by guest
http://jvi.asm.org/
sequences crucial for balanced retroviral RNA splicing have been identified in avian sarcoma viruses, namely, asuboptimal 3' ss (3, 16, 20, 21) and anegative regulator ofsplicing (NRS)
that can function in cis to decrease splicing of both RSV and cellular introns (1, 27, 28).
Tostudy the basis for the inefficientsplicing observed for the HIV-1
tatlrev
intron, we have created chimeric introns using splice sites of the humanP-globin
first intron and the HIV-1 tat/rev intron.Si analysis of total RNA from transfected COS-7 cells reveals that the tat/rev intron 3' ss, like that of RSV, is inefficient. Mutagenesis experiments indicate that the tat/rev intron 3' ss can be rendered efficient either by the introduction of an efficient branchpoint sequence 30 nucleotides (nt) up-stream of the AG dinucleotide or by improvement of the polypyrimidine tract. Subsequent experiments carried out withthetat/rev intron 5' ss in a heterologous context reveal that it
is efficiently utilized by the splicing machinery. Unlike the NRS-containing intron of RSV (1, 27, 28), the tat/rev intron does not appear to contain any cis-acting sequences that modulate splicing efficiency. Therefore, it appears that the tat/rev intron is inefficiently spliced simply because of the absence ofefficient signals within the 3' ss. Differences in the regulation of splicing between HIV-1 and RSV may have important implications for the fate of inefficiently spliced RNA with respect to nuclear export.
MATERIALS AND METHODS
Construction ofplasmids. The parent construct, pSV3,was constructed as follows. The EcoRI-BamHI fragment of pEE14 (a generous gift from Celltech Ltd.) carrying the SV40 early polyadenylation signal was inserted into the respective sites of pBluescript SK(+) and (BlSK; Stratagene). Subsequently, the BamHI and XbaI sites were eliminated by simultaneous re-striction enzyme digestion, blunting the protruding 5' ends with Klenow fragment, and religating. The blunt-ended Sall-XbaI fragment of Bl-CAT containing the chloramphenicol acetyltransferase (CAT) open reading frame was then inserted into the unique EcoRV site upstream of the SV40 early
polyadenylation signal. Finally, the SV40 early promoter,
contained onaXhoI-HindIII fragment, and a modified human ,B-globin intron (IVS-1), contained on a 214-bp
HindIll-BamHI fragment, were inserted simultaneously into theXhoI
andBamHI sites, generating pSV3.
HIV-1 sequences spanning the tat/rev intron 3' ss (at nt 8377 [the nucleotide numbering system used throughout cor-responds to that of the Hxb2 complete genome sequence in GenBank, accession no. K03455]) were derived from the Hxb2 proviral clone. The 321-bp HT fragment from the HindIII site atnt 8141 to the TaqIsite at nt8462 was blunted and inserted into the EcoRV site ofBlSK in the T7 orientation, generatingBl-HIVSAHT. The CTfragment
(nt.8333
to8462) was subcloned from Bl-HIVSA HT by PCR with the ClaIsenseprimer,5'-TlTTTGCTGTACTATCGATAGTGAATAG
AGT-3' (nt 8317 to 8346), and the T3 primer of BlSK, 5'-CTCGGAATTAACCCTCAC-3'. The 152-bp
ClaI-BamHI
fragment of the PCR-amplified DNA was inserted into BlSK, generating Bl-HIVSACT.
pSVHTSB and pSVCTSB were generated by inserting the
SalI-BamHI fragments of Bl-HIVSA HT and Bl-HIVSA CT, respectively, into the unique Sall andBamHI sites of pSV,B.
pSVCBSB was generated by PCR as follows. pSVCTSB was
used as template DNA for PCR amplification with the T7
primerofBlSK, 5'-AGTGAATTGTAATACGAC-3', and the
SA-Bam antisense primer, 5'-GGATCCGGGTCTGAAAC
GATAATG-3' (nt 8364 to 8381). TheXhoI-BamHI fragment
ofpSV,B was subsequently replaced by theXhoI-BamHI
frag-ment of the PCR-amplified DNA, generating pSVCBSB. The 2.4-kb fragment spanning the
tatlrev
intron 5' ss (at nt 6045) was derived from Bl-env SX C/X (6a). This construct contains a modified SalI-XhoI fragment of Hxb2 (nt 5786 to 8897) inwhich theHindIllsite at nt 8141 was converted to an XbaI site by site-directed mutagenesis (as outlined by Bio-Rad). This XbaI site was converted to a SalI site by the addition of aSall linker.pSVenvo3
wasconstructed by inserting the 2.4-kbSall fragment into the unique Sall site ofpSV,BSA,
a modifiedpSV, inwhich the 3-globin 5' ss had been deleted.
pSVHIVfi was constructed as follows. A 159-bp fragment
spanning the tat/rev intron 5' ss was obtained by PCR ampli-fication with the 5'HIVSD sense primer, 5'-CCAAGCTT GAGCTCATCAGAACAGTC-3' (nt 5999 to 6016) and the 3'HIVSD antisense primer, 5'-GGGAATTCTGATTACTAT
GGACCA-3' (nt 6128 to 6142). The 116-bp HindIII-EcoRI
fragment of the PCR-amplified DNA was subcloned into BlSK, generating Bl-envSD. The SV40 early promoter, con-tained on an XhoI-HindIII fragment, was inserted into the
XhoI and HindIII sites of Bl-envSD. The EcoRI site was subsequently converted to aSallsite by the addition of aSall
linker, and the KpnI-SalI fragment of the resulting construct was inserted into the KpnI andSallsites of pSV3, generating
pSVHIV,B.
Introduction of the branchpoint and polypyrimidine tract mutations into the HT fragment. For introduction of the ,3-globin branch point, mutagenesis was carried out entirely in vitro by three sequential PCRs as follows. In the firstreaction,
a fragment was amplified from BI-HIVSA CT with the muta-genic primer 3-BP, 5'-ATAGTGAATACACTGACGCAGG
GATATTC-3' (nt 8333 to 8361), and the T3 primer. The
,3-BP
primercontains fourmismatches (underlined)which createan exact copy of the reported ,B-globin branchpoint sequence, CACTGAC (32). The expected site of the RNA branch formation (shown in boldface type) coincides with an A residue 30 nt upstream of the AG dinucleotide of the HIV-1 tat/rev intron 3' ss. An aliquot of the resulting 229-bp PCR product was used as a pseudoprimer in conjunction with the
P-PCR1
primer,5'-CTITAAGTTGGTGGTGAGG-3' (a sense primercomplementarytosequences5' of theP-globin
5' ss), ina secondPCRto amplify a 488-bp fragment from pSVHTSB.
To increase the yield of amplified DNA, 1/10 of the second
PCRproduct was further amplified by PCR with the 1-PCR1
and T3 primers. The SalI-BamHI fragment of pSV3 was subsequentlyreplacedby theSalI-BamHIfragment of the final
PCRproduct, generating pSVHTSB*.
To introduce mutations into the polypyrimidine tractof the
tat/rev intron, a strategy identical to that outlined above was employed, the only modification being that the mutagenic
primer polypyr,
5'-GGATATTCTC
CTIfl'7'l'Cl'lTI7CAGAC3', was utilized in place of the 1-BP primer. Consequently, purine residues at positions -7, -10, -13, and -16 relative to the AG dinucleotide of the 3' ss (shown inboldface type) were changed to thymidine (underlined). These mutations resulted in the generation of an uninterrupted polypyrimidine tract of 17 pyrimidines. The SalI-BamHI fragment from the final PCR was inserted intopSV,B to generate pSVHTSBPPt.
To ensure that no other alterations to the
sequence
had occurred during the amplification cycles, all mutations were confirmed bychain-terminating DNAsequencing (35).Cell culture and transfection. COS-7 cells (ATCC CRL1651) were maintained in Iscove's modified Dulbecco's
mediumsupplemented with 10% fetal bovine serum. Culture dishes (diameter, 100 mm) were seeded with 106cells,and the cells weretransfected24 h later by the DEAE-dextran method
on November 9, 2019 by guest
http://jvi.asm.org/
as
previously
described(9). Forty-eighthoursposttransfection, the cells were washed twice in phosphate-buffered saline,gently scraped off thedishes,and pelleted.The cellpellet was
resuspended in 4 M guanidine thiocyanate-25 mM sodium
citrate (pH 7.0)-0.5% Sarkosyl-0.1 M ,B-mercaptoethanol. TotalRNAwasextractedaspreviouslydescribed(6).After the final precipitation, RNApelletswere washedwith 70%
etha-nol, dried, and redissolved in diethylpyrocarbonate-treated
water.RNAwasquantitated by determiningthe
A160
(anA2,60
value of 1 is 50 ,ug/ml).
SI nuclease protection analysis. DNA fragments used as
probes
werefrom either the SallortheHindlIl siteupstream of the 3' ss to the PvuII site within the CAT open readingframe. Todistinguish reannealedprobe from protected probe
resulting
from unspliced RNA, DNA fragments containedheterologous,
BlSK-derived DNA sequences upstream of the3'ss.Theprobefor the
3-galactosidase
controlwascontiguouswitha 3' portionof the lacZgene-fromthe PvuII sitetothe
EcoRIsite ofpCH110(Pharmacia;nt2929to3286)-and also contained
heterologous
sequences to distinguish reannealedprobe
from protected probe resulting from hybridization toRNA. Each DNA fragment was radiolabelled at the 5' end
with
[y-32P]ATP
by using T4 polynucleotide kinase (34).End-labelledDNAprobe
(approximately
2 x 103cpm;specificactivity, approximately
0.5 x106 cpm/,ug)
wasaddedtototal RNA(5
big)
in 30RlI
ofhybridizationbuffer {80%formamide;40 mM PIPES
[piperazine-N,N'-bis(2-ethanesulfonic
acid)],pH 6.8; 400 mM NaCl; 1 mM EDTA}. The nucleic acidwas
denatured at
75°C
for 10 minand allowedtohybridize for 16h at 50°C. Following hybridization, samples were rapidly
cooled to
4°C. Single-stranded
nucleic acid was digested byadding
300 ,u ofS1digestion
mix(50mMsodiumacetate [pH4.5],
280 mM NaCl, 4.5 mM ZnSO4, 20pLg
of denatured,sonicated salmon sperm DNA perml, 1,000UofS1 nuclease per
ml)
andincubating
at 25°C for 2 h. The reaction was terminatedbythe additionof 5[.l
of 500 mM EDTA and 20 ,ugofEscherichia coli tRNA. Proteinswere removed by
phenol-chloroform
(1:1) extraction,
and the aqueousphasewas etha-nolprecipitated.
Thedried nucleic acidpelletwasresuspendedin 80% formamide-10mM EDTA, heated at85°Cfor 5 min,
and
subjected
toelectrophoresison a5% polyacrylamide-8Murea
gel.
Theprotected DNAfragmentswerevisualized eitherby autoradiography
on Kodak XAR film or by using aphos-phorimager (Molecular Dynamics).
Quantitationofphosphor-imager-derived
data was carried out by using Image Quantsoftwareasdescribed
by
the manufacturer(MolecularDynam-ics).
RESULTS
Construction of pSV,I for examination of the
efficiency
of HIV-1splice
sites. Inordertofacilitate thestudy
of heterolo-goussplice
siteefficiency,
thepSV,B plasmid,
which containsamodified human
P-globin
intron(IVS-1)
upstreamofthe CAT openreading frame,
was constructed(Fig. IA).
The modified,B-globin
intron contains severalunique
restriction enzyme sites into whichheterologous splice
sites can be inserted toanalyze
theirefficiency
ofutilization. Inaddition,
the3-globin
intron contains several AUG codons(data
notshown)
such that translation of the RNA into functional CATprotein
isdependent
upon the removal of the intron. This featureprovides
areadily quantitated
marker for geneexpression
(Table
1).
S1
analysis
revealed that theP-globin
intron isefficiently
excised frompre-mRNA
in transfected COS-7 cells(Fig. 1B,
lane8;
Table1).
Analysis
wascarriedoutwithaprobe
spanning
the 3' ss,yielding protected products
of 302 and 206nt corresponding to the unspliced and spliced forms of the RNA, respectively. Furthermore, experiments using a probe spanning the entire CAT open reading frame of pSV3 indi-catedthat no cryptic splice sites within the CAT sequences are utilized (data not shown). This efficient splicing results in the accumulation of aconsiderable amount of spliced pSV,BRNA (Fig. 1 B, lane 8) and, in turn, high-level expression of CAT enzymeactivity (Table 1). Thus, the
pSVP3
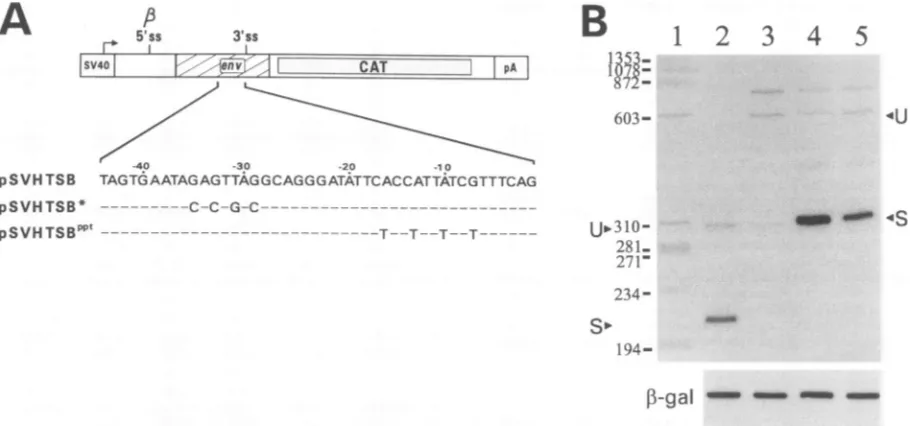
constructprovides a convenient system to examine the basis for the inefficient splicing of the tat/rev intron of HIV-1.Thetat/revintron 3'ssisintrinsicallyinefficient. Inorderto studytheefficiency of thetat/revintron 3' ss,the SB serieswas constructed(Fig. IA). In this series, the 3-globin 3'ssofpSV3 was replaced by env sequences spanning the tat/rev 3' ss by insertion intothe Sall and BamHI sites. The largestfragment tested(designated HT) contains 236 nt5' and 85 nt 3' of the tat/rev 3' ss. To testwhetherflanking sequences modulate the
efficiencyof the 3' ss, either 5' sequences or both 5' and 3'
sequences were deleted (designated CT and CB fragments, respectively). Analysis of theRNAgenerated frompSVHTSB revealed that thetat/revintron 3' ssis utilized (Fig. 1B, lane 9), but at an extremely low level relative to utilization of the
3-globin3' ssofpSV3 (Fig. 1B, lane 8). The level of splicing
observedwith thepSVHTSB chimeric construct is comparable tothe ratio ofunsplicedand splicedviral RNAs derived from otherHIV env expression vectors (13, 19, 25). These results indicate that the requirements for suboptimal splicing are located within the 321-bp HT fragment. Examination of the
pSVCTSBand pSVCBSBconstructsrevealed that deletionof
flanking sequences does not increase the efficiency of the
tat/revintron 3' ss(Fig. IB, lanes 10 and 11). In fact, compar-ison of the ratio ofspliced to unspliced RNA indicates that
splicingofpSVCTSBandpSVCBSB transcripts is less efficient
(or undetectable in the case of pSVCBSB) than that of
pSVHTSBtranscripts. Therefore,the results obtained with the
SB series indicate that the tat/rev intron 3' ss is functional, albeit at a very poor efficiency. Moreover, requirements for
suboptimal splicingappeartobeconfinedtotheregion that is
expected to contain the signals for splicing (i.e., the branch
point, the polypyrimidine tract, and the AG dinucleotide).
Consistent with the hypothesis that the tat/rev intron 3' ss is
inefficientlyrecognized by the host splicing apparatus,wehave
alsodetermined that the tat/rev intron 3' ss cannotcompete in ciswith efficient splice sites suchas thatof
3-globin
(42a).Thetat/rev intron3'ssisrendered efficientbythe introduc-tion of the 1-globinbranch pointor mutation of the polypy-rimidine tract. To examine the tat/rev intron 3' ss in more
detail,the sequencescomprisingthe 3'ssfrom several different
HIV-1 proviral clones were compared (Fig. 2). The striking observation is the presence ofan extremelyconserved region between 20 and 39 ntupstreamof the AG dinucleotide ofthe tat/rev intron3' ss. It is within this region that the A residue involved in RNAbranch formation is expectedto be located
(18). Although several potential A residues are found within
this region, none are contained within a context bearing
similarityto the weakly conserved 5'-UNCURAC-3'
branch-point consensus of
higher
eukaryotes(18).
In addition to theconservation of sequence in the anticipated
branchpoint
re-gion, there is also conservation of interruptions within the
polypyrimidinetract.These findingssuggest
(i)
that inefficientutilization of the tat/rev intron 3' ss may be due to a poor
branchpoint region and/or polypyrimidine tract and
(ii)
thatthere is an evolutionary constraint on the virus to maintain these suboptimal splicing signals.
To testthe hypothesisthat inefficient
splicing
of the tat/revintron3'ssis duetoeitherapoorbranchpoint
region
orapooron November 9, 2019 by guest
http://jvi.asm.org/
5'ss 3'ss
II
I S X B
r.
CAT
IF
hi
lSV40
I
I
CA
IP
H S B
3'ss\
8141 1 A46
SB HT
3'ss
8333 1 8462
'SB CT
3'ss
8333\ 18381
,SB CCB
B
1
2
3 4 5
6
7
8 9
1011
1353- 1078-
872-
603- 310-281.
271- 234-pSVCB
194-c
p
5'ss
3'ss
tV401
l CAT pAI
* 683 683 466
P-gal
control
-* 538 344 279
* 287 287 185
[image:4.612.81.529.81.430.2]pSVHTSB pSVCTSB pSVCBSB
FIG. 1. (A) SchematicrepresentationofpSV13 and itsSBderivatives. AllconstructscontaintheSV40early promoterandearlypolyadenylation signal(pA).TheCATopenreadingframeisdepicted byadotted box. NumbersaboveHIV-1 sequences(HT, CT,and CBfragments [hatched boxes]) spanningthetat/rev intron 3'ss arefrom the Hxb2 numberingsystem in GenBank accessionno. K03455.Splicesitesare indicated by vertical lines. 1B 5' ss, 3-globin 5'ss;,B3'ss, ,B-globin3'ss.Thetat/revintron 3' ssis labelled 3'sswithin theenvsequence. Importantrestriction
enzymesitesarealso shown.H, HindIll; S, SalI;X, XbaI; B,BamHI. The SBconstructswerederivedbyinsertion ofHIV-1sequencesintothe
unique SaIandBamHI sites ofpSV,B. (B)S1nucleaseprotection analysisof total RNA from COS-7 cellstransfected with the SBconstructs.Lane 1contains1/10theamountofprobeused forS1 protection analysisinlanes5, 9,and 10; lane 2contains1/10the amountofprobeused forS1 protection analysisin lanes6 and8;lane3contains1/10theamountofprobeused forS1 protection analysisin lanes7and 11. Lane 4 contains
radiolabelledmarkers. A5-jxg aliquotof total RNA fromCOS-7 cells transfected withpSVI (lane 8),pSVHTSB (lane 9), pSVCTSB (lane 10), andpSVCBSB (lane 11)wasused in theS1 protectionreactions.Asanegative control,total RNA frommock-transfected COS-7 cells(lanes 5,
6,and7)wasincubated withprobes anddigestedwithS1 nuclease.The ,B-galactosidase (1-gal)control S1 protectionreactionscarriedout in
parallel usinga,B-galactosidase-specific probearealsoshownbelow thelarge gel.Numbersontheleft indicate molecular sizes(inbasepairs). (C)
Schematicrepresentation of theexpected protectionpattern for the SB constructsoutlined in panel A, indicatingthe sizes of theprobes and
protected fragments corresponding to the unspliced and spliced RNAs derived from the constructs tested. The wavy line corresponds to
heterologoussequenceusedtodiscriminatebetweenunsplicedRNAandreannealedprobe.
polypyrimidinetract,several mutationsweregenerated.Inthe
first instance, the reported ,B-globin branchpoint sequence
CACTGAC(32) wasintroduced intopSVHTSB by mutagen-esis to generate pSVHTSB* (Fig. 3A). This construct is identical to pSVHTSB exceptthat theenvsequences contain
altered basesatpositions -29, -31, -33, and -35 relativeto
the AG dinucleotide of the 3' ss. The sequence
5'-_iAGQT
_A£i-3'atpositions -35 to -29within the highly conserved region was converted to the ,B-globin branchpoint sequence
shown above. This particular location within the conserved region was chosen for several reasons: (i) only four point
mutations (underlined)wererequiredtocreatean exactcopy
ofthe f-globin branchpointsequence,(ii) thesequenceresides within the -18 to -38 distance from the AG dinucleotide
commonly found for RNA branch formation, (iii) no A residues within the highly conserved region are altered, and (iv) the expected branchpoint A residue (shown above in boldface) coincides withanaturallyoccurringAresidue30nt
upstreamofthe AGdinucleotideofthetat/revintron3' ss.Si analysis (Fig. 3B,lane4)revealedthat this minor alteration of theprimarynucleotide sequence,whichintroduces anefficient
branch point, dramaticallyincreasesthelevelofspliced RNA observed. Comparison betweenpSVHTSB* andpSV,Bof the percentageoftotal RNA that is of the splicedform indicates thatsplicingofthepSVHTSB* transcriptis asefficientasthat
ofthepSV13 transcript (Table 1).Thehigherratioofsplicedto unsplicedRNAobserved forpSVHTSB*than forpSV1 is due bothto ahigherlevel ofspliced product (94.17versus 81.2%)
A
pSVP
S HpSVHT
pSVCT
Construct:
- m1 =
_-
on November 9, 2019 by guest
http://jvi.asm.org/
TABLE 1. Analysis of CATactivity and RNA species generated by
pSVf andits HTSB derivatives'
CAT/,B- RNAanalysisd
Construct" galactosidase
Spliced/unspliced
activity'
~
ratio %SplicedpSV3 21.44 ± 15.96 4.35 ± 0.53 81.20± 1.78 pSVHTSB 0.47 ±0.18 0.56± 0.09 40.57± 6.00 pSVHTSB* 9.73 ± 1.27 16.30 ± 1.89 94.17 ± 0.68 pSVHTSBPPt 6.27 ±0.88 6.92± 0.30 87.37 ± 0.51
aResultsareaveragesforthreeindependentsetsof transfections. Standard
deviations between the three experimentsarealso indicated.
bEach construct wascotransfected withthe
I-galactosidase
expressionplas-midpCH110(Pharmacia).
'Parallel assaysfor CAT and ,B-galactosidase activitieswere performed as
previouslydescribed(34). Numbersrepresent % conversion of CAT substrate
normalizedtotheA420obtained for the respective ,B-galactosidase assay. dQuantitation of S1RNAprotection experimentssuch as that showninFig. 3Bwascarriedoutby usingaphosphorimagerasdescribedbythemanufacturer
(Molecular Dynamics).
andto aslightlylower absolute level ofpSVHTSB* unspliced
RNA (Table 1 and Fig. 3B). Therefore, four simple point
mutations within an evolutionarily conserved region
encom-passingthe putative site of RNAbranch formation are
suffi-cient to render the otherwise inefficient tat/rev intron 3' ss
highly efficient.
Inordertoevaluate the contribution of the polypyrimidine
tract to the overall splicing inefficiency observed, the four
purine residues interrupting the polypyrimidine tract were
convertedtoTresidues bysite-directedmutagenesis, resulting in thegenerationofanuninterrupted polypyrimidinetract 17
nt long(Fig. 3A). Analysis ofthis constructalso revealed an
increase in utilization of the tat/rev intron 3'ss(Fig.3B, lane 5; Table1). Therefore,itcanbeconcluded that the fourpurines
thatinterruptthepolypyrimidinetractof thetat/rev intron 3'ss
contributetothe overall
inefficiency
of this site. Theobserva-tion that thesteady-statelevels ofunsplicedRNAdonotdiffer between thepSVHTSBconstructand its derivatives indicates that the differences in levels of spliced RNA cannot be attributedtodifferencesinthestabilityof theunsplicedRNA.
Rather, it would appear that if the RNA is unable to be
efficiently
spliced, it isdegraded in the nucleus.Thetat/rev intron 5' ss is
efficiently
utilized in a heterolo-gouscontext.The demonstration that the tat/rev intron 3'ssisinefficientis similar topreviousfindingswith the RSVsystem
(3,
20, 21). Moreover, the branchpoint region has also beenimplicated
in splicing inefficiency in the RSV system (16).Further work with RSV has identifiedan additionalcis-acting
element, NRS,
that functions todownregulate splicing
effi-ciency within the context of both RSV and heterologous
introns (1, 27, 28). To test for the presence of analogous
sequenceswithin theenvregionof HIV-1, thepSV, construct was used to examine theefficiency of the tat/rev intron 5' ss aloneorinthecontextofmostof theenvcoding region (Fig.
4A).
The construct with the tat/rev intron 5' ss,pSVHIVI,
contains 17 and 99
bp
of HIV-1 sequences upstream and downstream of the 5' ss,respectively.
Toaddress thepossibility
that theefficiencyof the tat/rev intron 5' ssisdownregulated by
distinct, cis-acting elements outside the
116-bp
fragment,
alarger portionof the HIV-1 genome
spanning
the 5'ss wasalsotested. pSVenv,B contains a 2.4-kb
fragment
of the HIV-1 genome which includes 258bpupstream of the 5' ssandmost of the tat/rev intron.Si
analysis
of the RNA from cells transfected with these constructs(Fig.
4B)
demonstrated that thetat/rev intron 5' ssisat leastasefficientasthe3-globin
5'HXB2
RF SF2 BRU
BH10
ELI MAL MN
PV22
Z2Z6
.39 -20
GLU CUUUCU AUAGU GAAUAGAGUUAGGCAGGGAU UUCACCA AUCG UCAG
GUGCUUUCU AUAGUG AAUAGAGUUAGGCAGGGAUA UCACCA UC U CAG GUGCUUUCU AUAGU GAAUAGAGUUAGGCAGGGAUACUCACCA GUC U CAG
GUACUUUCU AUAGUGAAUAGAGUUAGGCAGGGAUA UCACCA UCGU CAG
GUACUUUCUGUAGUGAAUAGAGUUAGGCAGGGAUA UCAC AU UCGU CAG
GUGCUUUCU UUAGU AAAUAGAGUUAGGCAGGGAUACUCACCU GUCGUU CAG
GUGCUUUCUUUAGU AAAUAGAGUUAGGCAGGGAUA UCACCUC GUCGU GCAG
GACUUUCU UAGUGAAUAGAGUUAGGCAGGGAUA UCACC AU GUCGU GCAG
G |ACUUUCU UAGU AUAGAGUUAGGCAGGGAU UCAC A U AUCGU CAG
GCUUUCU UAGU AAUAGAGUUAGGCAGGGAU CACCU U CAG
Val Lou Ser fie Val Asn Arg Val Arg Gin GlyF Tyr S.rS,rPro Lou Phe Gin
Lou Leu
Val
FIG. 2. Comparison of the nucleotide sequences comprising the
tat/rev intron 3' ss of various HIV-1 proviral clones. Sequences conserved in all 10proviral clones are boxed. At the extreme 3' end of each sequence is the AG dinucleotide that defines the 3' end of the tat/revintron. Immediatelyupstreamis the somewhat conserved poly-pyrimidinetractpreceded byalarge conserved region between 20 and 39 nt upstream of the AG dinucleotide. It is within this area that the critical A residue involved in formation ofthe RNA branch point is expected to reside. However, noobvious homologyto the 5'-UNCU-RAC-3' branchpoint consensus is detected within this region. The amino acid composition of the portion of the env gene product encoded by the nucleotide sequence is also shown. All HIV-1 proviral sequenceswereobtained from the GenBank data base.
ss, whether alone (Fig. 4B, lane 4) or in the context of the tat/revintron (Fig. 4B, lane 5). Therefore, unlike the inefficient
splicing in the RSV system, inefficient splicing of the tat/rev
intron cannot be ascribed to the action of cis-acting repressive sequenceswithin the intron.
DISCUSSION
Previous studies with simple retroviruses such as RSV have demonstrated that a suboptimal 3' ss and the presence of an NRSwhich downregulates the utilization of the 5' ss contrib-ute to the inefficient excision of introns within the viral RNA (1, 3, 16, 20, 21, 27, 28). That is, in order for their primary transcriptstoevade the cellularsplicing machinery most of the time, these simple retroviruses have evolved anintronic orga-nization whichcomprisesanintrinsically inefficient3' ss and a
distinct, cis-acting intron element which downregulates the
efficiencyofanotherwise efficient 5'ssbyacurrently unknown
mechanism.
Inaneffort to determine the basisfor theinefficientsplicing
of the tat/rev intron, chimeric introns composed of ,B-globin
and tat/rev intron splice sites were examined. The results obtained indicate that ifcis-actingsequenceswhich modulate splice site efficiencyarepresentwithin the
tatlrev
intron, theymust be context dependent. Deletion mapping demonstrated that, in the context of the constructs examined, the
require-ments for suboptimal splicing are intrinsic to the region
expectedtocompriseall of thesignalsof the
tatlrev
intron 3' ss, since removal of both 5' and 3' flanking sequences did not result inan increase insplicedRNA. Infact, analysisof the SB constructssuggeststhat sequences 3' of the AG dinucleotide of the 3' ss act toaugmentsplicing,
given that deletion ofthese sequences in pSVCBSB dramatically reduces the level ofsplicedproductrelativetothat obtainedwith either
pSVHTSB
orpSVCTSB (Fig. 1). This role ofexonsequencesinsplicing
is similartothatobserved in other systems(45), includingRSV
(16). Furthermore, the HIV-1 env sequences implicatedhere
(i.e.,nt8382to8462)encompassapurine-rich region
(data
notshown) similar to other exon sequences that have been
re-portedto affectsplicing
efficiency (45).
This observationsug-gests that these downstream
"splicing
enhancer" sequenceson November 9, 2019 by guest
http://jvi.asm.org/
[image:5.612.55.299.99.178.2]T2'
Isv4Of
i-i
i
CATpA
I
-40 -30 -20 -10
pSVH TSB TAGTGAATAGAGTTAGGCAGGGATATTCACCATTATCGTTTCAG
pSVHTSB*----
C-C-G-C---pSVHTSB"t-T---T--T--T
--T-B
603- Uw3l0-281l 271-
234-S14
194-1
2
3
4
5
*U
[image:6.612.85.540.79.292.2],<-gal
_
FIG. 3. Analysis oftat/rev intron3'sscontainingeitherbranchpointorpolypyrimidine tractmutations. (A)Schematicrepresentationofthe
constructs. ThepSVHTSB construct (Fig. 1A)was mutagenized by an invitro PCR-based method asdescribed in Materials and Methods to generatepSVHTSB*orpSVHTSBPPt.ThepSVHTSB* constructis identical topSVHTSB exceptfor the fourpointmutationsin theexpanded region (dashes denote unaltered bases).The AG dinucleotide that defines the 3' end of the intron is shownattheextreme3' endof thesequence. The three G-to-C mutations and the T-to-G mutationatpositions -29, -33, -35,and -31, respectively,relativetotheAGcreateanexact copy
ofthereported ,B-globin branchpointsequence,5'-CACTGAC-3'.Consequently,theAresidueatposition -30istheexpectedsite of RNA branch
formation in the case ofpSVHTSB*. The exactsite of RNA branch formation for the authentic tat/rev intron 3' ss in the context ofeither
pSVHTSBorthe HIV-1genomeis unknown. Toassesswhether thepolypyrimidinetractcould alsoplayarole indeterminingtheextentofsplicing
of thetat/revintron,thetractwasmutagenizedasindicatedtocreateacontinuous stretch of17pyrimidines, generatingpSVHTSBPPt. 3,,B-globin.
(B) S1nucleaseprotection analysis.A5-,g aliquotof total RNA from COS-7 cells transfected withpSVf (lane 2), pSVHTSB (lane 3), pSVHTSB* (lane 4), andpSVHTSBPPt (lane 5)wasused in the Si protectionreactions. Theprobeused for RNAanalysisof cells transfected withpSV,B is
identicaltothat used inprevious experiments (Fig. 1B),theprobeused forpSVHTSBis identical to that usedpreviouslyforpSVHTSB (Fig. 1),
and theprobesused forpSVHTSB*andpSVHTSBPPtaresimilartothat used forpSVHTSBexcept for thepointmutationsdescribed above.The
protectionpatternsforpSVHTSB, pSVHTSB*,andpSVHTSBPPt areasdepictedinFig.IC.Unspliced (U)andspliced(S)forms of the RNAs areindicated,thoseonthe leftreferringtoRNA derived frompSVI3 and thoseontheright referringtoRNAs derived frompSVHTSBandits
derivatives. Atthe bottom of thefigurearethe results obtaineduponparallelincubation withprobeto,B-galactosidase (,8-gal)mRNA. Numbers onthe left indicate molecular sizes(inbasepairs).
from diverse transcripts may have similar mechanisms of action.
Subsequent analysis of the region comprising the tat/rev
intron 3' ss revealed a striking conservation of sequences
between 20 and 39 nt 5' of the AG dinucleotide of the 3' ss
amongseveral HIV-1proviruses (Fig. 2). Althoughthisregion
encodesaportionof theenvprotein, giventhehighmutation rateof retroviruses and the redundancy of thegenetic code,
onewouldexpectsomevariation within the third base ofsome
codons within this region. The observed 100% conservation suggests another function for this region encompassing the putativesite of RNA branch formation. Basepairingbetween the branchpoint sequence on the pre-mRNA and the U2
snRNA is important for binding of the U2 snRNP to the pre-mRNA and subsequent biochemical reactions (32,48,49) andmaybeacriticalstepinthesplicingprocess(40).Because
thetat/revintron 3'ssdoesnotcontainanobviousbranchpoint
sequence, efficient base pairing with the RNA component of
the U2 snRNP maybe a rate-limiting step in splicingof the
tat/rev intron, resulting in inefficient splicing. Consistent with
this hypothesis, introduction ofan exactcopy of the 3-globin
branchpoint sequence within the highly conserved, putative branchpoint region converted the intrinsically inefficienttat/rev
intron 3' ssintoahighly efficient splice site. Thisdemonstrates
that theinefficiencyof thetat/revintron 3'ssis due,atleast in part, to the absence of a strong branchpoint consensus
se-quence and that the observed sequence conservation in this region of the HIV-1 genome maybe due to an evolutionary
constraint to maintain asuboptimal branch point, as in RSV (16). Thus, maintenance of a suboptimal branchpoint region
may be a common theme among retroviruses. Subsequent
examination of thepolypyrimidine tractof the
tatlrev
intron 3'ssalso revealedaconservation inthedisruptionof thetractby purine residues (Fig. 2). Mutations which converted these purines to thymidine resulted in a dramatic increase in the
efficiency of splicing (Fig. 3). However, the effect of the polypyrimidine tract mutation was marginally less than that
obtained with the introduction of the ,3-globin branch point (the branchpoint mutant yielded 94% spliced RNA, as
op-posedto87% splicedRNAobserved with thepolypyrimidine tract mutant). The introduction ofan efficient branch point
wouldappeartoprovideasignal thatcanfunctiondespitethe
suboptimal nature of the polypyrimidine tract. Similarly, im-provementof the polypyrimidine tractincreases splicing effi-ciency despite the absence of a consensus branchpoint
se-quence. Theincrease in splicing efficiency observed with the pSVHTSB* and pSVHTSBPPt mutants, in addition to ruling
outthepossibilitythatheterologous3'ss arenotfunctional in pSV3, demonstrates the absence of any other distinct cis-acting negative sequences within this region. Therefore, it
appears that a poor branchpoint region and a suboptimal
A
5i2sson November 9, 2019 by guest
http://jvi.asm.org/
p p
5'5s
3's,
r
H SX I
B
872-1
CAT |PA872-A
|I
I1 603-l
3'#o
v CAT I771 IAp
I
6027 SX
5ss
11027tfl4
1
2
3
4
5
_ t%S''I ,0
310-281. 271'
234-5Vsv
pSVenvp /. >
FIG. 4. Analysis of thetat/revintron 5'ss.(A) Schematic representation of theconstructs.pSVHIV,Band pSVenvl werederived by replacing
the,-globin (,B) 5' ssof pSV3 with portions of the HIV-1genomespanning thetat/revintron 5'ss.Theinitiation codon oftheenvopenreading
frame(hatched box) is indicated byanasterisk.(B) S1nucleaseprotection analysis.The end-labelled DNA probe used for analysis of theconstructs spanned the 3'ssandwasidenticaltothat used for pSV,B. Lane 1containsradiolabelled markers.A5-,ug aliquot of totalRNAfromCOS-7cells transfectedwithpSV,B (lane3), pSVHIV,B (lane 4), and pSVenv,B (lane 5)wasused intheS1 protection reactions.Asanegative control,total RNA
frommock-transfected COS-7 cells (lane 2)wasincubated with probe and digested with S1asnormal.Bands arisingfromS1-resistant hybrids
correspondingtounspliced(U) and spliced (S) RNAareindicated byarrows. Numbersonthe leftindicate molecular sizes (in base pairs).
polypyrimidinetractboth contributeto the inefficient splicing ofintrons containing thetat/rev intron 3' ss.
Havingidentifiedelements within thetat/revintron 3'ssthat are responsible for the inefficient splicing observed,we then
focused attention on other elements of the tat/rev intron. In particular, it was of interest to examine the efficiency of the
tat/rev intron 5' ss and to test for the presence ofnegative,
cis-acting intron elements analogoustothe NRSsequencesof
RSV (1, 27, 28).Uponexamination of theHIV-1tat/revintron 5' ssinaheterologouscontext,nosuch elementwasdetected.
Splicing ofchimeric intronscontaining thetat/rev intron 5' ss
with either minimal flanking sequences or the 5' ss in the context ofmostof the
tatlrev
intronwasatleastasefficientasthat of the ,-globin intron (Fig. 4). Moreover, there is no
evidence for the existence elsewhere in the tat/rev intron of cis-acting negative regulatory elements thatmodulatesplicing efficiency. From the dataregardingthetat/revintron 5' ssand
3'ssandtheireffectsonsplicing efficiency, itcanbe concluded
that theonlybasis forinefficientsplicingof thetat/revintronis the inefficientsignalspresentwithin the 3'ss.While itappears
that HIV-1 andRSV share somerequirements for inefficient
splicing (i.e., a suboptimal branchpoint region), their
respec-tiveunspliced RNAs differdramaticallyintheircapacitytobe transportedto thecytoplasm (8, 43, 46).
Different mechanisms for the achievement of suboptimal splicing in simple retroviruses (e.g., RSV) and HIV-1 could explain the observed differences in the metabolism of their respective mRNAs. Bothspliced andunspliced RSV mRNAs
are constitutively transported from the nucleus to the cyto-plasm(43). However, inthecaseofHIV-1,RNAscontaining
an intacttat/rev intron aresequestered within the nucleus in the absence of the viralprotein, Rev(12, 13, 19, 25). Experi-ments using subgenomic constructs of HIV-1 have demon-strated that theenvregioncontainsall thesignalsnecessaryfor nuclear sequestrationof viral mRNA observedin the absence of Rev (12, 13, 19, 25). There are currently two theories to
explainhowthese HIV-1 RNAsareretained in the nucleus: (i) partial recognitionofsplicesitesbycomponentsof the
splice-osome, resulting in the nuclear entrapment of pre-mRNA withinspliceosome complexes (4, 5, 23), and(ii) thepresence
ofdistinct, cis-acting repressive sequence (CRS) elements (7,
24, 33,36, 38).Ifsequestration of theHIV-1 RNA is the result of entrapment into spliceosome complexes because of the inefficiency of the splicing signals, then there must exist differences between HIV-1 and RSV that permit unspliced RNA of the latter to avoid the entrapping process. The
observationthatsplicinginefficiency in the RSVsystemisalso partly due to an inefficient branch point (16) suggests the
presence of either positive RNA export signals in RSV or
negative signals in HIV-1. To date, several distinct CRS-like elements have been identifiedthroughoutthe HIV-1transcript (7, 24, 33, 36, 38). Furthermore, the absence ofan NRS-like
elementwithintheenvregion ofHIV-1 may also explain the
observed difference in RNA metabolism. The NRS of RSV
mayfunctiontodownregulate recognition of the 5'ss.Failure to recognize the 5' ss mayprevent RSV primary transcripts from becoming committed to the splicing pathway, making them available fortransport tothecytoplasm. Support forsuch
amodel doesexist,sinceinactivationof the5'ssand 3'ssofan
intron has been shown to result in transport of unspliced mRNA into thecytoplasm (4, 5, 22).Inthecaseof the HIV-1
tat/revintron,the resultspresentedprovidestrong circumstan-tial evidence that boththe branchpointand thepolypyrimidine tract of the 3' ss interact poorly with the essential splicing components, U2 snRNP andU2AF, respectively. Poor
recog-nition of the polypyrimidine tract by U2AF implies that the recently described, ATP-independent early (E) splicing
com-mitment complex (29) composed of the pre-mRNA, Ul snRNP, and U2AF may not form efficiently with the tat/rev
intron of HIV-1. This implication is inconsistent with the splice-site-mediated model for nuclear retention of HIV-1 structuralmRNA.Instead,onthebasis of the data outlined in this paper, it is tempting to suggest that sequences distinct from the HIV-1tat/revsplicing signals playarole inmediating nuclear retention of structural mRNA. Indeed, recentresults indicate that the signals for suboptimal splicing and nuclear retentioncanbesegregated inthecase of HIV-1 (42a).
Insummary, the data presentedin this paperindicate that the balancedsplicingof the HIV-1tat/revintronis duesolelyto
the actionofsignalswithin the 3'ssand theneighboringexon.
There is evidence that a purine-rich sequence within the 3' exon acts to increase recognition of the tat/rev intron 3' ss
A
psvp
epSVHIV,B
on November 9, 2019 by guest
http://jvi.asm.org/
[image:7.612.115.500.79.235.2]despite
thesuboptimal
natureof boththe branchpointand thepolypyrimidine
tract; deletion of thepurine-rich regionrenders the tat/rev intron 3' ss completely nonfunctional. Having identified the basis for inefficientsplicing
of the HIV-1 tat/revintron,
ourinterest is nowfocusedonestablishingthe mecha-nismfor the nuclearsequestration
of HIV-1 structuralprotein mRNA. To test the various possibilities (some of which are mentioned above),chimericconstructscomprisingportionsof the RSV and HIV-1 genomes are being tested to identify sequence elements that underlie the observed difference innucleocytoplasmic transport of theirrespective mRNAs.
ACKNOWLEDGMENTS
A. Staffa issupported by anNSERC student scholarship, and A. Cochrane is the recipient of an FRSQ chercheur boursier. This researchwassupported bygrantsfrom the Medical Research Council of Canada.
WethankP.Harakidas and the other members ofourlaboratoryfor theirhelp andsupportthroughout thecourse of thiswork,P. Liston and D. J.Briedis forkindly providingaccess to HIVsequencesfrom
GenBank,andN. H.Acheson for his adviceandsuggestions.
REFERENCES
1. Arrigo, S., and K. Beemon. 1988. Regulation of Rous sarcoma
virusRNAsplicingandstability.Mol.Cell. Biol. 8:4858-4867. 2. Arya, S. K., C. Guo, S. F. Josephs, and F. Wong-Staal. 1985.
Trans-activator gene of human T-lymphotrophic virus type III
(HTLV-III).Science229:69-73.
3. Berberich, S. L., and C. M. Stoltzfus. 1991. Mutations in the
regionsof the Rous sarcomavirus3'splicesites:implicationsfor
regulationofalternativesplicing.J. Virol. 65:2640-2646.
4. Chang, D. D., and P. A. Sharp. 1989. Regulation by HIV Rev
dependsuponrecognitionofsplicesites.Cell59:789-795.
5. Chang, D. D.,and P.A.Sharp. 1990. MessengerRNA transport
and HIVrevregulation.Science249:614-615.
6. Chomczynski, P.,and N.Sacchi.1987.Single-stepmethod of RNA isolation byacidguanidinium thiocyanate-phenol-chloroform
ex-traction. Anal. Biochem. 162:156-159. 6a.Cochrane,A.Unpublisheddata.
7. Cochrane,A.W.,K.S.Jones,S.Beidas,P.J. Dillon,A. M.Skalka, and C. A. Rosen. 1991. Identification and characterization of
intragenicsequenceswhichrepresshuman immunodeficiency
vi-russtructural geneexpression.J.Virol. 65:5305-5313.
8. Coffin,J. M. 1990. Retroviridae and their replication, p.
1437-1500. In B. N. Fields and D. M. Knipe (ed.), Virology. Raven Press,NewYork.
9. Cullen,B. R. 1988. Useofeukaryotic expression technologyinthe functional analysisof clonedgenes. Methods Enzymol. 152:684-704.
10. Cullen,B. R.1991. Humanimmunodeficiencyvirusas aprototypic
complexretrovirus. J. Virol. 65:1053-1056.
11. Dayton,A.I.,J.G.Sodroski,C. A.Rosen,W. C.Goh,and W. A. Haseltine. 1986. The transactivator gene of the human T-cell
lymphotrophic
virus type III is required for replication. Cell 44:941-947.12. Emerman, M., R. Vazeux, and K. Peden. 1989. The rev gene product of the human immunodeficiencyvirus affects
envelope-specificRNAlocalization.Cell57:1155-1165.
13. Felber,B. K.,M. Hadzopoulou-Cladaras,C.Cladaras, T. Cope-land, andG.N.Pavlakis. 1989.Therevproteinof HIV-1 affects
the stabilityand transport of the viral mRNA. Proc. Natl. Acad.
Sci. USA 86:1495-1499.
14. Felber, B. K., and G. N. Pavlakis. 1993. Molecular biology of HIV-1: positive and negative regulatory elements important for virusexpression.AIDS7:S51-S62.
15. Fisher,A.G.,M. B.Feinberg,S. F.Josephs, M. E. Harper, L. M. Marselle,G.Reyes,M. A.Gonda,A.Aldovini,C.Debouk, R. C. Gallo, and F. Wong-Staal. 1986. The transactivator gene of HTLV-III is essential for virus replication. Nature (London) 320:367-371.
16. Fu, X.-D.,R. A.Katz,A. M. Skalka, andT.Maniatis. 1991. The
role of branchpoint and 3'-exon sequences in the control of balancedsplicingof avianretrovirus RNA. Genes Dev.5:211-220. 17. Ge, H.,andJ. L.Manley. 1990. Aprotein factor,ASF, controls cell-specificalternativesplicingofSV40earlypre-mRNAin vitro. Cell 62:25-34.
18. Green, M. R. 1991. Biochemical mechanisms ofconstitutive and regulated pre-mRNA splicing.Annu. Rev. CellBiol.7:559-599. 19. Hammarskjold,M.-L.,J. Heimer,B.Hammarskjold,I.Sangwan,
L.Albert,and D. Rekosh. 1989.Regulationof human immunode-ficiency virus env expression by therev gene product. J. Virol. 63:1959-1966.
20. Katz,R.A.,M.Kotler, and A. M.Skalka. 1988.cis-acting intron mutations that affect the efficiency of avian retroviral RNA splicing: implicationformechanismsof control. J.Virol. 62:2686-2695.
21. Katz, R. A., and A. M. Skalka.1990. Control ofretroviral RNA splicing through maintenance of suboptimalsplicing signals. Mol. Cell. Biol. 10:696-704.
22. Legrain, P., and M. Rosbash. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57:573-583.
23. Lu,X., J.Heimer,D.Rekosh,and M.-L.Hammarskjold. 1990.Ul small nuclear RNA plays a direct role in the formation of a rev-regulated human immunodeficiency virus env mRNA that remainsunspliced. Proc. Natl.Acad. Sci. USA87:7598-7602. 24. Maldarelli,F., M. A.Martin, and K. Strebel. 1991.Identification
of posttranscriptionally active inhibitory sequences in human immunodeficiency virustype1 RNA: novel levelof gene regula-tion. J. Virol. 65:5732-5743.
25. Malim, M. H., J. Hauber, S.-Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA.Nature (London) 338:254-257.
26. Maniatis,T.1991. Mechanismsof alternative pre-mRNAsplicing. Science 251:33-34.
27. McNally, M. T., and K. Beemon.1992. Intronic sequences and 3' splice sites control Rous sarcomavirus RNA splicing. J. Virol. 66:6-11.
28. McNally, M. T., R. R. Gontarek, and K. Beemon. 1991. Charac-terization of Rous sarcoma virus intronic sequences that nega-tively regulate splicing. Virology185:99-108.
29. Michaud,S., and R. Reed. 1993. A functional associationbetween the 5' and 3'splice sites is established inthe earliest
prespliceo-somecomplex (E) in mammals. GenesDev.7:1008-1020. 30. Moore, M. J., C. C. Query, and P. A. Sharp. 1993. Splicing of
precursors to mRNA by the spliceosome, p. 303-357. In R. F. Gesteland and J. F.Atkins (ed.), The RNA world. Cold Spring Harbor LaboratoryPress, Cold Spring Harbor,N.Y.
31. Muesing, M. A., D. H. Smith, and D. J. Capon. 1987.Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell 48:691-701.
32. Nelson,K.K., and M. R.Green. 1989. Mammalian U2 snRNPhas
a sequence-specific RNA-binding activity. Genes Dev. 3:1562-1571.
33. Rosen,C. A., E. Terwilliger, A. Dayton, J. G. Sodroski, and W. A. Haseltine. 1988. Intragenic cis-acting art gene-responsive se-quencesof thehuman immunodeficiencyvirus. Proc. Natl. Acad. Sci.USA85:2071-2075.
34. Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: alaboratorymanual, 2nd ed. Cold Spring Harbor Labo-ratoryPress, Cold Spring Harbor, N.Y.
35. Sanger,F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467.
36. Schwartz,S., M. Campbell, G. Nasioulas, J. Harrison, B. Felber, and G. Pavlakis. 1992. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independentgagexpression. J. Virol.66:7176-7182. 37. Schwartz, S.,B. K.Felber, D. M. Benko, E.-M.Fenyo, and G. N.
Pavlakis. 1990.Cloning and functional analysis of multiply spliced mRNAspeciesofhumanimmunodeficiency virus type 1. J. Virol. 64:2519-2529.
38. Schwartz, S.,B. K. Felber, and G. N. Pavlakis. 1992. Distinct RNA
on November 9, 2019 by guest
http://jvi.asm.org/
sequencesin thegagregion of humanimmunodeficiencyvirustype 1 decreaseRNA stabilityandinhibit expressionin the absence of
Rev protein. J. Virol. 66:150-159.
39. Smith, C. W. J., J. G. Patton, and B. Nadal-Ginard. 1989.
Alternative splicing inthecontrol ofgeneexpression. Annu. Rev.
Genet.23:527-577.
40. Smith, C.W.J., E. B. Porro,J. G.Patton, and B. Nadal-Ginard.
1989. Scanning from an independently specified branch point
definesthe 3' splice site of mammalianintrons. Nature (London) 342:243-247.
41. Sodroski,J.,W. C. Goh, C. Rosen, A. Dayton, E. Terwilliger, and
W. A. Haseltine. 1986. Asecondpost-transcriptional transactivator
gene required for the HTLV-III replication. Nature (London) 321:412-417.
42. Sodroski,J.,C. A. Rosen, and W.A.Haseltine. 1984.Tranis-acting transcriptional activationofthe longterminalrepeatof human T
lymphotropic viruses in infectedcells. Science 225:381-385. 42a.Staffa, A., andA. Cochrane.Unpublisheddata.
43. Stoltzfus,C. M. 1988. Synthesis andprocessingof aviansarcoma
retrovirus RNA.Adv. Virus Res. 35:1-38.
44. Terwilliger, E., R. Burghoff, R. Sia,J. Sodroski, W. Haseltine, and
C. Rosen. 1988. Theartgeneproduct of the human immunodefi-ciency virus is required for replication.J.Virol.62:655-658. 45. Watakabe,A., K. Tanaka, and Y. Shimura.1993.The role ofexon
sequencesinsplice site selection. Genes Dev. 7:407-418.
46. Wong-Staal,F. 1990. Humanimmunodeficiencyvirusesandtheir replication, p. 1529-1543. In B.N. Fields and D. M. Knipe(ed.), Fields virology. Raven Press, New York.
47. Wright, C.M., B. K.Felber,H.Paskalis, and G.N. Pavlakis. 1986. Expression and characterization of thetrans-activatorof HTLV-III/LAV virus. Science 234:988-992.
48. Wu,J., and J. L. Manley. 1989. Mammalian pre-mRNA branch
site selection by U2 snRNP involves base pairing. Genes Dev. 3:1553-1561.
49. Zhuang, Y., and A.M. Weiner.1989.Acompensatorybasechange in human U2snRNAcansuppress abranch site mutation.Genes
Dev. 3:1545-1552.
![FIG. 1.verticalboxes])signalenzymeuniqueprotectionradiolabelledparalleland6,protectedSchematicheterologous1 contains and (A) Schematic representation of pSV13 and its SB derivatives](https://thumb-us.123doks.com/thumbv2/123dok_us/1293837.82351/4.612.81.529.81.430/fig-verticalboxes-signalenzymeuniqueprotectionradiolabelledparalleland-protectedschematicheterologous-contains-schematic-representation-derivatives.webp)