Iran J Public Health, Vol. 47, No.8, Aug 2018, pp.1128-1136
Original Article
Four Non-functional
FUT1
Alleles Were Identified in Seven
Chinese Individuals with Para-Bombay Phenotypes
Wei LIANG
1,2, Feng CAI
3, Liang YANG
4, *Zhe ZHANG
5, *Zhicheng WANG
11. Dept. of Laboratory Medicine, Huashan Hospital, Shanghai Medical College, Fudan University, Shanghai, China 2. Dept. of Laboratory Medicine, The Second People's Hospital of Lianyungang City, Lianyungang, Jiangsu Province, China 3. Dept. of Laboratory Medicine, Shanghai Municipal Hospital of Traditional Chinese Medicine, Shanghai University of Traditional
Chinese Medicine, Shanghai, China
4. Laboratory Department of Center Blood Station Ningbo, Ningbo, Zhejiang Province, China 5. Health Bureau of Ningbo City, Ningbo, Zhejiang Province, China
*Corresponding Authors: Emails: ahwzc@126.com; nblwys@hotmail.com
(Received 19 Feb 2017; accepted 10 Jul 2017)
Introduction
The Bombay and para-Bombay phenotypes are characterized by the deficiency of H, A, and B blood group antigens on the red blood cell (RBC) (1). The ABO locus on 9q34 determines the A
and B antigens, while, α (1, 2)-fucosyltransferase genes FUT1 and FUT2 encode the H antigen, the precursor of A and B antigens. Both FUT1 and FUT2 gene encode α (1, 2)-fucosyltransferase and Abstract
Background: The para-Bombay phenotype is characterized by a lack of ABH antigens on red cells, but ABH substances are found in saliva. Molecular genetic analysis was performed for seven Chinese individuals serolog-ically typed as para-Bombay in Blood Station Center of Ningbo, Zhejiang Province, Ningbo, China from 2011 to 2014.
Methods: RBCs’ phenotype was characterized by standard serologic technique. Genomic DNA was se-quenced with primers that amplified the coding sequence of α (1, 2)-fucosyltransferase genes FUT1 (or H) and FUT2 (or Se), respectively. Routine ABO genotyping analysis was performed. Haplotypes of FUT1 were identi-fied by TOPO cloning sequencing. Phylogenetic tree of H proteins of different organisms was performed us-ing Mega 6 software.
Results: Seven independent individuals were demonstrated to possess the para-Bombay phenotype. RBC
ABO genotypes correlated with ABH substances in their saliva. FUT1 547delAG (h1), FUT1 880delTT (h2), FUT1 658T (h3) and FUT1 896C were identified in this study. FUT1 896C was first revealed by our team. The H-deficient allele reported here was rare and the molecular basis for H deficient alleles was diverse as well in the Chinese population. In addition, the FUT2 was also analyzed, only one FUT2 allele was detected in our study: Se357. Phylogenetic tree of the H proteins showed that H proteins could work as an evolutionary and
genetic marker to differentiate organisms in the world.
Conclusion: Molecular genetic backgrounds of seven Chinese individuals were summarized sporadic and ran-dom mutations in the FUT1 gene are responsible for the inactivation of the FUT1-encoded enzyme activity.
are closely linked on 19q13, showing 70% DNA sequence homology (2, 3), however, the biologi-cal role of them is distinct (4, 5), FUT1 is the H gene expressed mainly on the membrane of the human erythrocytes and FUT-2 is the Se gene expressed exclusively in the secretory glands and the digestive mucosa.
The para-Bombay phenotype is characterized by a non-functional FUT1 gene accompanied by an active FUT2 gene. The first mutant FUT1 gene was identified in an India individual who lacked the H enzyme and had no H antigens on erythro-cytes, which was a typical Bombay phenotype. To date, more than 43 silencing or weakening muta-tions have been described for FUT1 in the Blood Group Antigen Gene Mutation Database of the US National Center or Biotechnology Infor-mation. FUT1 gene determines the synthesis of H type 1 (following A/B antigens) adsorbed onto the membrane of RBC from the plasma, but the encoded enzyme activity by a deficient FUT1 gene is greatly abated, resulting in a lower amounts of H antigen (and A/B antigen) on the surface of RBC. In above situation, no matter the function of FUT2 gene is normal or not, H anti-gen (and A/B antianti-gen) is poorly expressed and can only be detected by adsorption-elution tests using proper the anti-H (and anti-A/B) reagents. The anti-H made from para-Bombay individuals usually shows a weaker reaction in the adsorp-tion-elution test compared with the anti-H from individuals with the Bombay phenotype, which usually shows strong reactive with a wide thermal range, whereas, it is less reactive and even does not react above room temperature for anti-H from para-Bombay individuals. This paper de-scribed the molecular genetic backgrounds of seven such Chinese individuals.
Materials and Methods
Blood samples and saliva samples
Six probands with the para-Bombay phenotypes were identified during pre-transfusion testing in the time-period 2011 to 2014. One proband was a volunteer donor at the Ningbo Blood Station of
Zhejiang Province in China, whose erythrocytes showed the rare phenotype with a cell and serum grouping discrepancy was suspected to be a para-Bombay individual. Overall, 5 mL of peripheral blood was bled with ethylenediaminetetraacetic acid dipotassium (EDTA-2K) anticoagulant from each individual. Saliva samples were presented by all the suspected para-Bombay individuals as well. ABH antigens on erythrocytes and in saliva were examined as well.
Genomic DNA was extracted from whole blood samples using a DNA isolation kit (Qiagen, Hil-den, Germany) according to the manufacturer’s instruction. The DNA of peripheral blood from 110 randomly chosen Chinese individuals with normal ABO blood group phenotypes were iso-lated to assess the frequency of H allele in natural population. All the subjects signed the informed consents
Blood group serological studies
ABO serology was performed with standard se-rological techniques. The adsorption-elution test (6) was used to detect trace amounts of A/B an-tigens on red blood cells or H anan-tigens in sera. The haemagglutination inhibition test was em-ployed to detect whether ABH substances were present or not in saliva (6). Lewis blood group was also tested to know the secretory type. For routine testing, one drop of anti-A, -B (Shenxing, Shanghai, China), anti-H, anti-AB, anti-Lea and anti-Leb (Sanquin, Amsterdam, Netherlands) was placed in a tube and mixed with washed RBCs, respectively. After centrifugation, the results of haemagglutination were observed macroscopical-ly and microscopicalmacroscopical-ly. The human anti-A, B was prepared by our laboratory.
ABO genotyping
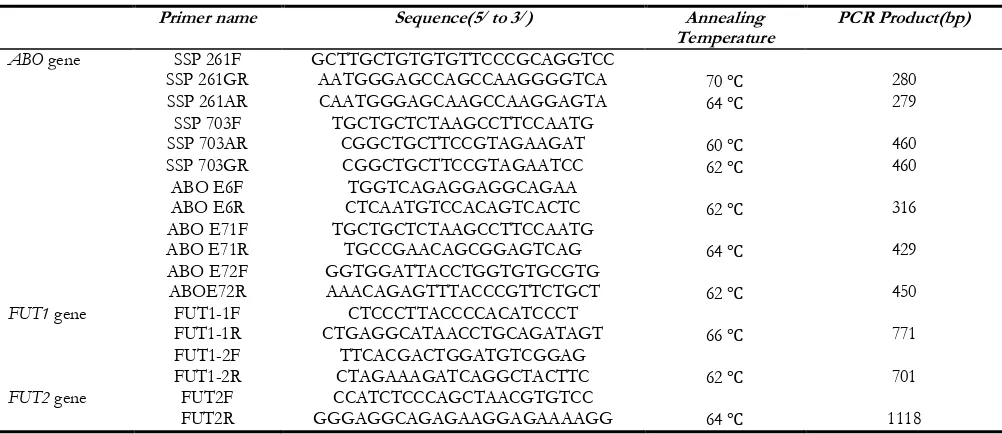
Table 1: Primers and PCR conditions used in the analysis of ABO, FUT1 and FUT2 genes
Primer name Sequence(5/ to 3/) Annealing
Temperature PCR Product(bp)
ABO gene SSP 261F GCTTGCTGTGTGTTCCCGCAGGTCC
SSP 261GR AATGGGAGCCAGCCAAGGGGTCA 70 ℃ 280
SSP 261AR CAATGGGAGCAAGCCAAGGAGTA 64 ℃ 279
SSP 703F TGCTGCTCTAAGCCTTCCAATG
SSP 703AR CGGCTGCTTCCGTAGAAGAT 60 ℃ 460
SSP 703GR CGGCTGCTTCCGTAGAATCC 62 ℃ 460
ABO E6F TGGTCAGAGGAGGCAGAA
ABO E6R CTCAATGTCCACAGTCACTC 62 ℃ 316
ABO E71F TGCTGCTCTAAGCCTTCCAATG
ABO E71R TGCCGAACAGCGGAGTCAG 64 ℃ 429
ABO E72F GGTGGATTACCTGGTGTGCGTG
ABOE72R AAACAGAGTTTACCCGTTCTGCT 62 ℃ 450
FUT1 gene FUT1-1F CTCCCTTACCCCACATCCCT
FUT1-1R CTGAGGCATAACCTGCAGATAGT 66 ℃ 771
FUT1-2F TTCACGACTGGATGTCGGAG
FUT1-2R CTAGAAAGATCAGGCTACTTC 62 ℃ 701
FUT2 gene FUT2F CCATCTCCCAGCTAACGTGTCC
FUT2R GGGAGGCAGAGAAGGAGAAAAGG 64 ℃ 1118
F: Forward primer, R: reverse primer, GR, AR: The reverse primer specified for an allele of ABO gene, whose certain site is base G, A, respectively
Sequencing of ABO exons 6 and 7
ABO exact genotypes were determined by sequenc-ing of exons 6 and 7of ABO gene, whose primers used, are listed in Table 1. DNA fragments were amplified with primers ABO-E6F and ABO-E6R for exon 6 or primers ABO-E71F, ABO-E71R, ABO-E72F and ABO-E72R for exon 7. In order to acquire clearer sequence diagrams, two pairs of primers were designed for exon 7. The 50μL reac-tion mixture contained 25μL 2×dNTP (TI-ANGEN, Beijing, China), 2.5 μL of each primer (Invitrogen, Life Technologies, USA), 200 ng of genomic DNA and water. After initial denaturation at 95 °C for 1 min, the reaction mixtures were sub-jected to 35 cycles of denaturation at 95 °C for 25 sec, followed by annealing at each optimal tempera-ture (Table 1) for 25 sec and extending at 75 °C for 45 sec, plus a final extension at 72 °C for 5 min. PCR products were separated on a 1.5% agarose gel (Biowest, Gene Company, Spain), all showed a single bright band, then the PCR products were purified and unidirectionally sequenced with an ABI BigDye Terminator Cycle Sequencing kit (Ap-plied Biosystems, CA, USA) and Universal DNA Purification Kit (TIANGEN, Beijing, China) ac-cording to the manufacturer’s instructions, respec-tively. The sequence data were analyzed by FinchTV1.4 software (Geospiza, Seattle, USA) and
the ABO genotypes were assigned according to the nucleotides at the polymorphic ABO positions. All the acquired nucleotides sequences were compared with standard ABO polymorphisms from the dbRBC of NCBI and each SNP or mutation was analyzed and documented in the ABO gene.
Sequencing of FUT1
Two DNA fragments covering the entire coding region (1098bp) were amplified to identify the mu-tations in the FUT1. The reagents and protocols used in the PCR were the same as the sequencing of ABO gene mentioned in the above section. The sequence data were analyzed by FinchTV1.4 soft-ware (Geospiza, Seattle, USA) and all achieved nu-cleotides sequences were compared with standard Hh polymorphisms from the dbRBC of NCBI, and every mutation in the FUT1 gene was analyzed, each FUT1 genotype was assigned at last.
Analysis of FUT1 haplotype
LB plates were selected randomly and screened using colony-PCR for each sample. Plasmid DNA of positive colony was extracted by a kit (TIANGEN Beijing, China) and used as tem-plates for the sequencing reaction. The PCR products were sent to Shanghai Sunny Biotech-nology Co., Ltd (Sunny, Shanghai, China), where all the following experiment steps were finished.
Sequencing of FUT2
To analyse the genotype of FUT2, the whole cod-ing region (1118bp) of FUT2 was amplified uscod-ing
the primers (Table 1). The primer design and PCR amplification of FUT2 were performed (7).
Phylogenetic analysis
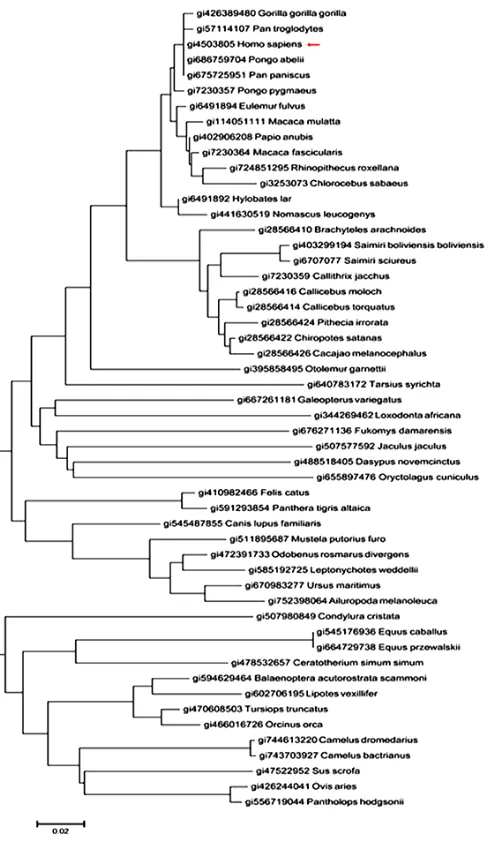
Human sapiens H protein (gi4503805) sequence as query sequence was pasted in the text area of BlastP, and 52 organisms who express the H pro-teins were searched out, every protein sequence was downloaded in FASTA format. The evolu-tionary history (Fig. 1) was inferred using the Neighbor-Joining method (8).
Fig. 1: Evolutionary relationships of taxa
The evolutionary distances were computed using the Poisson correction method (9) and the evolu-tionary analyses were conducted in MEGA6 (10).
Results
Serological results and ABO genotypes
The ABH substances on RBCs could not be de-tected using direct agglutination, even all the rea-gents, polyclonal, monoclonal anti-sera and the lectin Ulex europaeus (anti-H) were chosen to perform such experiment (Table 2). However, the microscale A and/or B antigens on red cells were detected by the absorption-elution assay. The presence of ABH substances in saliva was consistent with their Le (a–b+) phenotypes.
The analysis of the FUT1 gene
Three different mutations (h1, h2 and h3) were detected in the six individuals with the para-Bombay phenotypes using DNA sequencing based on the entire FUT1 coding region. The genotypes of heterozygous (h1h3) or homozygous (h1h1, h2h2) were identified (Table 2), according to the nomenclature for non-functional FUT1 alleles (11). However, for the case 7, two hetero-zygous mutations of the FUT1,
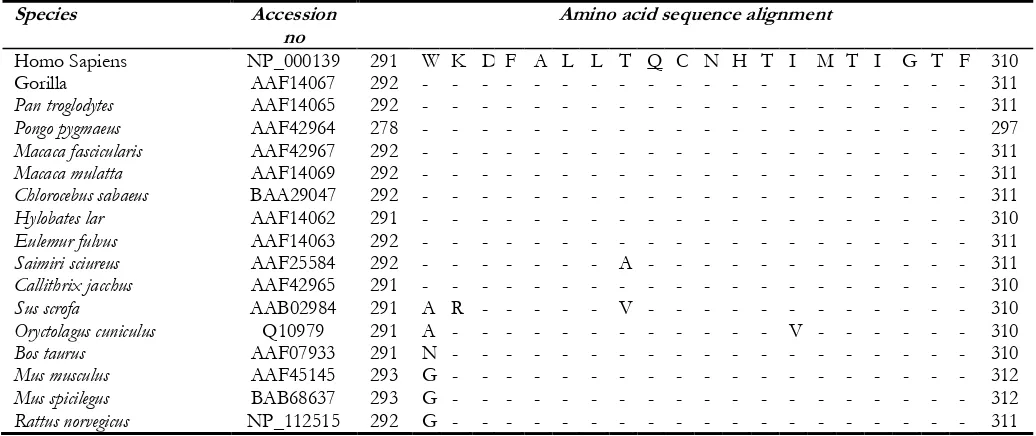
547-552AGAGAG/AGAG, and 896T/C were identi-fied by our team (12). Analysis of sequences ho-mologous to human FUT1 showed that Gln299 was conserved in the FUT1 enzymes of 16 other mammals reported to date (Table 3), which sug-gested that Gln299 of the human FUT1 enzyme may be important in maintaining the biological function.
The analysis of the FUT2 gene
The relevant ABH antigens were detected in the saliva for each individual, which showed that an active FUT2 gene existed in each individual. The homozygous mutation 357T was observed in each individual by direct DNA sequencing com-pared with the reference sequence (GenBank ac-cession no. U17894) in the coding region. The 357C>T variant of FUT2 did not result in an amino acid change, are common in Asian popula-tions (13).
Phylogenetic analysis
Phylogenetic tree was portrayed, showing that H proteins could work as an evolutionary and ge-netic marker to differentiate organisms in the world.
Table 2: Phenotypes and genotypes of 7 Chinese para-Bombay individuals
No Haemagglutination Absorption
-elution Antigens in saliva
Anti-H In serum
Genotypes Para-Bombay
Phenotype
A B H Lewis A B A B ABO FUT1 FUT2
1 - - - a-b+ - + - + + B101
/O01 /h1 h1 Se
357
/Se357 B
2 - - - a-b+ + - + - + A102
/O02 /h3 h1 Se
357
/Se357
A
3 - - - a-b+ + + + + + A102
/B10 1
h1
/h1 Se
357
/Se357 AB
4 - - - a-b+ + - + - + A101
/O01 /h3 h1 Se
357
/Se357
A
5 - - - a-b+ + - + - + A102
/O02 /h2 h2 Se
357
/Se357
A
6 - - - a-b+ + - + - + A102
/O01 /h3 h1 Se
357
/Se357
A
7 - - - a-b+ - + - + + B101
/O02 /h? h1 Se
357
/Se357 B
Table 3: Amino acid sequence alignment for FUT1 enzyme
Species Accession
no Amino acid sequence alignment
Homo Sapiens NP_000139 291 W K D F A L L T Q C N H T I M T I G T F 310
Gorilla AAF14067 292 - - - 311
Pan troglodytes AAF14065 292 - - - 311
Pongo pygmaeus AAF42964 278 - - - 297
Macaca fascicularis AAF42967 292 - - - 311
Macaca mulatta AAF14069 292 - - - 311
Chlorocebus sabaeus BAA29047 292 - - - 311
Hylobates lar AAF14062 291 - - - 310
Eulemur fulvus AAF14063 292 - - - 311
Saimiri sciureus AAF25584 292 - - - A - - - 311
Callithrix jacchus AAF42965 291 - - - 310
Sus scrofa AAB02984 291 A R - - - V - - - 310
Oryctolagus cuniculus Q10979 291 A - - - V - - - 310
Bos taurus AAF07933 291 N - - - 310
Mus musculus AAF45145 293 G - - - 312
Mus spicilegus BAB68637 293 G - - - 312
Rattus norvegicus NP_112515 292 G - - - 311
Dashes symbolize amino acid sequences identity with the human sequence. The affected amino acid in the FUT1 896C allele is underlined
Discussion
In the present study, we detected seven individu-als; all of them possessed the para-Bombay phe-notype, having the distinct genetic background, respectively. Four non-functional FUT1 alleles were tested by DNA sequencing based on the entire FUT1 coding region, including three re-ported defective FUT1 alleles: FUT1 547delAG (h1), FUT1 880delTT (h2), FUT1 658T (h3) and a novel FUT1 allele, FUT1 896C (13). Both alleles' h1 and h2 are two-base deletions: the AG dele-tion is located at nucleotides 547–552 for h1 and the TT bases are deleted at nucleotides 880–882 for h2. The h3 allele contains a C658 to T mis-sense mutation, which results in a change from Arg to Cys at amino acid position 220. These mutations were also reported in individuals with the para-Bombay phenotypes in other places (11,14). FUT1 896 C was first revealed by our team. The H-deficient allele reported here was, as expected, rare in the Chinese population and the molecular basis for H deficient alleles was diverse as well. In addition to the FUT1, the FUT2 was also analyzed, only one FUT2 allele was detected in our study: Se357. Se357 allele was very common
in the Asian populations (7, 14-16). FUT2 gene analysis results were consistent with the subjects' secretor status. Different ethnic and/or geo-graphic mutations are revealed for the FUT2 gene and some of the mutations could result in a non-secretor phenotype. The prevalent synony-mous mutation for FUT2 gene is 357C>T in Asian populations compared with counterpart, the nonsense mutation 428G>A in the African and Caucasian populations (17). “The relatively high allele frequency for some of the FUT2-null alleles is likely an evolutionary advantage when the soluble and/or mucosal H antigens are absent, and the presence of H determinants on mucosal surfaces may be more biologically im-portant than their cellular analogs, various reports of the increased resistance to infection by a wide range of pathogens in individuals of the nonse-cretor phenotype supported the observations (18-21).”
be as high as 1:347 (22). In the whole Japanese population, the incidence of Bombay and para-Bombay conjectured is approximately one in two or 300000 (23). There are more para-Bombay phenotypes than Bombay in the Chinese popula-tion. Data showed that the incidence of FUT1 mutations were 1/8000-1/10000, 1-15620 in Taiwan and Hong Kong, respectively (7,11). To date, more than 43 effective mutations have been documented for FUT1. The mutation, giv-ing rise to Bombay phenotype was first described (24), FUT1 725T>G, together with the deletion of the FUT2 gene has been detected only in sub-jects from subcontinental Indian (25). Another preferment mutation is the FUT1 349C>T, usu-ally found on the island of Reunion, moreover, FUT1 547delAG(h1), FUT1 880delTT(h2), FUT1 658T(h3) mutation was found mainly in Chinese population (14, 16, 26). FUT1 695A, FUT1 990delG, FUT1 721C mutation was prevalent in Japanese (23) and so on. The mutation of FUT1 gene is closely related to the geographical regions, demonstrated by this study.
In contrast, non-functional FUT2 mutations are keeping at a relative steady frequency, about 20% in most populations. In European and African populations, the most prevalent nonsense muta-tion is 428G>A, with an allele frequency of 0.47 and 0.416, respectively (27). In Asian popula-tions, the allele harboring both the synonymous mutation 357C>T and the inactivating mutation 385A>T is the main cause of the nonsecretor phenotype with a frequency of 0.406 (28, 29). The inactivation of the FUT1 gene happened af-ter FUT2 gene inactivation, as all of the Bombay and nonsecretor para-Bombay individuals had the same inactivated FUT2 allele but possessed dis-tinct inactivated FUT1 alleles (23), according to our study, there might be some specific selective advantage on the individuals with the mutant FUT2 alleles, but some selective disadvantage on the individuals with the mutant FUT1 alleles. FUT2 mutations were more ethnically specific and may be used as anthropologic markers (27, 30).
Conclusion
Four non-functional FUT1 alleles (h1, h2, h3, FUT1 896C) were identified in seven Chinese individuals with para-Bombay phenotypes and on the same Se357/Se357 haplotype background. As the para-Bombay phenotype is rare in the natural population, it may bring troubles in clinical blood transfusion, blood typing and so on; this article would contribute to understanding the special blood group not only in theory but also in prac-tice.
Ethical considerations
Ethical issues (Including plagiarism, Informed consent, misconduct, data fabrication and/or fal-sification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
This study was financially supported by grants from the National Natural Science Foundation of China (NSFC 81270650), the Shanghai Municipal Commission of Health and Family Planning (No. 20134450), Lianyungang Science and Technology Bureau Project (SH1526) and Bengbu Medical College Research Project (BYKY17182)
Conflict of interest
The authors declare that there is no conflict of interests.
References
1. Zhang A, Chi Q, Ren B (2015). Genomic analy-sis of para- Bombay individuals in south-eastern China: the possibility of linkage and disequilibrium between FUT1 and FUT2. Blood Transfus, 13(3): 472-477.
APOC2, D19S7 and D19S9. Ann Hum Genet, 55(Pt 3): 225-233.
3. Reguigne-Arnould I, Couillin P, Mollicone R et al (1995). Relative positions of two clusters of human alpha-L-fucosyltransferases in 19q (FUT1-FUT2) and 19p (FUT6-FUT3-FUT5) within the microsatellite genetic map of chromosome 19. Cytogenet Cell Genet, 71(2): 158-162.
4. Oriol R, Danilovs J, Hawkins BR (1981). A new genetic model proposing that the Se gene is a structural gene closely linked to the H gene. Am J Hum Genet, 33(3): 421-431.
5. Le Pendu J, Cartron JP, Lemieux RU, et al (1985). The presence of at least two different H-blood-group-related beta-D-gal alpha-2-L-fucosyltrans- ferases in human serum and the genetics of blood group H substances. Am J Hum Genet, 37(4): 749-760.
6. Matsushita M, Otani K, Sakamoto Y, et al (2015). Increase in Alkaline Phosphatase Ac-tivity after High-Fat Meal Ingestion is Corre-lated to the Amount of ABH Substances in Saliva. Rinsho Byori, 63(5): 543-547.
7. Yip SP, Chee KY, Chan PY, et al (2002). Molec-ular genetic analysis of para-Bombay pheno-types in Chinese: a novel non-functional FUT1 allele is identified. Vox Sang, 83(3): 258-262.
8. Saitou N, Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 4(4): 406-425.
9. Zuckerkandl E, Pauling L (1965). Evolutionary divergence and convergence in proteins. Ed-ited in Evolving Genes and Proteins by Bryson V and Vogel HJ, Academic Press. New York, 97-166.
10. Tamura K, Stecher G, Peterson D, et al (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol, 30(12): 2725-9.
11. Yu LC, Yang YH, Broadberry RE, et al (1997). Heterogeneity of the human H blood group alpha(1,2)fucosyltransferase gene among para-Bombay individuals. Vox Sang, 72(1): 36-40. 12. Liang W, Xu H, Liu YY, et al (2015). Molecular
genetic analysis of para-Bombay phenotype in Chinese persons: a novel FUT1 allele is iden-tified. Transfusion, 55(6 Pt 2): 1588.
13. Blumenfeld OO, Patnaik SK (2004). Allelic genes of blood group antigens: a source of human mutations and cSNPs documented in the Blood Group Antigen Gene Mutation Database. Hum Mutat, 23(1): 8-16.
14. Cai XH, Jin S, Liu X, et al (2011). Molecular ge-netic analysis for the para-Bombay blood group revealing two novel alleles in the FUT1 gene. Blood Transfus, 9(4): 466-468.
15. Wang B, Koda Y, Soejima M, et al (1997). Two missense mutations of H type al-pha(1,2)fucosyltransferase gene (FUT1) re-sponsible for para-Bombay phenotype. Vox Sang, 72(1): 31-35.
16. Xu X, Tao S, Ying Y, et al (2011). A novel FUT1 allele was identified in a Chinese individual with para-Bombay phenotype. Transfus Med, 21(6): 385-393.
17. Kelly RJ, Rouquier S, Giorgi D, et al (1995). Se-quence and expression of a candidate for the human Secretor blood group al-pha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem, 270(9): 4640-9.
18. Storry JR, Johannesson JS, Poole J, et al (2006). Identification of six new alleles at the FUT1 and FUT2 loci in ethnically diverse individu-als with Bombay and Para-Bombay pheno-types. Transfusion, 46(12): 2149-2155.
19. Ali S, Niang MA, N'doye I, et al (2000). Secretor polymorphism and human immunodeficiency virus infection in Senegalese women. J Infect Dis, 181(2): 737-739.
20. Thorven M, Grahn A, Hedlund KO, et al (2005). A homozygous nonsense mutation (428G-->A) in the human secretor (FUT2) gene pro-vides resistance to symptomatic norovirus (GGII) infections. J Virol, 79(24): 15351-5. 21. Kindberg E, Hejdeman B, Bratt G, et al (2006).
A nonsense mutation (428G-->A) in the fu-cosyltransferase FUT2 gene affects the pro-gression of HIV-1 infection. AIDS, 20(5): 685-9.
22. Wagner FF, Flegel WA (1997). Polymorphism of the h allele and the population frequency of sporadic nonfunctional alleles. Transfusion, 37(3): 284-290.
of Bombay and para-Bombay individuals that inactivate H enzyme. Blood, 90(2): 839-849. 24. Bhende YM, Deshpande CK, Bhatia HM, et al
(1994). A "new" blood-group character relat-ed to the ABO system. 1952. Indian J Mrelat-ed Res, 99: 3p.
25. Koda Y, Soejima M, Johnson PH, et al (1997). Missense mutation of FUT1 and deletion of FUT2 are responsible for Indian Bombay phenotype of ABO blood group system. Bio-chem Biophys Res Commun, 238(1): 21-25. 26. Yan L, Zhu F, Xu X, et al (2005). Molecular
ba-sis for para-Bombay phenotypes in Chinese persons, including a novel nonfunctional FUT1 allele. Transfusion, 45(5): 725-730. 27. Liu Y, Koda Y, Soejima M, et al (1998).
Exten-sive polymorphism of the FUT2 gene in an African (Xhosa) population of South Africa. Hum Genet, 103(2): 204-210.
28. Henry S, Mollicone R, Fernandez P, et al (1996). Molecular basis for erythrocyte Le(a+ b+) and salivary ABH partial-secretor phenotypes: expression of a FUT2 secretor allele with an A-->T mutation at nucleotide 385 correlates with reduced alpha(1,2) fucosyltransferase ac-tivity. Glycoconj J, 13(6): 985-993.
29. Kudo T, Iwasaki H, Nishihara S, et al (1996). Molecular genetic analysis of the human Lew-is hLew-isto-blood group system. II. Secretor gene inactivation by a novel single missense muta-tion A385T in Japanese nonsecretor individu-als. J Biol Chem, 271(16): 9830-7.