Copyright 0 1989 by the Genetics Society of America
Y
Chromosome Evolution in the Subgenus
Mus (Genus Mus)
Priscilla
K.Tucker,' Barbara K.
Lee
and Eva
M.
Eicher
The Jackson Laboratory, Bar Harbor, Maine 04609Manuscript received November 30, 1987 Accepted for publication January 20, 1989
ABSTRACT
A 305 base pair DNA sequence isolated from the Y chromosome of the inbred mouse strain C57BL/10 was used to investigate the pattern and tempo of evolution of Y chromosome DNA
sequences for five species in the subgenus M u s , including M u s spretus, Mus hortulanus, Mus abbotti, Mus musculus and Mus domesticus. Variation in hybridization patterns between species was character- ized by differences in fragment lengths of both intensely and faintly hybridizing fragments, whereas variation in hybridization patterns within species was characterized primarily by differences in
fragment lengths of faintly hybridizing fragments. Phylogenetic analyses were conducted based on fragment size variation within and among species. Phylogenetic relationships inferred from these analyses partly agree with the phylogenetic relationships obtained from biochemical and mitochondrial DNA data. We conclude that a set of DNA sequences common to the Y chromosomes of a closely related group of species in the subgenus Mus has evolved rapidly as reflected by sequence divergence and sequence amplification.
I
N the classical model of sex chromosome evolution, heteromorphic sex chromosomes are hypothesized to have evolved from a pair of homologous chromo- somes by the suppression of crossing over between the sex chromosome precursors and the subsequent loss of gene function on the sex chromosome that becomes the Y chromosome (MULLER 1914). OHNO (1967) recalled this hypothesis to explain vertebrate sex chro- mosome evolution including the evolution of the mammalian X and Y chromosomes. T h e X and Y chromosomes of mammals exhibit extreme hetero- morphism and, with rare exception, recombination between them is limited to a small region (SOLARI1974). Thus, the majority of the Y chromosome is monosomic and inherited from father to son.
Recent molecular investigations of the human (GOODFELLOW, DARLING and WOLFE 1985) and mouse Y chromosomes (SINGH, PURWM and JONES 1981 ; JONES and SINGH 198 1; PHILLIPS et al. 1982; EICHER,
PHILLIPS and WASHBURN 1983; LAMAR and PALMER 1984; BISHOP et al. 1985; NISHIOKA and LAMOTHE 1986; NALLASETH and DEWEY 1986) identified DNA sequences specific to or enriched on the Y chromo- some. Additionally, using Y-specific or Y-enriched re- peated DNA sequences as probes, restriction fragment length polymorphisms have been identified among Y chromosomes of human populations (CASANOVA et al. 1985) as well as among Y chromosomes of laboratory inbred mouse strains and wild mice (LAMAR and PAL-
MER 1984; BISHOP et
d .
1985; NISHIOKA and LA- MOTHE 1986; EICHER et al. 1989). For example, twoArbor, Michigan 48 109.
Genetics 122: 169-179 (May, 1989)
' Current address: Museum of Zoology, University of Michigan, Ann
DNA sequences, AC11 (NISHIOKA and LAMOTHE
1986) and YBlO (EICHER et al. 1989), isolated from the Y chromosomes from two inbred mouse strains hybridize to genomic DNA from males of some species in the subgenus M u s and distinct hybridization pat- terns characterized by differences in fragment length and hybridization intensity were observed for each species. These data suggest that Y chromosome-spe- cific DNA sequences rapidly evolve in closely related species.
In this report we present additional evidence for the rapid evolution of DNA sequences found exclu- sively on the mouse Y chromosome from an investi- gation of the tempo and mode of Y chromosomal sequence change at both the species and population levels. We also investigated how a paternally inherited character contributes to an understanding of the phy- logenetic relationships within the genus Mus. We used pYB 10 (EICHER et al. 1989) as a probe in combination with eight restriction enzymes to identify species-spe- cific patterns of hybridization from a large sample of mice in the subgenus Mus, including Mus spretus, Mus hortulanus (= M. spicilegus, BONHOMME 1986), Mus abbotti (= M . spretoides or M. macedonicus, BONHOMME
1986), M u s musculus and Mus domesticus. Throughout this study we recognize M. musculus and M. domesticus
as distinct species rather than subspecies or semispe- cies based on data from the hybrid zone investigations of these two taxa (HUNT and SELANDER 1973; SAGE, WHITNEY and WILSON 1986; SAGE et al. 1986).
MATERIALS AND METHODS
170 P. K. Tucker, B. K.
cies, number of individuals sampled, original collecting lo-
calities and source of taxa used in o u r study are listed in Table 1.
DNA preparation: High molecular weight mouse ge- nomic DNA was prepared from either frozen tissue or frozen nuclear pellets. D N A from liver, kidney, spleen, and testis was prepared following the method of JENKINS et al.
(1982). Frozen nuclear pellets were incubated a t 65" over- night in 10 ml of extraction buffer (50 mM Tris, pH 8.0,
100 mM EDTA, 100 mM NaCI, 1% SDS) and 0.5 ml of proteinase K (10 mg/ml) in 10 mM Tris, pH 7.5, prior to
extraction with phenol following the method of JENKINS et al. (1982).
Restriction endonuclease digestions and Southern blot preparation: Restriction enzyme digests of genomic DNAs were performed following the procedures of the supplier BRL (Bethesda Research Laboratories, Inc.). Ten micro- grams of genomic D N A were digested for 4 hr and separated by size in 1% agarose at 30 V for up to 20 hr. DNA was transferred to Zeta-Probe nylon membranes (Bio-Rad) in
0.4 M NaOH overnight without pretreatment (REED and MANN 1985).
DNA labeling The probe pYB10, derived from the Y
chromosome of C57BL10 (EICHER et al. 1989), was labeled with [a-"PI-dCTP (Amersham) following MANIATIS,
FRITSCH and SAMBROOK (1 982) using the T4 polymerase kit
supplied by BRL. The specific activity of labeled D N A for
all experiments was greater than 0.5 X lo9 cpm/pg.
Hybridization conditions and autoradiography: Nylon membranes were prewashed at 65" for 1 hr in 0.1 % SSC and 0.5% SDS and prehybridized at 65" for 4 hr in 100 ml of 4 X SSCP [ 1 X SSCP = 121 mM NaCI, 15 mM Nan citrate, 15 mM Na2HP04, 5 mM NaH2P04], 10 X Denhardt's solu-
tion and 1 % SDS. Membranes were hybridized overnight at 65" in 20 ml of 4 X SSCP, 2 X Denhardt's solution, 1% SDS, 0.2 pg/ml denatured sonicated salmon sperm DNA, and approximately 1.0 X lo6 cpm/ml denatured radioac- tively labeled probe. Following hybridization, the nylon membranes were rinsed twice in 2 X SSC/O. 1 % SDS at room temperature for 15 min and three times in 0.1 X SSC/O. I %
SDS at 65" for 30 min. The stringency conditions at 65",
0.1 X SSC, and a 39% G-C content (determined from DNA sequence analysis of YB10, EMBL accession number
X 12900) were calculated to be approximately 98% (SHAW
et al. 1984). The membranes were exposed to Kodak XAR- 5 film by varying lengths of time at room temperature or at
-70" with Dupont Cronex Lightening Plus intensifying screens.
Analysis of autoradiographs: The approximate size of each restriction fragment was ascertained using a custom- ized computer program written for the Apple Ile and a HIPAD digitizer (Houston Instrument). Phylogenetic anal- yses using parsimony (PAUP, version 2.4, SWOFFORD 1985) were conducted for t w o sets of taxa using restriction frag- ments lengths as characters. Fragments of equal molecular weight generated by each restriction enzyme were assumed identical between taxa. Each individual taxon was scored for the presence or absence of all restriction fragments identified in a given set of taxa. For all phylogenetic analyses, variation in copy number atnong shared restriction frag- ments was not scored as separate characters and thus the phylogenetic trees produced represent minimal estimates of
the variation among Y chromosomes. In cases where diver- gent taxa shared fragments of identical size, DNAs from these taxa were electrophoresed on the same gel to identify more accurately shared fragments. Genomic DNAs from all wild caught M . domesticus and M . abbotti were analyzed twice and, with t w o exceptions (Table l ) , genomic DNAs from a
L .ee and E. M. Eicher
minimum of two laboratory raised mice from each taxa were analyzed.
RESULTS
Species-specific patterns of hybridization were ob- served when Southern blots containing male genomic
DNAs from M . abbotti, M. hortulanus, M. spretus, M. musculus, a n d M. domesticus (Table 1) were probed with pYB10. A total of 180 different restriction frag- ments were identified by single digests of male ge- nomic DNA using eight different restriction enzymes, representing four (HaeIII, TaqI), five ( H i n f l ) a n d six
(BglII, EcoRI, HindIII, PvuII, PstI) base cutters. No hybridization was observed when Southern blots con- taining female genomic DNAs from M. abbotti, M. hortulanus, M. spretus, M. musculus a n d M. domesticus (Table 1) were probed with pYBlO (data not shown). This verifies the observation by EICHER et al. (1989) that YBlO sequences are found exclusively o n t h e Y
chromosome in these five species. T h e hybridization patterns observed with PstI (Figure 1) typify all single digests using the eight restriction enzymes listed
above. Three basic trends emerged. First, there is overall less hybridization to DNA from M . spretus and M. hortulanus than to M. abbotti, M. domesticus a n d M. musculus. Second, for six enzymes, HaeIII, TagI, BglII,
HindIII, PvuII a n d PstI, a t least one restriction frag- ment is shared among all five species and these shared fragments typically vary in intensity of hybridization among species. Third, between species variation is characterized primarily by the presence and absence
of both intensely a n d faintly hybridizing restriction fragments, whereas within species variation is charac- terized primarily, but not exclusively, by the presence and absence of faintly hybridizing restriction frag- ments. Intensely hybridizing fragments, including
shared fragments that vary i a hybridization intensity between species, are tabulated for each enzyme and each species in Table 2.
We conducted a phylogenetic analysis (PAUP, ver- sion 2.4, SWOFFORD, 1985) of these five Mus species using the 180 restriction fragments as characters. TWO
minimum length trees were produced from 170 apo-
morphic (derived) characters (Figure 2). This analysis substantiates the observed species-specific patterns of hybridization because each species branch, including M . spretus, M. hortulanus, M. abbotti, M. musculus a n d M. domesticus, is clearly defined by a series of restric- tion fragments including the majority of intensely
hybridizing fragments. Within species variation of
YB 10 sequences, characterized primarily by faintly hybridizing restriction fragments, was also observed for each species.
T h e phylogenetic analysis provided an objective method to infer relatedness among taxa. In both trees
Y Chromosome Evolution in Mus 171
TABLE 1
Species, number of individuals sampled, original collecting localities and source of taxa used in the analysis of RFLP's in the genus Mus (subgenus Mus)
Taxa Number F, M" Original collecting localityb Source'
M. spretus 4 , 4 * *
4 , 4 * *
M. hortulanus 4 , 4 * *
4 , 4 * *
M. abbotti 1 , 2 * *
M. m. musculusd 3 , 4 * *
4, 6* 8 , 8 * *
4, 4* 0, 1 0, 1 0, I * 0, I * 0, 1 0, 3* 0, 1 * 0, 1* 0, 2* 0, 1* 0, 8* 0, 2 0, 1
0, 5 0, 5* 0, 1 * 0, 1 *
0, 1 0, 2* 0, 1* 0, 5*
M. m. castaneus 4 , 4 * *
M . m. molossinus 4 , 4 * *
0, 1*
M. domesticusd 5, 5*
4, 4 4, 4 4, 4 4 , 4 4 , 4 4, 4 4 , 4
8 , 8 * *
4, 4 4, 4* 0, 1" 0, 1
0, 3*
0, 2* 0, 1* 0, 2*
0, 2*
0, 2*
0, 1 *
0, 1 *
0, 1 * 0, I * 0, 1
0, 1
0, 2
Spain: Cadiz Prov.; Puerto Real Morocco: Azrou
Austria: Burgenland Prov.; 6 km ENE Halbturn Yugoslavia: Serbia; 20 km N Pancevo
Yugoslavia: Macedonia; 6 km NW Gradsko Denmark: Skive
Denmark: Viborg Co.; Vejrumbro
Czechoslovakia: Moravia; Studenec, approx. 35 km W Brno Czechoslovakia: Slovakia; Sladeckovce, approx. 50 km E. Bratislava Yugoslavia: Serbia; Belgrade
Austria: Burgenland Prov.; Halbturn Austria: Braunau am Inn bezirk; Braunau Austria: Braunau am Inn bezirk; Ranshofen A. Austria: Braunau am Inn bezirk; Ranshofen B. Austria: Braunau am Inn bezirk; Nofing F.R.G.: Bavaria; Freising Kreise, Giggenhausen F.R.G.: Bavaria; Freising Kreise, Freising F.R.G.: Bavaria; Freising Kreise, Achering F.R.G.: Bavaria; Freising Kreise, Dornhaselbach F.R.G.: Bavaria; Freising Kreise, Rudlfing F.R.G.: Bavaria; Freising Kreise, Tuntenhausen F.R.G.: Bavaria; Freising Kreise, Massenhausen F.R.G.: Bavaria; Erding Kreise, Schwaig F.R.G.: Bavaria; Erding Kreise, Sonnendorf F.R.G.: Bavaria; Erding Kreise, Gut Wildschwaig F.R.G.: Bavaria; Erding Kreise, Hogersdorf F.R.G.: Bavaria; Erding Kreise, Tittenkofen F.R.G.: Bavaria; Traunstein Kreise, Traunstein F.R.G.: Bavaria; Rottal-Inn Kreise, Mitterskirchen F.R.G.: Bavaria; Rottal-Inn Kreise, Simbach Thailand: Chonburi Province, Chonburi Japan: Kyushu
Japan: Okinawa, Naha U.S.A.: Texas; College Station
U.S.A.: Maryland; Upper Marlboro, University of Maryland To- bacco Farm
Seed shed Chemical shed Drying barn Tool shed
USA.: Maryland; Queen Anne Co., Ridgely, J.J. Downs U.S.A.: Maryland; Davidsonville, Haven's Farm U.S.A.: Maryland; Davidsonville, Sanner's Farm U.S.A.: Maryland; Queen Anne Co., Centreville U.S.A.: Delaware; Lewes
U.S.A.: California; Bouquet Canyon
Great Britain: Berkshire; 2 mi SW Abingdon, Culham College Spain: Barcelona; 0.5 km W San Fausto
Egypt: Faiyum Governate; AI Faiyum Depression Egypt: Giza Governate; near Bashtil
Israel: Jerusalen Italy: lsole Eolie; Lipari Italy: Apennine; Molise Italy: Bergamo; Orobie Italy: Laatsch
Italy: Tuscany; Pisa
Italy: Sondrio Prov.; Morbegno Greece: Ellas
Yugoslavia: Dalamatia; Metkovic
F.R.G.: Bavaria; Freising Kreise, Thalhausen F.R.G.: Bavaria; Freising Kreise, Kammerberg
1 1 1 1 4 1 1 1 1 , 3 4 4 4 3 4 4 4 4 4, 3 4 4, 3 4 4 4, 3
4, 3 4 4 4 4 4 4, 3
1 1, 3
172 P. K . Tucker-, B. K . Lee a n d E. M. Eicher
TABLE 1
Continued
-,.. .l\<l .. Sumher I-. A10 Origin;d c o k - c t i n g l o m l i t v b Source‘
.\I. domps/irusd 0. 18* F.K.G.: Bactria; I;rrising Kreise, Gesselthauscn A.
0 . 9* F.R.G.: IkIcwia; Freising Kreise. Gesselthausen H. 0. I * F.K.G.: lhvaria: Freising Kreise. Gesselthatrsen 0 , I6* F .K .G .: lktvaria: Freising Kreise, Xeufidlrn
0, :3* F.K.G.: lhvaria: Freising Kreise. Eherspoint 0, I F.K.G.: Iklvaria; Frcising Krrise, Achering
0 , 8* F.K.G.: 13avari:I; Augshrg Kreise, Augshurg
0. I F . K . G . : Ikwari:~; Ihchatt Kreise, F.herst,ach
0, I * F.R.G.: R;lden-M’~rrtternl,erg; Niirenberg Zoo
0. 2* F . K . G . : R;~den-M’~Irttcrnberg; l‘iibingen
0, 1 F.R.G.: Kheinl;md P f i ~ l z ; Neustadt 0 . 1 * F.K.G.: Hessen; Heidelberg
0. 1 F . K . G . : Holstein; Schleswig, Hrlgoland Ishnd 0 , t i * F . K . G . : 13avaria: Freising Kreise. “assenhausen
4. 3
4 , 3
4 4. 3
4. J 4, 3
4 4. 3 4 4 4
3 3 3
0, 1 Austria: Uraunnn an1 Inn bezirk: Kanshofcn A. 4
0. 1 Switzerland: \’;~ud <>Inton; Givrins, near Nyon 4
.\I. (1. hrtvirmtris 3.48: Morocco: 7 km Nib’ Azrou 1 . 3
AI. d . posrhiavinus 4 * 4 , Switrerl;~nd: Grisons Canton: Zalende. Posclliavo 1
.\,I. d. prarttxus 4 , 4 * Morocco: T a f i l a l t 0;lsis. Erfoutl 1
,\4. dompstirus ssp. 4, 4 * * Egypt: Abu K ~ I H . ; I S ~ I 1
J, J* Italy: Sondrio Prov.; Tiratlo 1
’
I,oc.;tIities listed represent the original collecting localities of either wild caught individuals or of Iahoratory raised descendents of wild ” 1; = fen1;Ile: \I = ~nale.caught intlivitlu;lls ~ r s e t l i n this stctdy.
‘ Sources include h1)oratory raised individuals provided by MICHAEL POTTER ( 1 ) . EVA M. EICHER (2). o r KICHARI) SAGE (3) and wild caught individttals provitlrd b y R I C H A R D SAGE (4) or PRISCII.I.A T U C K E R ( 5 ) . Laboratory raised individu;1ls from PWI-I-KR and EICHER prohibly rc*l)resent ;I single Ychrotnosotne for e;Ich locality a s no variation i n YRI 0 sequences was detected among the individuals s;unlpletl from any givrn locality.
d Species identification of mice from Austria ; ~ n d F.R.G. are based on their Ychromoson~ type.
I ) o t ~ h l r asterisks identify DNAs from taxa that were cut w i t h eight restriction enzymes and single asterisks identify taxa that were cut with
?- l’he Y chronmsonles of mice from Lipari, Molise, ; ~ n d Orohie were placed on a C57BI.6(] txtckground prior to this study.
five restriction enzymes. DNAs from a l l other taxa were cut w i t h ;I nlininlum of two enzymes.
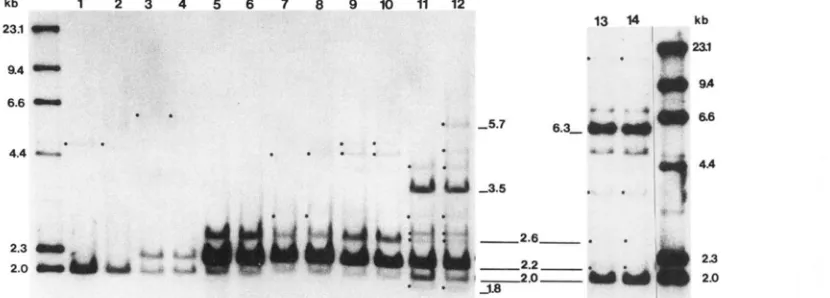
I‘l(;llRE 1 .-Acltol;ltliogr~~~)l~s of niak genonlic 1)Y.i cut w i t h Pstl ;und p r o b t d w i t h p Y B I O . Outside Ianes are Iamhd;r-Hindlll molecular wcighl n~;~rkct.s. h n p l r s include the follo\ving species: I;me I = .\I. sprptus. S p i n ; lane 2 = A!. sprrtus. Morocco; lane 3 = M . hortulanus.
/\ustri;l: I;IIIC 4 = .\I. Irortrclanus, Yugoslavia: lane 5 = ,\I. musrulus musrultts. Denmark; lane 6 =
M.
m. mtcscttlus, CzecI1oslav;lkia; lanes 7 andX = .\I. m. rno1ossinu.r. J a p a n ; lanes $1 ;Ind 1 0 = .\I. m. rastantus, Thailand: lane 1 I = .\I. domrstirus. Egypt; I;mc I2 = M . domrsticus, U.S.A.;
and lanes I3 ; ~ n d I4 = .\I. ahhotti, Yugoslavia. 1.ow copy nurnher fragments are identified b y closed circles. A 2.0-kb Pstl fragment shared anlong a 1 1 tasa, is found i n higher copy nunlhcr i n ’11. sprttus and .\I. ahhotti. The differcvcc i l l intensity of hybridization between the two sample5 of.\l. sprptus is due t o a n untlerlo;ltling of lane 2. A 2 . 2 4 ) Pstl and ;I 2.6-kt) Pstl fragment shared ;Inlong M. ahhotti, M. musrulus and .\I. rlom~.rtirrrs urics i l l copy number actx)ss tasa. Two Pstl high copy ntrmher fr;lgnlents, 3.5-kh and ;I 6.3-kb i n length. ;Ire unique to M.
domp.rtirfrs and .\I. abbotti respectively. h w copy nun,ber fragments are either species-specific (the 1.8-kh psll fr;lgment o f i\4. dOmPStiCUS) or unique t o a populatiotl w i t h i n ;I species (the 5.7-kl) Pstl fragment of ,\I. dompstirus-U.S.A.).
Y Chromosome Evolution in Mus 173
TABLE 2
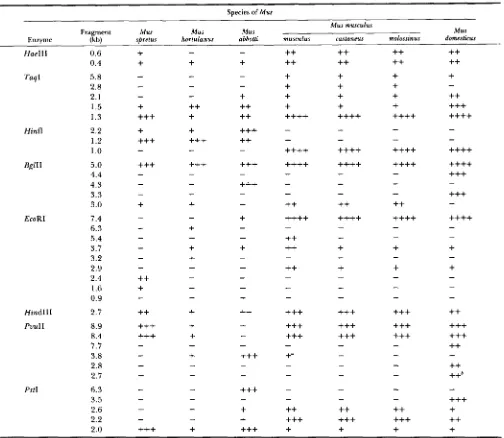
Presence and absence of restriction fragments found in multiple copies in at least one of five species in the genus MUS
~~
Species of Mus
Fragment Mus Mus Mus
Mus musculus
Mus
Enzyme (kb) spretus hortulanus abbotti musculus castaneus molossinus domesficus
Hinfl
Bgl I I
EcoRI
Pstl
Haelll 0.6
0 . 4
Tag I 5.8
2.8 2.1 1.5 1.3 2.2 1.2 1 .0 5.0 4.4 4 . 3 3.3 3.0 7.4 6.3 5.4 3.7 3.2 2.9 2.4 1.6 0.9
Hind111 2.7
PUUI I 8 . 9 8.4 7.7 3.8 2.8 2.7 6.3 3.5 2.6 2.2 2.0 -
+
--
+
++
++
+++
++
-+++
-+++
-
-
+
-
+
- -+
-
+
+
++
+
-
-
+++
--
+++
-
+
+
+++
++
++
+
+
+
+
++++
--
++++
++++
-
-
-
++
++++
-++
++
-
++
--
-+++
+++
+++
-+"
- - --
++
+++
+
++
++
+
+
+
+
++++
-
-
++++
++++
-
-
-
++
++++
-+
-
-
+
--
-+++
+++
+++
-
- --
-
-
++
+++
+
++
++
+
+
+
+
++++
-
-++++
++++
--
-++
++++
-+
-
-
+
-
-
-
+++
+++
+++
-
-
-
-
-
-
++
+++
+
++
++
+
-
++
+++
++++
--
++++
++++
+++
-+++
-
++++
-
-+
-+
-
- -++
+++
+++
++
-
++
++b-
+++
+
++
+
T h e presence or absence of this fragment varies within the subspecies M . m. musculus.'
T h e presence o r absence of this fragment varies within the species M . domesticus.A [-I in each column indicates the absence of a specific fragment for a given species. A [+I in each column indicates the presence of a fragment. Multiple [+]'s indicate presumed copy number differences based on intensity of hybridization between species for a given fragment.
A greater number of [+]'s indicates the presence of a greater number of copies for a fragment of a specific size. T h e specific number of
[+]'s, however, is not a strict measure of the degree of amplification. Low copy number fragments are not listed in this table.
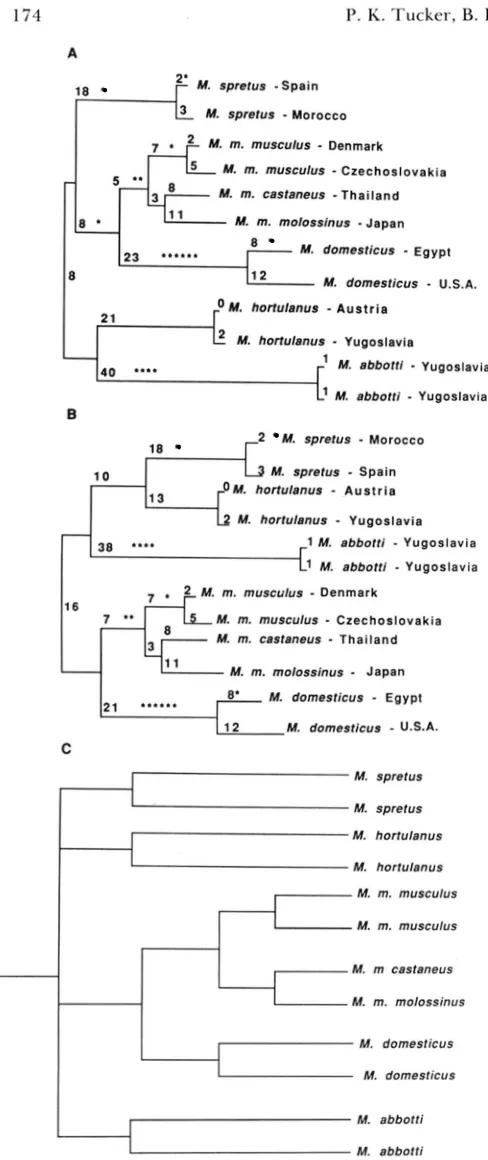
to M . domesticus, indicating that these three subspecies comprise a monophyletic group. T h e relationships among M . spretus, M . hortulanus and M . abbotti differ between the two trees. In tree A (Figure 2A) M . abbotti and M . hortulanus are more closely related to each other than they are to M . spretus or to M . musculus
and M . domesticus. In tree B (Figure 2B) M . spretus and M . hortulanus are more closely related to each other than they are to M . abbotti. When more than one minimum length tree is produced in a pylogenetic analysis a consensus tree can be constructed using only concordant data from the minimum length trees. A
consensus tree (Figure 2C) was constructed from trees A and B. In this tree M. spretus, M . hortulanus, and
M . abbotti are as distantly related from each other as they are from M . musculus and M . domesticus.
To characterize the evolution of YBlO sequences further, a more extensive analysis of within species variation was conducted for M . musculus and M . do- mesticus. Genomic DNA from male mice of 2 4 geo- graphically isolated populations of M . musculus and 30 geographically isolated populations of M . domesti-
174 P. K . Tucker, B. K . Lee and E. M. Eicher
18
m. musculus
-
Denmark-
5.*
8 M. m. musculus-
Czechoslovakia 3 M. m. castaneus -Thailand-
8 .-r(l
M. m. molosslnus -Japan23 ...a 8 M. domesticus
-
Egypt 8 M. domesticus-
U.S.A.2 1 o M. hortuknus
-
Austria 2 M. hortulanus-
Yugoslavia-
4 0....
'
M. abbotti-
Yugoslavia'
M. abbottf-
YugoslaviaB
18 2 M. spretus
-
Morocco 1 0 M. spretus-
Spain13 OM. hortulanus
-
Austria M. hortulanus-
Yugoslavia-
38
....
1 M. abbottl-
Yugoslavia 1 M. abbottl-
Yugoslavia16
.
musculus
-
Denmark M. m. mUSCulus-
Czechoslovakia M. m. cast8neus-
Thailand-
M. m. molosslnus-
Japan 8. M. domesticus-
Egypt 21...
M. domestlcus
-
U.S.A.C
M. spretus M. spretus M. hortulanus M. hortulanus M. m. musculus M. m. musculus
I
M. M. m m. caslaneus molossinus1
M. domesticusM. domesticus
M. 8bbOtti M. abbottf
FIGURE 2.--E\~olutionary trees constructed by a phylogenetic analysis using parsimony (PAUP). Trees A and B represent the most parsimonious trees found from an ;nlalysis of restriction frag- ment length polymorphism data using pYBI0. The trees were rooted at the midpoint of greatest patristic distance. The length of each tree is 188 ; ~ n d the consistency index, CI (FARRIS 1970). is 0.899. The CI. calculated a s "the sum of the ranges of a11 characters i n the data divided b y the number of evolutionary changes on the tree (i.e.. tree length)" (BROOKS, O'GRADY and WIIXY 1986). is an indicator of the amount of homoplasy (parallel, reversal, or conver- gent evolutionary events) i n a tree. A high CI reflects minimal homoplasy. The number of characters (restriction fragments) that
FIGURE 3.-An autoradiograph ot male genomlc DNAs troni geographically isolated populations of M . domesticus digested with Tagl and probed w i t h pYBI0. The outside lane is a lambda-Hindlll molecular weight marker. Samples include mice from the following localities: lane 1 = Centreville, U.S.A.; lane 2 = College Station.
U.S.A.; lane 3 = Nurenberg. F.K.G., lane 4 = Heidelberg, F.K.C.; lane 5 = Ellas, Greece; lane 6 = Zalende. Switzerland (identified as M . d. poschiauinus); lane 7 = Iipari, Italy; lane 8 = Orobie, Italy; lane 9 = Molise, Italy; lane 10 = Azrou, Morocco (identified as M .
d. brevirostris): lane 1 I = Erfoud, Morocco (identified as M . d.
pradexfus); lane 12 = Abu Rawash, Egypt. Mice from Zalende.
Orobie, Molise and Lipari are from Robertsonian metacentric populations. Low copy number fragments are identified by closed circles. All M . domesticus sampled share four high copy number
Tag1 fragments that are I . 3 kb, 1.5 kb, 2.1 kb and 5.8 kh in length.
W i t h the exception of a 6.6-kb Tag1 fragment. the variation within
M . domesfirus is characterized by nunlerous low copy number frag- ments.
blots of these DNAs were probed with pYBlO (Table 1). In this more extensive second analysis a total of 185 fragments were identified by single digests of genomic DNA. T h e hybridization patterns observed when male genomic DNA was digested with TaqI (Figure 3) typify the patterns seen with the other restriction enzymes and substantiate the initial obser- vation that within species variation is characterized primarily by faintly hybridizing restriction fragments. T h e populations of M . musculus sampled represent three recognized subspecies, 1) two populations of M . m. musculus from Denmark, two populations from Czechoslovakia, three populations from Austria and eleven populations from T h e Federal Republic of Germany, 2) one population of M . m. castaneus from Thailand and 3) two populations of M . m. molossinus from Japan. T h e populations of
M.
domesticus sampled included four of the currently recognized subspecies.Y Chromosome Evolution in Mus 175
However, because the subspecific designations of M . domesticus are disputed (FERRIS et al. 1983; BON- HOMME et al. 1984), our samples were identified only to species with the exception of M . d. brevirostris from Morocco, M . d . praetextus from Morocco and M . d. poschiavinus from Switzerland and Italy (Table 1). T h e remaining samples of M . domesticus are from one locality from Great Britain, one from Spain, three from Egypt, one from Israel, six from Italy, one from Greece, ten from the Federal Republic of Germany and three from the United States. An initial investi- gation of Y chromosome variation in M . domesticus
samples from Delaware and Maryland (Table 1) indi- cated no variation among Y chromosomes of these mice and thus the Y chromosome of M . domesticus
from these localities was represented by a single local-
ity (Maryland: Queen Anne Co.; Centreville). A subset of the M . musculus and M . domesticus
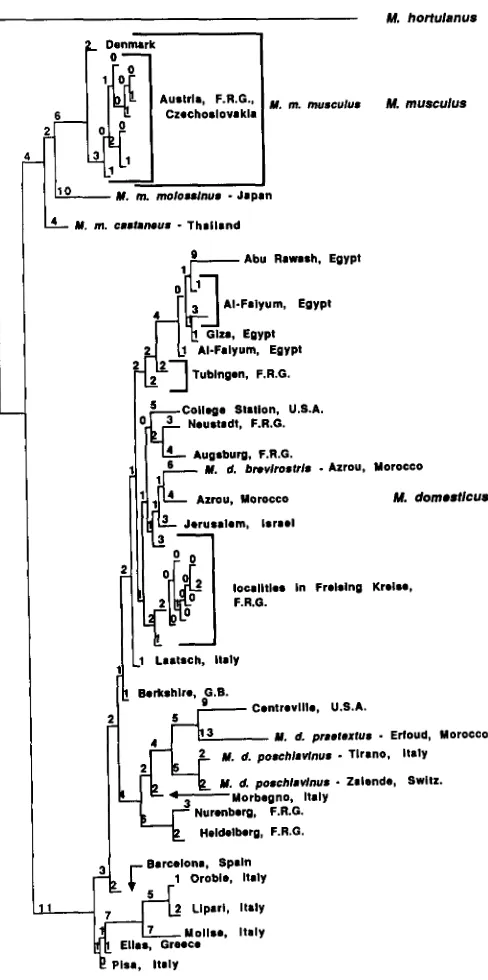
samples reflecting the variation found among Y chro- mosomes was used in a phylogenetic analysis and a series of minimal length trees were produced from
11
7
apomorphic characters of the 185 characters (restriction fragments) identified. T h e most com- monly observed arrangement of taxa among ten trees analyzed is represented in Figure 4. T h e lower con- sistency index for this tree indicates that a large num- ber of characters represented convergent, parallel, or reversal events. These primarily include restriction fragments found within the M . domesticus group. Pre- sumably, fragments of identical molecular weight, identified as shared fragments, may have evolved in- dependently within the M . domesticus lineage. M . mus- culus and M . domesticus fell into two discrete groups in all trees analyzed indicating that each of these two species comprise a monophyletic group. In addition, the arrangement of taxa within the M . musculus group remained the same, whereas subgroups of taxa within the M . domesticus group varied in position along the length of the tree in each tree analyzed.In the M . musculus group two distinct groups were found within the subspecies M . m. musculus. These include mice from Denmark and from central Europe. As in the first phylogenetic analysis (Figure 2), M . m. molossinus and M . m. castaneus are distinct subgroups within the M . musculus group. In contrast to the first analysis, M . m. molossinus is more closely related to M . m. musculus than is M . m. castaneus.
Relationships among mice within the M . domesticus
group varied among the trees analyzed, although cer- tain subgroups of mice were consistently defined by four or more restriction fragments. Such subgroups include 1) populations from Orobie, Lipari and Molise in Italy having Robertsonian metacentric chromo- somes; 2) Robertsonian chromosome bearing popula- tions from Tirano, Italy, and Zalende, Switzerland, identified as M . d. poschiavinus; 3) a group of mice
I M. hortulanus
Czrchoslovskls m. musculus
r M. m. csstansua
-
ThallandAbu Rawa8h. Egypt
M. musculus
AI-Falyum, Egypt GIzB, Egypt Tubingen, F.R.G.
M. d. bnvlroshls
-
Azrou, MoroccoAzrou, Morocco M. domortlcus
Jerusalam, Isrssl
localltlas In Frslmlng Krelsa, F.R.G.
Barcelona. Spsln 1 Orobls, Italy
FIGURE 4.-An evolutionary tree constructed by a phylogenetic analysis using parsimony. This tree is one in a series of heuristic trees produced from the restriction fragment length polymorphism data using pYB10. Mus hortulanus was used as an outgroup. The tree length is 249. The consistency index is 0.526. The number of characters (restriction fragments) that determine the length of each horizontal branch is given.
from Egypt; 4) a mouse identified as M . d. praetextus
176 P. K. Tucker, B. K. Lee and E. M. Eicher
lack of variation observed among M. musculus, in particular and, among M. m. musculus from Europe. Finally, a sample of M. musculus and M. domesticus
mice collected along a 160-km transect in the Federal Republic of Germany and Austria (the Austria-F.R.G. sample of M. musculus and the Freising-Kreise sample of M. domesticus) were included in this analysis and minimal variation involving one or two fragments was identified among Y chromosomes within either spe- cies.
DISCUSSION
EICHER et al. (1989) have suggested that YBlO sequences arose on the Y chromosome of the ancestor common to M. spretus, M. abbotti, M. hortulanus, M. musculus and M. domesticus as these are the only species in the subgenus Mus that have YB 10 sequences exclu- sively on the Y chromosome. They also suggested that the species-specific patterns of hybridization observed with YBlO resulted from both sequence divergence and sequence amplification of the Y chromosome dur- ing subsequent speciation events. In this paper we have further investigated the evolution of YBlO and adjacent Y chromosome sequences in these five species at both the species and population levels. We found that Y chromosome differences between species are characterized by variation in both intensely and faintly hybridizing fragments whereas Y chromosome differ- ences within species are characterized primarily by variation in faintly hybridizing fragments.
Variation in hybridization intensity of YBlO se- quences could be due to either differences in copy number or to differences in sequence homology. How- ever, we interpret the observed differences in intensity of hybridization among restriction fragments as pri- marily reflecting variation in copy number of YBlO sequences, intensely hybridizing fragments being high copy number fragments and faintly hybridizing frag- ments being low copy number fragments. T h e reason for this interpretation results from two observations. No new fragments were observed when M. spretus, M . hortulanus, M. abbotti, M. musculus and M. domesticus
DNAs were hybridized with pYBlO at lower strin- gency conditions and hybridization intensity increased for all restriction fragments across taxa following a decrease in stringency conditions (EICHER et al. 1989). We, thus, interpret variation in intensity of hybridi- zation among equal sized (shared) fragments as reflect- ing amplification of YBlO sequences. Additionally, we suggest that presumed high copy number fragments that are unique to a given taxon reflect sequence divergence and amplification of Y chromosome se- quences whereas presumed low copy fragments unique to a given taxon reflect sequence divergence such as single restriction site changes. In general, we conclude that interspecies variation of YB 10 and ad-
jacent sequences is characterized by both sequence divergence and sequence amplification, whereas in- traspecies variation is characterized primarily by se- quence divergence.
We also performed phylogenetic analyses to deter- mine whether and at what taxonomic level a paternally inherited repeated DNA sequence contributes to our understanding of the phylogenetic relationships in the genus Mus. Ostensibly, the use of a multiple copy DNA sequence for phylogenetic analyses is problem- atic. Restriction fragments of a repeated sequence cannot be mapped to specific sites and thus it is impossible to ascertain whether the presence or ab- sence of each restriction fragment represents an in- dependent evolutionary event. For example, different size restriction fragments can be produced by site changes or by amplification events. In the latter case, different size fragments generated by different restric- tion enzymes may actually reflect only a single ampli- fication event.
In spite of this problem, there is strong evidence to suggest that the YBlO repeat is phylogenetically in- formative. This is evidenced from the similarities be- tween the phylogenetic trees depicted in Figure 2 and phylogenies of the same taxa constructed from a va- riety of data sets. For example, a consensus of the two trees depicted in Figure 2 is concordant with trees generated from mitochondrial DNA data of FERRIS et al. (1983) with regard to the position of M . spretus, M. hortulanus and M. abbotti. In both the mitochon- drial DNA and Y chromosome analyses these taxa are as different from each other as they are from the M. musculus/M. domesticus lineage. This is in contrast to phylogenetic trees generated from biochemical data that place M. hortulanus and M. abbotti as sister taxa (BONHOMME et al. 1984) and a phylogenetic tree gen- erated from differences in the 5 ’ end of the 16s mitochondrial rDNA molecule (FORT et al. 1984; BONHOMME 1986) that places M. hortulanus (= M. spicilegus) as a sister taxa to the M. domesticuslM. musculus lineage. T h e position of M. m. castaneus
within the M. musculus cluster in all the phylogenetic trees (Figures 2, A and B, and 4) contrasts with its placement as a distinct lineage in phylogenetic trees generated from the mitochondrial DNA (FERRIS et al.
1983) but its position in the second phylogenetic analysis (Figure 4) is concordant with the phylogenetic relationships generated from biochemical data of
BONHOMME et al. (1984). In addition, the placement of M , m , musculus and M . m. molossinus as sister taxa relative to M. m. castaneus and M. domesticus in the second phylogenetic analysis (Figure 4) is in agree- ment with phylogenetic analyses generated from mi- tochondrial DNA data (FERRIS et al. 1983; YONEGAWA
Y Chromosome Evolution in Mus 177
ZUKI et al. 1986), T-lymphocyte differentiation anti-
gen types (KURIHARA et al. 1985; MORIWAKI et al.
1986) and &-microglobulin types of the class I major histocompatibility antigens (ROBINSON et al. 1984).
T h e placement of M. musculus and M. domesticus as sister taxa relative to M. spretus, M. hortulanus and M. abbotti agrees with phylogenetic trees generated from both the mitochondrial and biochemical data sets. Finally, the position of M. domesticus and M. musculus
as monophyletic taxa is in accordance with all the data sets in the above comparisons. We conclude that the similarities among the phylogenetic relationships gen- erated by the Y chromosome data and other data sets indicate that the paternally inherited repeated se- quence YBlO provides phylogenetic information within and among a closely related group of species. Inconsistencies among the trees generated by differ- ent sets of characters probably reflect the close evo- lutionary relationships of these taxa.
Variation of Y chromosomes among M . spretus, M. hortulanus, M . abbotti, M . musculus and M. domesticus
is indicative of evolution of this Y chromosome re- peated sequence within the last 3-5 million years as
M . spretus, M. hortulanus and M. abbotti are hypothe- sized to have diverged from M. domesticus and M. musculus 3-5 million years ago (FERRIS et al. 1983).
Variation of Y chromosomes within the M. domesticus
and M. musculus samples is indicative of a rapid rate of evolution of this Y chromosome repeated sequence. If M. musculus and M. domesticus have diverged within the last 1-2 million years (FERRIS et al. 1983), Y
chromosome change has taken place within both lin- eages at the population level since that time.
Greater within species variation among Y chromo- somes in the M . domesticus samples compared to the
M . musculus samples can be attributed to either dis- parities in sampling, as the major portion of our M. musculus samples are from central Europe and thus may represent a single migration of this species into that region from central Asia, or to differential mu- tation or fixation rates between these two species. It is interesting to note that most of the polymorphism for the t-complex of genes in European populations of wild house mice is found within M. domesticus and not within M. musculus (KLEIN, SIPOS and FIGUEROA
1984). This suggests that the evolution of the t-com- plex and the Y chromosome in these two species reflect common histories.
T h e divergent Y chromosomes of the Robertsonian metacentric chromosome populations of M. domesticus
sampled (all with greater than five Robertsonians) may reflect the reproductive isolation of these Robertson- ian populations from surrounding M. domesticus with the normal ( 2 N = 40) acrocentric karyotype (CA- PANNA et al. 1976). T h e similarity among the Y chro- mosomes of the Robertsonian metacentric chromo-
some populations from Orobie, Molise and Lipari is surprising for two reasons. These populations are geographically isolated from each other and, with the exception of two Robertsonian chromosomes shared between the Orobie and Molise populations, each carries a different arrangement of Robertsonian chro- mosomes (reviewed in SAGE 198 1). T h e similarity of YB 10 sequences among these populations suggests that they have a common origin.
T h e inability to distinguish mice identified as M. d. domesticus or M. d. brevirostris using the YBlO se- quence corroborates the findings of the biochemical and mitochondrial DNA data (SAGE 1978, 1981; BON-
HOMME et al. 1978; BRITTON and THALER 1978; FER-
RIS et al. 1983). Although the biochemical and mito-
chondrial data suggest otherwise, our single sample identified as M. d. praetextus is clearly distinguishable on the basis of its Y chromosome type from other M .
domesticus and may reflect a real subspecific differ- ence. Additionally, a M . domesticus Y chromosome type identified in North America is distinct from the Y chromosome types sampled in Europe and the Medi- terranean region. Presumably, this Y chromosome type exists in an, as yet, unidentified population of M. domesticus in Europe, assuming that the presence of
M . domesticus in N. America is associated with Euro- pean colonization.
Documentation of the rapid rate of evolution of a Y chromosome specific repeated DNA sequence in a group of closely related species presents many intrigu- ing questions. For example, does a DNA sequence have a unique mode of evolution by virtue of its isolation on a monosomic chromosome? T h e exclusive occurrence of YB 10 sequences on the Y chromosome in the subgenus Mus and its restriction on the Y chromosome to a region that does not undergo recom- bination with the X chromosome (EICHER, PHILLIPS and WASHBURN 1983) precludes mechanisms of am- plification that involve homologous (or nonhomolo- gous) chromosome exchange. Amplification of this sequence could occur by unequal exchange between sister chromatids of the Y chromosome or by a process of duplicative transposition (ORGEL and CRICK 1980; DOOLITTLE and SAPIENZA 1980) limited in this in- stance to the Y chromosome. Are other Y chromosome specific DNA sequences evolving in a like manner,
i.e., is sequence amplification and divergence a general phenomenon of the mammalian Y chromosome?
178 P. K. Tucker, B. K. Lee and E. M. Eicher
maternally inherited mitochondrial
DNA.
Con- versely, such rapid evolution of Y chromosomeD N A
sequences may preclude their use for phylogenetic studies over broad taxonomic groups.
We especially thank RICHARD D. SAGE of the Museum of Ver- tebrate Zoology, University of California, Berkeley, and MICHAEL POTTER at the National Institute of Health for providing frozen tissue from wild mice for this study. Wild mice from POTTER were made available through National Cancer Institute contract N 0 1 - CB-25584. We are indebted to RODNEY L. HONEYCUTT and WARD C. WHEELER of the Museum of Comparative Zoology, Harvard University, for invaluable assistance in running the phylogenetic analyses and for many helpful discussions concerning the phyloge- netic data. We also thank the following people for reviewing the manuscript: RODNEY L. HONEYCUTT of Harvard University, and EDWARD H. BIRKENMEIER and JOSEPH N. NADEAU of T h e Jackson Laboratory. This publication was supported by grants HD07065 and GM20919 from the National Institutes of Health. T h e National Institutes of Health is not responsible for its contents nor do they necessarily represent the official views of that agency.
L I T E R A T U R E CITED
BISHOP, C. E., P. BOURSOT, B. BARON, F. BONHOMMEand D. HATAT, 1985 Most classical Mus musculus domesticus laboratory mouse strains carry a Mus musculus musculus Y chromosome. Nature 315: 70-72.
BONHOMME, F., 1986 Evolutionary relationships in the genus Mus.
Curr. Top. Microbiol. Immunol. 127: 19-34.
BONHOMME. F., J. BRITTON-DAVIDIAN, L. THALER and C. TRIAN-
TAPHYLIDIS, 1978 Sur I’existence en Europe de quatre
groupes d e souris (genre Mus I.) du rang espece e t semi-espece demontree par la genetique biochemique. C. R. Acad. Sci. Paris 287: 631-633.
BONHOMME. F., J. CATALAN, J. BRITTON-DAVIDIAN, V. M. CHAP-
M A N , K . MORIWAKI, E. NEVO and L. THALER, 1984
Biochemical diversity and evolution in the genus Mus. Biochem. Genet. 22: 275-303.
BRITTON, J., and L. THALER, 1978 Evidence for the presence of two sympatric species of mice (Genus Mus L.) in southern France based on biochemical genetics. Biochem. Genet. 1 6 213-225.
BROOKS, D. R., R. T . O’GRADY and E. 0. WILEY, 1986 A measure of the information content of phylogenetic trees, and its use as an optimality criterion. Syst. Zool. 35: 571-581.
CAPANNA, E . , A. GROPP, H . WINKING, G. NOACK and M.-V. CIVI- TELLI, 1976 Robertsonian metacentrics in the mouse. Chro- mosonm 58: 341-353.
CASANOVA, M., R. LEROY, C. BOUCEKKINE, J. WEISSENBACH, C. BISHOP, M . FELLOUS, M. PURRELLO, G. FIORI and M. SINIS-
CALCO, 1985 A human Y-linked DNA polymorphism and its
potential for estimating genetic and evolutionary distance. Sci- ence 230: 1403- 1406.
DOOLITTLE, W. F., and C. SAPIENZA, 1980 Selfish genes, the phenotype paradigm and genome evolution. Nature 2 8 4 601-
603.
EICHER, E. M., S . J. PHILLIPS and L. L. WASHRURN, 1983 T h e use of molecular probes and chromosomal rearrangements to par- tition the mouse Y chromosome into functional regions, pp. 57-7 1. In Recombinant D N A and Medical Genetics, edited by A. MESSER and I . H. PORTER. Academic Press, New York.
EICHER, E. M., K. W. HUTCHISON, S. J. PHILLIPS, P. K. TUCKER and B. K. LEE, 1989 A repeated segment on the mouse Y chromosonle is composed of retroviral-related, Y-enriched, and Y-specific sequences. Genetics 122: 181-192.
FARRIS, J. S., 1970 Methods for computing Wagner trees. Syst. Zool. 1 9 83-92.
FERRIS, S. D., R. D. SAGE, E. M. PRAGER, U . RITTE and A. C. WILSON, 1983 Mitochondrial DNA evolution in mice. Ge- netics 105: 681-721.
FORT, P., F. BONHOMME, P. DARLU, M. PIECHACZYK, P. JEANTEUR and L. THALER, 1984 Clonal divergence of mitochondrial DNA versus populational evolution of nuclear genome. Evol. Theory 7: 8 1-90,
GOODFELLOW, P., S. DARLING and J. WOLFE, 1985 T h e human Y chromosome. J. Med. Genet. 22: 329-344.
HUNT, W. G., and R. K. SELANDER, 1973 Biochemical genetics of hybridization in European house mice. Heredity 3: 11-33.
JENKINS, N. A., N. G. COPELAND, B. A. TAYLOR and B. K. LEE,
1982 Organization, distribution and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromo- somes of Mus musculus. J. Virol. 43: 26-36.
JONES, K. W., and L. SINGH, 1981 Conserved repeated DNA sequences in vertebrate sex chromosomes. Hum. Genet. 58: 46-
53.
KLEIN, J., P. SIPOS and F. FIGUEROA, 1984 Polymorphism of t- complex genes in European wild mice. Genet. Res. 44: 39-46. KURIHARA, Y., N. MIYASHITA, K . MORIWAKI, M. L. PETRAS, F.
BONHOMME, W. S. CHO and S. KOHNO, 1985 Serological survey of T-lymphocyte differentiation antigens in wild mice. Immunogenetics 22: 2 1 1-2 18.
LAMAR, E. E., and E. PALMER, 1984 Y-encoded, species specific DNA in mice: evidence that the Y chromosome exists in two polymorphic forms in inbred strains. Cell 37: 17 1-1 77. MANIATIS, T., E. F. FRITSCH and J. SAMBROOK, 1982 Molecular
Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory,
Cold Spring Harbor, N.Y.
MORIWAKI, K., N. MIYASHITA, H. SUZUKI, Y. KURIHARA and H. YONEKAWA, 1986 Genetic features of major geographical isolates of Mus musculus. Curr. Top. Microbiol. Immunol. 127: 55-61,
MULLER, H. J., 1914 A gene for the fourth chromosome of Drosophila. J. Exp. Zool. 17: 325-336.
NALLASETH, F. S., and M. J. DEWEY, 1986 Moderately repeated mouse Y chromosomal sequence Families present distinct types of organization and evolutionary change. Nucleic Acids Res. 14: 5295-5307.
NISHIOKA, Y., and E. LAMOTHE, 1986 Isolation and characteriza- tion of a mouse Y chromosomal repetitive sequence. Genetics 113: 417-432.
OHNO, S., 1967 Sex chromosomes and sex-linked genes, pp. 1-
192 in Monographs on Endocrinology, Vol. 1 , edited by A. LAR-
HART, T. MANN, L. T . SAMUELS and J. ZANDER. Springer-
Verlag, New York.
ORGEL, L. E., and F. H. CRICK, 1980 Selfish DNA: the ultimate parasite. Nature 284: 604-607.
PHILLIPS, S. J., E. H. BIRKENMEIER, R. CALLAHAN and E. M. EICHER, 1982 Male and female mouse DNAs can be discriminated using retroviral probes. Nature 297: 241-243.
REED, K . C., and D. A. MANN, 1985 Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 13: 7207- 722 1.
RORINSON, P. J., M. STEINMETZ, K. MORIWAKI and K. FISCHER- LINDAHL, 1984 Beta-2 microglobulin types in mice of wild origin. Immunogenetics 2 0 655-665.
SAGE, R. D., 1978 Genetic heterogeneity of Spanish house mice
(Mus musculus complex), pp. 5 19-553 in Origins of Inbred Mice,
edited by H . C . MORSE 111. Academic Press, New York. SAGE, R. D., 1981 Wild mice, pp. 39-90 in TheMousein Biomedical
Research Vol. I: History, Genetics and Wild Mice, edited by H. L.
FOSTER, J. D. SMALL and J. G. Fox. Academic Press, New York.
Y Chromosome Evolution in Mus 179 analysis of a hybrid zone between domesticus and musculus
mice (Mus musculus complex): hemoglobin polymorphisms. Curr. Top. Microbiol. Immunol. 127: 75-85.
SHAW, G . M., M. A. GONDA, G. H. FLICKINGER, B. H. HAHN, R. C. GALLO and F. WONG-STAAL, 1984 Genomes of evolutionarily divergent members of the human T-cell leukemia virus family (HTLV-I and HTLV-11) a r e highly conserved, especially in pX. Proc. Natl. Acad. Sci. USA 81: 4544-4548.
SINGH, L., I. F. PURDOM and K. W. JONES, 1981 Conserved sex chromosome-associated nucleotide sequences in eukaryotes. Cold Spring Harbor Symp. Quant. Biol. 45: 805-8 13.
SOLARI, A. J., 1974 T h e behavior of the XY pair in mammals. Int. Rev. Cytol. 3 8 273-317.
SUZUKI, H., N. MIYASHITA, K. MORIWAKI, R. KOMINAMI, M. MUR- AMATSU, T . KANEHISA, F. BONHOMME, M. L. PETRAS, 2. Yu and D. Lu, 1986 Evolutknary implication of heterogeneity of the nontranscribed spacer region of ribosomal DNA repeat- ing units in various subspecies of Mus musculus. Mol. Biol. Evol. 3: 126-137.
SWOFFORD, P. L., 1985 PAUP version 2.4. Illinois Natural History Survey, Champaign, IL.
YONEGAWA, H., 0. GOTOH, Y. TAGASHIRA, Y. MATSUSHIMA, L.4. SHI, W. S. CHO, N. MIYASHITA and K. MORIWAKI, 1986 A hybrid origin of Japanese mice M . m. molossinus. Curr. Top. Microbiol. Immunol. 127: 62-67.