Rapid and Concerted Evolution
of
Repeat Units in
a
Balbiani Ring Gene
U.
Lendahl,* H. Saiga,t'*
C.
Hoog,* J.-E. Edstromt and
L.
Wieslander*.'
*Department of Molecular Genetics, Medical Nobel Institute, Karolinska Znstitutet, S-104 01 Stockholm, Sweden, ?Department of Genetics, The Wallenberg Laboratory, University of Lund, S-220 07 Lund, Sweden, and *Department of Molecular Biology,
University of Occupational and Environmental Health, 1-1 Zseigaoku, Kitakyushu, 807 Japan Manuscript received March 3, 1987
Revised copy accepted May 30, 1987
ABSTRACT
The Balbiani ring (BR) genes in the midge Chironomus, a genus belonging to Diptera, code for large secretory proteins, used to construct the larval tube. The 15-23-kb long core block in each gene consists of an array of tandemly arranged approximately 200-bp long repeat units, where a single repeat unit is composed of a constant and a subrepeat region. In order to investigate the evolutionary fate of highly repetitive coding DNA, the BRl core block in Chironomus pallidivittatus was characterized and compared to the orthologous core block in the sibling species Chironomus tentans. We find that the 75-100 repeat units in the B R l y core block have evolved in an unusual fashion. In all repeat units the constant regions display an expected high degree of homology between the two species, 94% at the nucleotide level. In contrast, the subrepeat regions in all repeat units have diverged concertedly, both as to length, number and sequence of the subrepeats. The observed changes in all repeat units of the core block probably have occurred after speciation of C. pallidivittatus and C. tentans. These findings demonstrate that a tandemly reiterated coding sequence can rapidly and concertedly convert into a related sequence, much in the same way as has been described for satellite DNA.
EPETITIVE DNA more frequently seems to be
R
subject to dramatic evolutionary changes than nonrepetitive DNA. This is most conspicuously dem- onstrated in noncoding satellite DNA sequences, where millions of bp can rapidly evolve by introduc- tion of identical mutations into all the tandemly ar- ranged, short repeats (LEWIN 1983). However, also in genes that are internally repetitious or repeated as a number of gene copies, e.g., in sgs (MUSKAVITCH and HOGNESS 1982), silk fibroin (SPRAGUE et al. 1979), collagen (YAMADA et al. 1980) and globin genes (HILL et al. 1985) dramatic changes like length polymor- phisms or deletions of entire genes do occur.To study the correlation between extreme repeti- tion in coding sequences and the rate and mode of evolution we have analyzed recent evolutionary diver- gence in a family of internally repetitive genes, the Balbiani ring (BR) gene family in the dipteran genus Chironomus. T h e BR genes encode very large struc- tural proteins, -1
O6
daltons (EDSTROM, RYDLANDER and FRANCKE 1980; RYDLANDER, PIGON and EDSTROM1980; KAO and CASE 1985), which are the main con- stituents of the larval tube, an extracorporeal struc- ture necessary for feeding and protection of the aquatic larva (GROSSBACH 1977). BR genes have been characterized at the molecular level in three Chirono- mus species: C. tentans, C. pallidivittatus and C. thummi.
T h e sequence data presented in this article have been submitted to the EMBL/GenBank Data Libraries under the accession number Y00615.
To whom all correspondence should be sent. Genetics 117: 43-49 (September, 1987)
All BR genes are highly repetitive and have a common architecture (see Figure 1). Each gene contains a 15- 23-kb long core block, built from one type of tandemly arranged repeat unit, 150-270 bp in length (PUSTELL et al. 1984; WIESLANDER et al. 1984 and references therein). A repeat unit consists of a constant region and a subrepeat region; the latter is built from shorter, tandemly arranged subrepeats. Constant regions are 4 9 4 5 % conserved at the nucleotide level between the different BR genes, while subrepeat regions differ both in composition, length and number of subre- peats. T h e contemporary hierarchically repetitive BR genes have probably evolved from a short common primordial sequence akin to part of a constant region by duplications and stepwise remodelings, creating successively more complex repetitive coding se- quences (SUMECI, WIESLANDER and DANEHOLT 1982; C. HOOc and L. WIESLANDER, unpublished data). Duplicative translocations have during evolution re- sulted in the limited number of BR genes, which are located in separate chromosomal loci. T h e BR genes thus exemplify how sequence duplication has played an important role in intragenic evolution as well in the evolution of gene families (LI 1983).
sions (BEERMANN 1955). Moreover, if crossed under laboratory conditions they give viable offspring. Nu- cleotide sequence analysis has shown that the orthol- ogous BR2a, BR2P and BR6 core blocks are virtually identical in the two species. By contrast, the C. palli- divittatus B R l a core block, which presumably is lo- cated in a separate gene in the BRl chromosomal region (GALLER et al. 1984), lacks an equivalent in C. tentans, probably due to a recent deletion of the corresponding core block in C. tentans (LENDAHL and WIESLANDER 1985). This unexpected difference stim- ulated us to search for further rapid evolutionary events, correlated with the extreme degree of repeti- tion in the BR genes.
In this report we describe the cloning and charac- terization in C. pallidivittatus of the equivalent to the BRly core block in C. tentans (WIESLANDER, SUMEGI and DANEHOLT 1982). We find that the large core block has evolved in an extraordinary way. All repeat units have diverged concertedly, but whereas constant regions retain an anticipated high degree of homol- ogy, subrepeat regions have diverged to such an ex- tent that direct homologies are very low. These ob- servations bring new information on the evolution of the entire BR gene family and on how highly reiter- ated coding sequences rapidly can evolve in a complex and satellite DNA like fashion.
M A T E R I A L S A N D M E T H O D S
Isolation of DNA and RNA: C. pallidivittatus and C. tentans DNA was extracted according t o SUMEGI, WIESLAN-
DER a n d DANEHOLT (1 982). Total R N A a n d 7 5 s RNA from salivary glands was prepared as described by GALLER et al. (1985) and RYDLANDER, PICON a n d EDSTROM (1980), re- spectively.
Cloning procedures: A cDNA library in pBR 322, rep- resenting C. pallidivittatus 7 5 s RNA, was screened (GRUN-
STEIN a n d HOCNESS 1975), using the cDNA insert from t h e C. tentans B R l y clone pCt21 (WIESLANDER, SUMECI a n d DANEHOLT 1982) as probe. Four positively hybridizing re- combinants were selected for sequence analysis. B R l y sub- repeat region specific subclones for C. pallidivittatus a n d C.
tentans were obtained by subcloning a 68-bp DNA segment from pCt2 1 [positions 15 1-2 18 in WIESLANDER, SUMECI a n d DANEHOLT (1 982)] a n d a 120 bp DNA segment from pCp20 (positions 53-172, Figure 2) into M13 m p 8 (MESSING a n d VIEIRA 1982) a n d GEM-1 (MELTON et al. 1984) vectors.
D N A sequencing: Purified C. pallidivittatus cDNA inserts were progressively shortened by exonuclease Ba13 1 , blunt ended, recloned into t h e M 1 3 m p 8 vector a n d sequenced (SANGER a n d COULSON 1978), using 35S-dATP.
Filter hybridization techniques: T e n micrograms of total genomic C. pallidivittatus o r C. tentans DNA were digested with the restriction enzymes indicated, u n d e r conditions suggested by the manufacturer (New England Biolabs). All DNA concentrations were determined by fluorography (LA-
BARCA a n d PAICEN 1980). T h e DNA fragments were sub- jected t o separation in 1% agarose gels a n d blotted to nitrocellulose filters (SOUTHERN 1975). T h e filter-bound DNA was hybridized t o 32P-labeled in vitro synthesized RNA (MELTON et al. 1984; LENDAHL a n d WIESLANDER 1987)
...._ ._...
m2 0-
BR2 -B
BR6 I
-
C U
FIGURE 1.-Principle structure of the C. tentans BR gene core blocks. (Top) Part of a BR core block with tandemly arranged repeat units is illustrated. (Below) In higher magnification, repeat units from the BRIT, BR2a, BR2P and BR6 core blocks are depicted. The different repeat units are drawn in the same scale to illustrate the small length differences. The C. tentans BRIT, BRZa, BR2@ and BR6 core blocks are located in separate genes. In each case approximately 75-100 tandemly arranged repeat units of one type form the core block. A repeat unit is composed of two regions: a constant region (c) and a subrepeat region (sr). The constant region lacks internal repetition and is the most conserved element, as compared between the different paralogous core blocks (homolo- gies at the nucleotide level ranging from 49 to 85% between B R l y , BR2a, BR2P and BR6 in C. tentans). The subrepeat region is in all repeat unit types built from a number of tandemly arranged short subrepeats. Due to the difference in both nucleotide sequences, length and number of subrepeats between the different core blocks direct sequence homologies are much lower in the subrepeat re- gions.
corresponding t o the subrepeat regions of C. tentans a n d C . pallidivittatus B R l y genes, at 4 7 " in 5 X S E T (1 X S E T is 1 5 0 mM NaCI, 30 mM Tris-HCI (pH 8.0), 1 mM EDTA), 5 0 % formamide. Following hybridization, t h e filters were washed, final wash 2 X 3 0 min in 0, 1 X SET, 6 0 " , a n d autoradiographed.
S1-like mapping experiments: In each S1-like mapping experiment total R N A from three salivary glands was used. T h e in vitro transcription of probes a n d t h e S l-like mapping were performed according to MELTON et al. (1984), with minor modifications (LENDAHL a n d WIESLANDER 1987).
RESULTS
Structure and genomic organization of the C. pal-
Zidivittutus homolog to the C. tentuns B R l y core
block: Four C. pallidivittatus cDNA recombinant plas- mids that hybridized to the repeat unit of the C. tentans B R l y core block were selected for detailed studies. T h e inserts were sequenced and in total the sequence of 925 bp was determined (Figure
2).
All four inserts are very closely related though unique, as demonstrated by the 17 base substitutions found when the inserts were aligned and compared. Of these 17base substitutions, 10 were silent mutations and one a conservative Lys + Arg substitution, indicating a selective pressure on the sequence.
Evolution Gene
pcp 2n: pcp 26: pcp 1: pcp 28:
pcp 20:
pcp 26:
pcP 1: pcp 28:
pcp 20:
pcp 26:
pcP 1:
pcp 28:
pcp 20:
pCp 26:
pcP 1: pcp 28:
Lyi Pro Ser Lyi Sar Cly Pro ArR Pro Ser Lyi Ser Cly Pro ArR Pro Scr Cln Scr c l y Pro Arg Pro Ser Lyi Ser Cly Pro Lya Pro
I I LY 1 I p a
1 M
Sar Thr Sar Cly Pro Arg Pro Ser Lye Ser Cly Pro Arg Pro Ser Lyi Set Cly Pry Lya P r o S.r Lyi S.r Cly Pro Arg Pro C l u Lyi
L V l ISer I I
I
C Y ~ Cly k r A h Met Aru Lyi Ala Clu Ala Clu I.ya Cva Ala Aru Ara Lya c l y Arg Phc A m Ala A m Lyi Cyi Arg Cy. Thr Scr A l e k r Scr
300
CGT MACCA AGC MA TCA G& CCT AGA CCA AGC AMTCA
FIGURE 2.-Nucleotide and amino acid sequences for the four C. pallidiuittahcs BRly cDNA inserts. T h e four clones are aligned in the constant regions. All substitutions are denoted at corresponding positions. Every tenth nucleotide position is indicated by a dot. Subrepeat regions are underlined and the borders between individual subrepeats are marked by short vertical lines.
bp sequence which is tandemly repeated a t least 8
times in the subrepeat region. According to Southern blot experiments the total repeat unit size does not exceed 300 bp (Figure 3, lane IC). Altogether, 75-
100 repeat units of the BRl type are present in the genome of C. pullidivittutus, determined from titration experiments using cloned material as standards (Fig- ure 3, lanes IC,
g
and h). T h e repeat units are tan- demly arranged, as revealed by the ladder pattern on Southern blots after partial cleavage with a restriction enzyme cleaving onceper
B R l y repeat unit (Figure3, lane lk).
Comparison of the homologous C. paZZidivittutus and C. tentuns
BRly
core block sequences: A com- parison of the homologous gene sequences from the two species revealed several interesting features (Fig- ure 4a). T h e two sequences were aligned in the con- stant regions, which are conserved t o 94% at the nucleotide level, thus explaining the hybridization using theC.
tentans B R l y probe in the initial screen- ing experiments. Among the 27 amino acids between the first and last cysteine residues in the constant region only one substitution was found (Lys + Asn) at position 231 in Figure 2. In situ hybridization experiments using one of the characterized C. pulli-divittutus clones, pCp 20, as
probe
resulted in hybrid- ization only to the BRl locus on C. pullidivittutus polytene chromosomes (data not shown). Based on its location in the BRI locus and the 94% conservation in the constant region between the two species we will designate the characterized C. pullidivittutus core block BR 1 y.In contrast t o the expected homologies in the con- stant regions, subrepeat regions in the two BR 1 y core
1
2
abc defghi
k
ab c defghi
2.60
0.50
0.16
FIGURE 3.-Analysis of the distribution of the B R l y core block subrepeat types in C. pa1lidivittatu.s and C. tentans by Southern blot hybridization. Lanes are as follows: 10 r g genomic DNA from C.
tentans (b) and from C. pallidivittatus (c). completely digested with
Rsal. T e n r g genomic DNA from C. pallidivittahcs (k), partially
RsuI digested. DNA from the subcloned C. fentans BRly 33-bp subrepeat type in amounts corresponding to one (e) and five (f) repeat unit copies in 10 a g genomic Chirnomus DNA. DNA from the subcloned C. pallidivittatus BRly21 bp subrepeat type in amounts corresponding to one (g) and five (h) repeat unit copies in 10 rg genomic Chironomus DNA. DNA from subcloned subrepeat regions representing the C. tentans BR2a core block (a), the C.
pallidivittatuc B R l a core block (d), and the C. tentans BRlB core block (i) (C. H M G and L. WIESLANDER. unpublished data). in amounts corresponding to five repeat units in 10 Pg genomic Chironomus DNA. T h e amounts of plasmids in all control experi- ments were calculated according to LENDAHL and WIESLANDER
(1985), taking into account the size of the subcloned subrepeat region, the vector and the size of the Chironomus genome. South- em blots from lane a t o k in the left part of the figure (1) were hybridized with the C. pal1idivittatu.s BRl y subrepeat region specific probe and lane a to i in the right part of the figure (2) with the C.
46
0 500 io00 1500 bp
a.
b.
3’ untronsloted
3’ exon mglm
I . I .
0
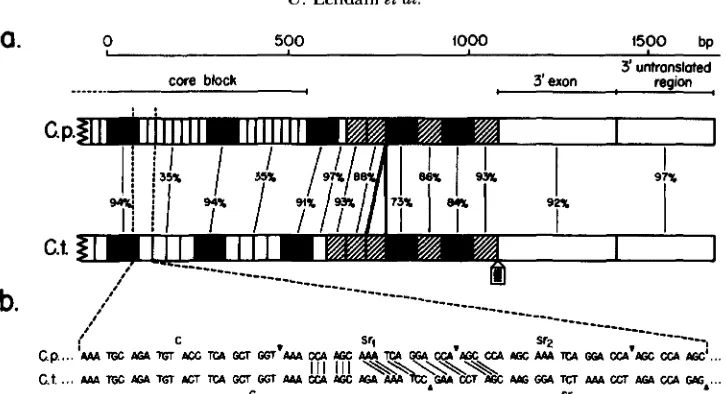
FIGURE 4.-a, Schematic comparison of the C. pallidivittatw and C. tentans BRly genes from the 3’ part of the core block to the poly A addition site. The two genes are perfectly aligned from the end of the core block to the poly A addition site except that C. tentuns contains an extra Cys-1 element at approximately position 750 (SAIGA et al. 1987; H%c, ENCBERC and WIESLANDER 1986). Likewise, the intron around position 1000 is not present in the C. pallidivittutus sequence, since this sequence is derived from a cDNA clone. Filled areas represent constant regions, short nonfilled areas subrepeat regions, hatched areas Cys-1 elements, dotted areas the intron and the two large open areas at the extreme right the 3‘ exon and the 3’ untranslated region. The homologies between C. fullidiuittatus and C. tentuns in the different regions are expressed as direct homologies at the nucleotide level since no deletions/insertions exist. In the core block the homologies between the subrepeat regions are calculated by comparing the four subrepeats of C. tentuns to six subrepeats in C. pallidivittatus, aligned in the 5 ’ ends. Homologies in the constant regions are calculated from the first to the last cysteine codons. b, Comparison of the nucleotide sequences from the constant/subrepeat region junction in the C . pallidivittutw and C. tentuns BRly core blocks. The sequences are aligned in the constant regions. The homologies between a 21-bp subrepeat in C. pallidivittutus and a 24-bp sequence on the border between the constant and subrepeat regions in C. tentuns are outlined. A vertical line indicates a conserved nucleotide. The borders between constant regions (c) and subrepeat regions and between individual subrepeats (sr) are represented by small triangles.
blocks are radically different. In fact, both the nucleo- tide composition and the length of the individual subrepeats as well as the number of subrepeats in the subrepeat region are different. In spite of the low direct sequence homology between the two subrepeat regions similarities do exist. All seven amino acids, RPSKSGP, encoded in the C. pallidivittatus subrepeat region are present in the 11 amino acid long encoded in the subrepeat of C. tentans, KPSKGSKPRPE, though in a different order. Furthermore, the total subrepeat region length is similar. This is due to a compensatory larger number, at least eight, of the short subrepeat in each subrepeat region in C. palli- divittatus compared to four copies in C. tentans.
We then investigated if each type of subrepeat region is restricted exclusively to one species, or if either of the two types is present in both species. Subclones specific to the two subrepeat regions were constructed and used as hybridization probes to Southern blots of genomic DNA from both species (Figure 3). Hybridization conditions were chosen such that no cross-hybridization to other BR gene se- quences was detectable (Figure 3, lanes a, d and i), while a single copy of the B R l y repeat unit in the genome would still have been detected (Figure 3, lanes e-h). T h e results demonstrated that the
2
1 -bp type of subrepeat region is found only in the C. pallidivittatus genome (Figure 3 , lane c) and that the 33 bp type isconfined to the C. tentans genome (Figure 3, lane b).
Heterogeneity within BR gene core blocks: We
also analyzed the degree of heterogeneity between repeat units within each of the B R l y core blocks. This was accomplished by hybridizing subrepeat region specific probes to their corresponding mRNA in an S1-like analysis (Figure 5, lane 1). All subrepeat re- gions in the core block are transcribed but only those that are identical to the probe sequence will form perfect RNase resistant hybrids. Shorter than full length hybrids are the result of transcribed subrepeat regions not completely homologous to the cloned probe. Here we observe that the heterogeneity in the C. pallidivittatus B R l y core block is greater than in the C. tentans B R l y core block, where fewer short bands were seen (Figure 5, lanes 1 and 2). As a comparison we also analyzed the heterogeneity be- tween the two species in the BR2 region, where two core blocks, BR2a and BR2B, have been shown to exist in separate genes (GALLER et al. 1985; C . HMG
BR Gene
c.p.
C.t.
1
34
2
34
M
FIGURE 5.-Analysis of the heterogeneity in the BRIT, BR2a and BR2@ core blocks in C. pallidivittatus and C. tentans. The hybrids of the S1-like mapping experiments were subjected to
separation in polyacrylamide-urea gels, which were dried on filter paper and autoradiographed. In lanes 1 , 3 and 4 to the left (C.p)
total RNA from C. pallidbittatus was used and in lanes 2 , s and 4 to the right (C.t) total RNA from C. tentans. As probes were used
34P-labeled in vitro transcripts corresponding to (1) 120 bp of the C. pallidivittatus B R l y subrepeat region, (2) 68 bp of the C. tentans
BR 1 y subrepeat region, (3) 50 bp of the C. tentans BR2@ subrepeat region and (4) 80 bp of the C. tentans BR2a subrepeat region. Lane (M) is a size marker (Sau 3A digested plasmid pBR 322 DNA) with the following fragment lengths, from top to bottom 105. 91, 781 75,46. 36, and 3 1 nucleotides.
i.e., in the band patterns no differences could be
discerned (Figure 5, lanes 3 and 4).
DISCUSSION
Evolution of the B R l y core
block
Several lines of evidence indicate that the diverged B R l r core blocks are orthologous sequences. First of all, they are 94% conserved in the constant regions. This means that they are considerably more similar to each other than to any other BR core block constant region, where homologies range from 49 to 85%. T h e B R l r core blocks are also present at the same chromosomal location in both species. T h e 3’ untranslated regions of the BRl genes ( H W , ENCBERC and WIESLANDER1986; SAICA et al. 1987) are in addition more closely related to each other than to the corresponding re-
gions in any other BR gene (C. Hijijc and L. WIES- LANDER, unpublished data). T h e two short regions between the core block and the 3’ untranslated re-
gion, the 3‘ block and the 3’ exon, are also highly homologous between the two BRl genes. However, in these particular regions the
C.
pallidivittatus BRl gene exhibits an even higher homology to the corre- sponding part of a BR2 gene in both species (SAICA etal. 1987; C. H W and L. WIESLANDER, unpublished data), probably due to a gene conversion event. Fi- nally, while all other BR gene core blocks except one are close to identical in both species (LENDAHL and WIESLANDER 1985), each B R l r core block, with its specific type of subrepeat region, is exclusively present in one species. In conclusion, we believe that the
C.
pallidivittatus and
C.
tentans B R l r core blocks are orthologous sequences that have diverged in a con- certed fashion late in evolution, presumably after speciation of the two sibling species.T h e evolutionary divergence has resulted in two B R l r core blocks with a pattern of alternating high and low homologies, in the constant and subrepeat regions respectively (Figure 4a). We envisage that the first step in the divergence was a transition from one subrepeat type to the other, somewhere in the core block. Such a local transition would only require three base pair substitutions, two of which are silent, and one in frame deletion to transform a 24 bp sequence at the
C.
tentans constant/subrepeat region junction to the 21 bp subrepeat ofC.
pallidivittatus (Figure 4b). T h e new subrepeat could then have duplicated locally to form a new subrepeat region, e.g., by slippage duringD N A
replication and repair, a mechanism known to operate in tandemly repeated shortD N A
sequences (FARABOUCH et al. 1978; JONES and KAFA-TOS 1982). Concomitant with the changes in the sub- repeat region, a juxtaposed constant region acquired base pair substitutions changing two codons.
As
the final phase in the core block evolution we believe that the new repeat unit, consisting of the new subrepeat region and the slightly modified constant region, spread by gene conversion(KLEIN
and PETES 1981)or unequal crossing over
(SMITH
1976) throughout the core block at the expense of the preexistingtype.
T h e spreading of the new subrepeat and constant region could however also have occurred in two in- dependent homogenization waves. T h e high degree of tandem repetition at the repeat unit level facilitates the aligning out of register necessary for unequal crossing-over to occur and structural observations in other BR genes support this view ( H W and WIS
larger heterogeneity in the C. pallidivittatus core block (Figure 5, lanes 1 and
2)
could argue for a more ancient origin, as could the presence in the 3‘ coding region in both species of the sequence CCAA- GCAAATCTGAACCAAGA (HOiic, ENCBERG and WIESLANDER 1986; SAIGA et al. 1987), which is very similar to the C. paElidivittatus type of subrepeat. On the other hand, such a shared sequence could instead be the founder of a more recently evolved C. pa2lidi- vittatus core block, especially since it is juxtaposed to a variant of a constant region. We also can not rule out the possibility that the two core blocks have di- verged to about the same extent, but in separate directions, from a common ancestor sequence present at speciation. Whichever alternative is correct, we demonstrate the fixation of a new subrepeat type within a subrepeat region and then its spreading to all subrepeat regions within the core block. T h e rapid and concerted divergence has furthermore led to a situation, where the coding B R l y core block has evolved considerably faster than the noncoding 3’ part of the gene. By contrast, in most other genes the noncoding regions evolve faster (YAFFE et al. 1985).Not only the B R l y core block has behaved evolu- tionarily anomalous, but also the C. pallidivittatus B R l a core block, which lacks a counterpart in C. tentans (LENDAHL and WIESLANDER 1985). This insta- bility in the BRl region is not paralleled in another BR region on the same chromosome; the BR2 region. In both the BR2a and BR2P core blocks individual repeat units are closely related in the two species (LENDAHL and WIESLANDER 1985). We now show that also the patterns of heterogeneity between repeat units in the BR2a and BR28 core blocks are conserved between C. pallidivittatus and C . tentans (Figure 5). T h e reason for this difference in evolutionary stability between two BR regions on the same chromosome remains obscure.
Implications for evolution of the gene family:
According to a recently proposed model for evolution of the BR gene family the ancestral BR gene was built from a simple, repetitive structure, consisting of a tandemly arranged 36 base pair primordial sequence, similar to the Cys-1 elements (C. HOOc and L. WIES- LANDER, unpublished data). Successive stepwise re- modelings of the repetitive sequences then gave rise to the contemporary, more complex repeats, at the expense of the preexisting ones. T h e model assumes that concerted changes have taken place during BR gene evolution and our data provide the experimental evidence that changes in all monomeric units of a long tandemly arranged repeat sequence actually do occur rapidly. Also the pattern of concerted divergence in the B R l y core block, i.e., that subrepeat regions evolve faster than constant regions, is reflected in the contemporary BR genes. Thus, in all paralogous BR
genes studied subrepeat regions are more diverged than constant regions (PUSTELL et al. 1984; WIESLAN- DER et al. 1984).
T h e pronounced changes in the B R l y core blocks over evolutionary short time periods suggest that BR genes functionally have been allowed to acquire dif- ferent forms and that their highly repetitive structure has been a driving force for the rapid and concerted divergence. However, for a number of reasons we assume BR genes to be subject to certain functional constraints. First, most mutations between individual repeat units are silent mutations. Second, in all sub- repeat regions, including the BRl y subrepeat region, codons for proline and charged amino acids are pre- ferred. Third, codons, e.g., cysteine, are highly con- served in constant regions, suggesting functional im- portance. Furthermore, the repeat unit length gen- erally seems to be kept within certain boundaries, which is true also for the two B R l y repeat units. Finally, the individual regulation of the BR genes during ontogeny also suggests that the different BR genes are at least to some extent functionally special- ized (LENDAHL and WIESLANDER 1987).
Concerted evolution of transcribed sequences has previously been observed in gene families with tan- demly arranged gene copies, e.g., in 5 s and ribosomal RNA genes, where the identity between individual gene copies is maintained within a species (ARNHEIM
1983). Our data show that also tandem repeats within a single gene can evolve in a concerted fashion. This should occur also in other genes with internal tandem repeats and the S antigen gene in different serotypes of Plasmodium falciparum appears to be one such example (COWMAN et aE. 1985). T h e evolution of the BR 1 y core block furthermore demonstrates that mu- tations can spread and rapidly become homogenized at successive levels, i.e., at subrepeat region and core block level, in a hierarchically repetitive gene.
We thank B. DANEHOLT for advice and support and A. BRAGES- SON, B. ERICSSON and E. LONG for technical assistance. We are indebted to E. VESTERBACK for typing the manuscripts. This work was supported by the Swedish Natural Science Research Council, the Swedish Cancer Society, the Wallenberg Foundation, Magnus Bergvalls Stiftelse, Gunvor and Josef Anirs Stiftelse and Karolinska Institutet.
LITERATURE CITED
ARNHEIM, N., 1983 Concerted evolution of multigene families. pp. 38-61. In: Evolution of Genes and Proteins, Edited by M. NEI and R. K. KOEHN. Sinauer Associates, Sunderland, Mass.
Cytologische Analyse eines Camptochiron- omus-Artbastards. I. Kreuzungsergebnisse und die Evolution des Karyotypus. Chromosoma 7: 198-259.
COWMAN, A. F., R. B. SAINT, R. L. COPPEL, G. V. BROWN, R. F. ANDERS and D. J. KEMP, 1985 Conserved sequences flank variable tandem repeats in two S-antigens of Plasmodium falci-
parum. Cell 40: 775-783.
Concom- BEERMANN, W., 1955
49
itant induction of a Balbiani ring gene and a giant secretory protein in Chironomus salivary glands. Chromosoma 81: 1 1 5- 124.
FARABOUGH, P., U. SCHMEISSNER, M. HOFER and J. H. MILLER,
1978 Genetic studies of the lac repressor. VII. On the molec- ular nature of spontaneous hotspots in the lac I gene of Esche-
richia coli. J. Mol. Biol. 1 2 6 847-863.
CALLER, R., L. RYDLANDER, N. RIEDEL, H. KLUDING and J.-E. EDSTROM, 1984 Balbiani ring induction in phosphate metab- olism. Proc. Natl. Acad. Sci. USA 81: 1448-1452.
CALLER, R., H. SAIGA, R. M. WIDMER, M. LEZZI and J.-E. EDSTROM,
1985 Two genes in Balbiani ring 2 with metabolically differ- ent 7 5 s transcripts. EMBO J. 11: 2977-2981.
The salivary gland of Chironomus (Diptera): a model system for the study of cell differentiation. pp. 147- 196, Vol. 8. In Results and Problems in Cell Dqferentiation,
Edited by W. BEERMANN. Springer, Berlin.
Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl. Acad. Sci. USA 72: 3961-3965.
HILL, A. S., R. D. NICHOLLS, S. L. THEIN and D. R. HIGGS,
1985 Recombination within the human embryonic zeta-glo- bin locus: a common zeta-zeta chromosome produced by gene conversion of the pseudo-zeta gene. Cell 42: 809-8 19.
Different evolutionary be- haviour of structurally related repetitive sequences occurring in the same Balbiani ring gene in Chironomus tentans. Proc. Natl. Acad. Sci. USA 81: 5165-5169.
A BR 1 gene
in Chironomus tentans has a composite structure: a large repet-
itive core block is separated from a short unrelated 3’-terminal domain by a small intron. Nucleic Acids Res. 14: 703-719.
JACKLE, U., J. C. DE ALMEIDA, R. GALLER, H. KLUDING, H. LEH-
RACH and J.-E. EDSTROM, 1982 Constant and variable parts
in the Balbiani ring 2 repeat unit and the translation terminator region. EMBO J. 1: 883-888.
JONES, C. W. and F. C. KAFATOS, 1982 Accepted mutations in a gene family: evolutionary diversification of duplicated DNA. J. Mol. Evol. 1 9 87-103.
A novel giant secretion poly- peptide in Chironomus salivary glands: implications for another Balbiani ring gene. J. Cell Biol. 101: 1044-1051.
Intrachromosomal gene con- version in yeast: a new type of genetic exchange. Nature 289: 144-148.
LABARCA, C. and K. PAIGEN, 1980 A simple, rapid and sensitive DNA assay procedure. Anal. Biochem. 102: 344-352.
LENDAHL, U. and L. WIESLANDER, 1985 Abrupt evolutionary change in the Balbiani ring gene family in two sibling species of Chironomus. J. Mol. Evol. 22: 63-68.
LENDAHL, U. and L. WIESLANDER, 1987 Balbiani (BR) genes exhibit different patterns of expression during development. Dev. Biol. 121: 130-138.
Organization of simple sequence DNA. pp. 381- 392. In: Genes, Edited by B. LEWIN. John Wiley & Sons, New York.
Evolution of duplicate genes and pseudogenes. pp, 14-37. In: Evolution of Genes and Proteins, Edited by M. NEI and R. K. KOEHN. Sinauer Associates, Sunderland, Mass. GROSSBACH, U. 1977
GRUNSTEIN, M. and D. S. HOCNESS, 1975
H i i i j ~ , C. and L. WIESLANDER, 1984
HMG, C., C. ENGBERG and L. WIESLANDER, 1986
KAO, W.-Y. and S. T. CASE, 1985
KLEIN, W. L. and D. PETES, 1981
LEWIN, B., 1983
LI, W.-H., 1983
MELTON, D. A., P. A. KRIEG, T . REBAGLIATI, K. ZINN and M. R. GREEN, 1984 Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12: 7035- 7056.
A new pair of M 1 3 vectors for selecting either strand of double-digest restriction fragments. Gene 1 9 269-275.
An expandeble gene that encodes a Drosophila glue protein is not expressed in variants lacking remote upstream sequences. Cell 29: 1041- 1051.
PUSTELL, J., F. C. KAFATOS, U . WOBUS and H. BAUMLEIN,
1984 Balbiani ring DNA: sequence comparisons and evolu- tionary history of a family of hierarchically repetitive protein- coding genes. J. Mol. Evol. 20: 281-295.
Sequences translated by Balbiani ring 75s RNA in vitro are present in giant secretory protein from Chironomus tentans. Chromosoma
SAIGA, H., C. GROND, E. R. SCHMIDT and J.-E. EDSTROM,
1987 Evolutionary conservation of the 3’-ends of members of a family of giant secretory protein gene in Chironomus
pallidivittatus. J. Mol. Evol. 25: 20-28.
The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 87: 107-1 10.
Evolution of repeated DNA sequences by unequal crossover. Science 191: 528-535.
Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol.
SPRAGUE, K. U., M. B. ROTH, R. F. MANNING and L. P. GAGE,
1979 Alleles of the fibroin gene coding for proteins of differ- ent lengths. Cell 17: 407-413.
A hierarchic arrangement of the repetitive sequences in the Balbiani 2 gene
of Chironomus tentans. Cell 3 0 579-587.
The Balbiani ring 2 gene
in Chironomus tentans is built from two types of tandemly
arranged major repeat units with a common evolutionary ori- gin. EMBO J. 2: 1169-1 175.
Evidence for a common ancestor sequence for the Balbiani ring 1 and Balbiani ring 2 genes in Chironomus tentans. Proc. Natl. Acad. Sci. USA 7 9 6956-6960.
WIESLANDER, L., C. HO&, J.-0. HO&, H. JORNVALL, U. LENDAHL and B. DANEHOLT, 1984 Conserved and non-conserved struc- tures in the secretory proteins encoded in the Balbiani ring genes of Chironomus tentans. J. Mol. Evol. 20: 304-312.
Highly conserved sequences in the 3’ untranslated region of mRNAs coding for homologous proteins in distantly related species. Nucleic Acids Res. 13: 3723-3737.
YAMADA, Y., E. AVVEDIMENTO, M. MUDRYJ, H. OHKUBO, G. Vo-
GELI, M. IRANI, l. PASTAN and B. CROMBRUGGHE, 1980 The
collagen gene: evidence for its evolutionary by amplification of a DNA segment containing an exon of 54 bp. Cell 22: 887- 892.
Communicating editor: W.-H. LI MESSING, J. and J. VIEIRA, 1982
MUSKAVITCH, M. A. T . and D. S. HOGNESS, 1982
RYDLANDER, L., A. PIGON and J.-E. EDSTROM, 1980
81: 101-113.
SANGER, F. and A. R. COULSON, 1978
SMITH, G. P . , 1976
SOUTHERN, E. M., 1975
98: 503-5 17.
SUMEGI, J., L. WIESLANDER and B. DANEHOLT, 1982
WIESLANDER, L. and U. LENDAHL, 1983
WIESLANDER, L., J. SUMEGI and B. DANEHOLT, 1982