Copyright © 2002, American Society for Microbiology. All Rights Reserved.
Dissemination of New Methicillin-Resistant
Staphylococcus aureus
Clones in the Community
Keiko Okuma,
1Kozue Iwakawa,
1John D. Turnidge,
2Warren B. Grubb,
3Jan M. Bell,
2Frances G. O’Brien,
3Geoffrey W. Coombs,
4John W. Pearman,
4Fred C. Tenover,
5Maria Kapi,
1Chuntima Tiensasitorn,
1Teruyo Ito,
1and Keiichi Hiramatsu
1*
Department of Bacteriology, Juntendo University, Tokyo, Japan1; Department of Microbiology and InfectiousDiseases, Women’s and Children’s Hospital, North Adelaide,2and Molecular Genetics Research Unit,
School of Biomedical Sciences, Curtin University of Technology,3and Department of
Microbiology and Infectious Diseases, Royal Perth Hospital,4Perth, Australia;
and National Center for Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, Georgia 303335
Received 25 April 2002/Returned for modification 21 June 2002/Accepted 6 August 2002
Multiple methicillin-resistantStaphylococcus aureus(MRSA) clones carrying type IV staphylococcal cassette chromosome mecwere identified in the community-acquired MRSA strains of both the United States and Australia. They multiplied much faster than health-care-associated MRSA and were resistant to fewer non-beta-lactam antibiotics. They seem to have been derived from more diverse S. aureus populations than health-care-associated MRSA strains.
Methicillin-resistant Staphylococcus aureus (MRSA), be-sides having established itself as a major hospital pathogen, is now beginning to prevail in the wider community as well (1, 3–5). However, we do not know if the subgroup of MRSA designated community-acquired MRSA (C-MRSA) share a common origin of derivation with the other subgroup of MRSA in hospitals, namely the health-care-associated MRSA (H-MRSA). The majority of H-MRSA strains carry one of the three types of staphylococcal cassette chromo-some mec (SCCmec) as the methicillin resistance determi-nant on their chromosomes (19, 22). However, members of our group have recently identified a novel SCCmec, desig-nated type IV, in the C-MRSA strains isolated at a Chicago children’s hospital (23). This raised a possibility that C-MRSA might have an origin of derivation distinct from that of H-MRSA, and type-IV SCCmeccould be its unique genetic marker (14). To further test this view, we now analyzed 23 well-characterized C-MRSA strains (2–4, 24–26, 28) whose sources of isolation were not associated with risk factors for H-MRSA infection (e.g., recent hospitalization, recent sur-gery, residence in a long-term care facility, drug use, etc.) (7, 11) and 12 Australian MRSA strains designated non-multire-sistant oxacillin-renon-multire-sistantS. aureus(NORSA) (9) and com-pared them with the representative H-MRSA strains. NORSA strains, though frequently isolated in hospitals, are considered to be the descendants of C-MRSA strains in Australia (10).
Table 1 shows that the majority of C-MRSA strains were susceptible to most of the non-beta-lactam antibiotics, as
NORSA strains are by definition (9). Although the non-multi-resistant nature of C-MRSA has been well recognized as a characteristic of C-MRSA (16), this was not without exception: strain 01083 was resistant to four non-beta-lactam antibiotics (Table 1). This indicates that though it is a rare occurrence, C-MRSA strains may also acquire resistance to non-beta-lactam antibiotics, presumably through exposure to the antibi-otics.
Table 1 also shows that C-MRSA/NORSA strains had rela-tively lower levels of oxacillin and imipenem resistance than H-MRSA strains (with the exceptions of N315 and 85/2082) (20). This indicated that they had the heterogeneous methicil-lin resistance phenotype, which was confirmed by population analysis (Fig. 1). MW2, a representative C-MRSA strain (2), possessed typical heterogeneous subpopulations of resistant cells, whereas the “truly” (i.e.,mecA-negative) methicillin-sus-ceptible strain 476, the putative progenitor strain of MW2 (see below), did not have the resistant subpopulations. Mu3, a typical H-MRSA strain, on the other hand, had a distinct pattern of resistance called homogeneous methicillin resis-tance. This implied that unlike H-MRSA strains, C-MRSA strains were not selected out by the exposure to these potent beta-lactam antibiotics used in the hospital, testifying fur-ther to their predominant propagation occurring in the com-munity.
C-MRSA/NORSA strains grew significantly faster than H-MRSA strains: the mean doubling times (8) of the former group of strains were 28.79 ⫾ 7.09 and 28.24 ⫾ 2.48 min, respectively, whereas that for the latter was 38.81 ⫾ 7.01 min (see Table 1). The difference was statistically significant (Pvalue of ⬍0.0001 byttest). This high growth rate may be a prerequisite in the absence of antibiotics for C-MRSA to achieve successful colonization of humans by outcompeting the numerous bacterial species in the environment.
* Corresponding author. Mailing address: Department of Bacte-riology, Faculty of Medicine, Juntendo University, 2-1-1 Hongo, Bunkyo-Ku, Tokyo, Japan, 113-8421. Phone: 81 (3) 5802-10410. Fax: 81 (3) 5684-7830. E-mail: hiram@med.juntendo.ac.jp.
4289
on May 15, 2020 by guest
http://jcm.asm.org/
TABLE 1. Genotyping and antibiogram of tested MRSA strains Category a Source b Strain name Coagulase isotype MIC (mg/liter) of c: SCC mec type d PFGE pattern MLST luk-PV g Doubling time (min) h Refer- ence(s) ERY KAN TOB GEN TET NOR OXA CZX IMP ST, allelic pro file e CC f C MN, US C1998000370 7 0.5 8 0.5 0.5 32 1 4 512 0.25 IVa J4 1, 1-1-1-1-1-1-1 1 ⫹ 27.89 3, 24 C ND, US C1999000529 7 0.5 4 0.5 0.25 0.5 1 8 >512 0.5 IVa J4 1, 1-1-1-1-1-1-1 1 ⫹ 26.38 3, 24 C MN, US C1999000193 7 0.5 8 0.5 0.5 0.5 1 4 >512 0.25 IVa J3 1, 1-1-1-1-1-1-1 1 ⫹ 26.73 3, 24 C ND, US MW2 7 0.5 4 0.25 0.25 0.5 1 8 >512 0.5 IVa J4 1, 1-1-1-1-1-1-1 1 ⫹ 28.67 2, 3, 24 C MN, US C2001001201 7 >512 8 0.25 0.25 0.5 1 8 >512 0.5 IVa J4 1, 1-1-1-1-1-1-1 1 ⫹ 27.4 3, 24 C MN, US C2001000101 7 0.5 8 0.5 0.5 0.5 1 4 >512 0.25 IVa J4 1, 1-1-1-1-1-1-1 1 ⫹ 27.25 3, 24 C MN, US C200100818 7 >512 8 0.5 0.25 0.5 1 16 >512 0.25 IVa J5 1, 1-1-1-1-1-1-1 1 ⫹ 27.72 3, 24 C Woo, AU A80 3355 4 0.5 4 0.25 0.25 0.5 2 16 >512 0.125 IVa H1 30, 2-2-2-2-6-3-2 30 ⫹ 26.22 25 C Woo, AU A82 3549 4 0.5 2 0.25 0.25 0.5 2 16 >512 0.125 IVa H3 30, 2-2-2-2-6-3-2 30 ⫹ 28.09 25 C Woo, AU A83 0528 4 0.5 4 0.25 0.25 0.5 2 16 >512 ⱕ 0.063 IVa H1 30, 2-2-2-2-6-3-2 30 ⫹ 27.08 25 C Woo, AU B82 6559 4 0.5 2 0.25 0.25 0.5 2 16 512 ⱕ 0.063 IVa H1 30, 2-2-2-2-6-3-2 30 ⫹ 27.57 25 C Woo, AU D82 1552 4 0.5 2 0.25 0.25 0.5 2 16 >512 0.125 IVa H1 30, 2-2-2-2-6-3-2 30 ⫹ 61.03 25 C Woo, AU E80 2537 4 0.5 4 0.25 0.25 0.5 2 8 >512 ⱕ 0.063 IVa H2 30, 2-2-2-2-6-3-2 30 ⫹ 26.97 25 C Woo, AU F81 0539 4 0.5 4 0.25 0.25 0.5 2 16 >512 ⱕ 0.063 IVa H1 30, 2-2-2-2-6-3-2 30 ⫹ 26.44 25 C Woo, AU 180 2552 4 0.5 2 0.25 0.25 0.5 2 16 512 0.125 IVa H3 30, 2-2-2-2-6-3-2 30 ⫹ 26.86 25 C Per, AU i M9N 3 >512 4 0.25 0.25 0.5 2 16 512 0.125 IVa I1 78, 22-1-14-23-12-53-31 298 ⫺ 26.75 26 C Per, AU i M33T 3 >512 4 0.25 0.25 0.5 2 16 512 0.125 IVa I2 78, 22-1-14-23-12-53-31 298 ⫺ 26.44 26 C Per, AU i W1S 1 0.5 2 0.5 0.25 0.5 0.5 2 32 0.063 New E2 45, 10-14-8-6-10-3-2 45 ⫺ 27.74 26 C Per, AU i C7N 7 >512 4 0.5 0.5 0.5 2 16 512 0.25 IVa J6 1, 1-1-1-1-1-1-1 1 ⫺ 30.26 26 C TN, US j 00215 7 0.25 4 0.5 0.25 0.5 0.5 64 >512 2 IVa E1 45, 10-14-8-6-10-3-2 45 ⫺ 27.71 28 C MS, US k 01083 3 64 >512 0.5 0.5 32 16 8 512 0.125 IVa A3 8, 3-3-1-1-4-4-3 8 ⫹ 27.78 4 C MS, US k 01093 5 16 512 0.5 0.5 0.5 2 16 >512 0.125 IVa F 72, 1-4-1-8-4-4-3 8 ⫺ 25.94 4 C MS, US k 01102 3 0.25 4 0.5 0.5 0.5 1 8 >512 0.25 IVb A2 8, 3-3-1-1-4-4-3 8 ⫹ 28.29 4 N Ade, AU 81 0342 3 0.25 4 0.5 0.5 0.5 8 21 6 ⱕ 0.063 New A1 8, 3-3-1-1-4-4-3 8 ⫺ 26.86 This study N Ade, AU 91 2572 7 0.5 4 0.25 0.25 0.5 1 4 >512 0.25 IVa J7 1, 1-1-1-1-1-1-1 1 ⫺ 27.15 This study N Ade, AU 91 2619 7 0.5 8 0.5 0.5 0.5 2 64 >512 2 IVa J7 76, 1-63-1-1-1-1-1 1 ⫺ 27.47 This study N Ade, AU WCH379 7 0.5 2 0.25 0.5 0.5 1 16 >512 4 IVa J8 1, 1-1-1-1-1-1-1 1 ⫺ 33.31 This study N Ade, AU 91 2574 7 0.25 1 0.25 0.25 0.5 128 8 256 ⱕ 0.063 New G 22, 7-6-1-5-8-8-6 22 ⫺ 29.52 This study N Ade, AU SAP260 2 >512 4 0.5 0.25 0.5 0.5 4 >512 0.25 IVa B 73, 1-4-27-4-12-1-10 5 ⫺ 26.64 This study N Per, AU 81 0937 7 0.5 2 0.25 0.5 0.5 1 4 >512 0.5 IVa J1 1, 1-1-1-1-1-1-1 1 ⫺ 25.29 This study N Per, A U 91 2125 7 0.5 4 0.25 0.5 0.5 0.5 16 >512 0.5 IVa J2 1, 1-1-1-1-1-1-1 1 ⫺ 30.25 This study N Per, AU 91 1703 3 0.25 2 0.125 0.25 8 1 16 512 0.125 IVa A2 8, 3-3-1-1-4-4-3 8 ⫺ 32.0 This study N Bri, AU 81 1238 7 0.5 2 0.25 0.5 0.5 1 8 >512 0.5 IVa J1 1, 1-1-1-1-1-1-1 1 ⫺ 26.06 This study N Bri, AU 91 2666 7 0.5 2 0.5 0.25 0.5 0.5 2 512 0.125 IVa J2 1, 1-1-1-1-1-1-1 1 ⫺ 26.65 This study N Dar, AU SAP411 6 ⱕ 0.125 4 0.25 0.5 32 1 4 128 0.125 IVa N 75, 36-3-43-34-39-52-49 S ⫺ 27.68 This study H Ade, AU 81 0508 4 >512 256 256 0.5 0.25 512 512 >512 128 II M2 36, 2-2-2-2-3-3-2 30 ⫺ 32.22 This study H Ade, AU 91 2231 4 >512 2 0.25 0.5 32 32 128 >512 16 III K4 239, 2-3-1-1-4-4-3 8 ⫺ 35.8 This study H Bri, AU 91 1573 4 >512 128 64 8 16 1 64 >512 4 III K2 239, 2-3-1-1-4-4-3 8 ⫺ 41.82 This study H Bri, AU 91 1575 4 >512 512 16 32 1 32 256 >512 32 III K6 239, 2-3-1-1-4-4-3 8 ⫺ 50.46 This study H Bri, AU 91 2145 4 >512 512 16 32 32 32 64 >512 16 III K1 239, 2-3-1-1-4-4-3 8 ⫺ 50.64 This study H Dar, AU SAP344 4 >512 4 0.25 0.5 32 32 256 >512 32 III K3 239, 2-3-1-1-4-4-3 8 ⫺ 34.51 This study H Per, AU 91 2118 4 >512 512 16 32 32 256 512 >512 64 III L 239, 2-3-1-1-4-4-3 8 ⫺ 38.45 This study H UK NCTC10442 3 0.125 1 0.125 0.5 128 1 256 >512 16 I D 250, 3-3-1-1-4-4-16 8 ⫺ 36.44 19 H JP N315 2 >512 >512 512 0.5 0.125 2 16 16 1 II C 5, 1-4-1-4-12-1-10 5 ⫺ 34.28 19 H NZ 85/2082 4 512 >512 8 64 128 2 32 >512 0.5 III K5 239, 2-3-1-1-4-4-3 8 ⫺ 43.53 19 H UK Strain 252 l 4 >512 128 128 0.25 0.5 >512 512 >512 64 II M1 36, 2-2-2-2-3-3-2 30 ⫺ 29.79 20, — m H UK MSSA476 7 0.5 4 0.5 0.5 1 1 0.5 8 ⱕ 0.063 J1 1, 1-1-1-1-1-1-1 1 ⫺ 27.66 — m aC, C-MRSA; N, NORSA; H, H-MRSA. bMN, M innesota; US, United States; ND, North Dakota; Woo, Wooloongabba, Australia, AU, Australia; Per, Perth, Australia; TN, Tennessee; MS, M ississ ippi; Ade, Adelaide, Australia; Bri, Brisbane, Australia; Dar, Darwin, Australia; UK, United Kingdom; JP, Japan; NZ, New Zealand. cMICs were determined by the NCCLS-based plate dilution method. Antibiotics: ERY, erythromycin; KAN, kanamycin; TOB, tobramycin; GEN, gentamicin; TET, tetracycline; NOR, nor floxacin; OXA, oxacillin; CZX, ceftizoxime; IMP, imipenem. Values in bold signify resistance to these antibiotics. dNew, new type of SCC mec possessing class C2 mec gene complex (see Fig. 2). eST, sequence type. fClonal complex, based on BURST (based upon related sequence types). S, singleton (not assigned to any clonal complex). gluk-PV genes encode Panton-Valentin leucocidin proteins. hDoubling time during exponential growth phase (optical density of 0.05 to ⬃ 1.0 at 660 nm) measured by using TN-2612 (Advantec Toyo Kaisha, Ltd., Tokyo, Japan) as previously described (8). iIsolated from aborigine. jIsolated from food-poisoning strain. kIsolated from state prison.
lE-MRSA16. m—
,http://www.sanger.ac.uk/Projects/S.aureus/.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:2.603.51.454.60.782.2]The MRSA genotype is the sum of the SCCmectype and the type of its recipient chromosome (12). First, by using multiple locus sequence typing (MLST), we identified the chromosome genotype of the test strains. Enright et al. reported that 356 of 359 MRSA strains from 20 countries were classified into only five clonal complexes (CCs), CC5,
CC8, CC22, CC30, and CC45, with the rest, three strains, possessing sequence types (STs) of rare occurrence (6). All the 11 H-MRSA strains used in this study were reasonably classified into three of these five CCs (Table 1). However, 35 C-MRSA/NORSA strains possessed 10 different STs that constituted one singleton (ST75) and seven CCs that, surpris-ingly, included all five H-MRSA CCs described above (Table 1).
Among the seven C-MRSA CCs, especially notable was CC1, which contained the internationally dominant C-MRSA strains; eight U.S. strains represented by MW2 and six Austra-lian strains belonged to this clonal complex. Remarkably, no H-MRSA strains belonged to this complex (6). Curiously, a highly virulent methicillin-susceptibleS. aureus(MSSA) strain, 476, whose whole genome sequence has been determined, be-longs to this complex (http://www.mlst.net/new/index.htm). MSSA 476 and two NORSA strains belonging to CC1 even shared an identical pulsed-field gel electrophoresis (PFGE) pattern (Table 1). Detailed comparison revealed that the only significant difference between the two chromosomes was the presence of type IV SCCmec in MW2, which in-dicated that strain 476 represented the progenitor MSSA strain from which MW2 was generated by acquiring type IV SCCmec.
The pattern of clonal distribution of the 35 C-MRSA/ NORSA strains was statistically distinct from that of 359 MRSA strains analyzed in a previous study plus 11 H-MRSA strains used in this study (P value of ⬍0.000001 by Fisher’s exact test). This clearly indicated that distinct clonal popula-tions were successfully propagated as C-MRSA/NORSA and H-MRSA, presumably through different selective pressures ex-erted on them, e.g., fast-growing S. aureus or S. epidermidis
strains for the former and exposure to multiple antibiotics for the latter.
The MLST data, despite the small number of tested strains, indicated that C-MRSA/NORSA strains were gen-erated fromS. aureus clones of much more diverse genetic backgrounds than expected. This was also supported by PFGE analysis (Table 1), which showed that the C-MRSA/ NORSA strains were classified into nine unrelated groups according to the criteria described by Tenover et al. (27). Moreover, these strains consisted of producers of as many as seven coagulase isotypes (Table 1). Since only eight coagu-lase isotypes are known among S. aureus strains isolated from various sources (18), this also supported the view that C-MRSA/NORSA represents diverseS. aureus genomes as the origin of derivation.
Next, we determined SCCmectypes by PCR typing of the
mecgene complex andccr gene complex as described previ-ously (19, 21). Table 2 and Fig. 2 show the nucleotide se-quences and locations of the primers used (15, 19, 21). In contrast to the heterogeneity of C-MRSA/NORSA chromo-somes demonstrated above, all except for three strains har-bored type IV SCCmec, and the remaining three harbored a novel SCCmec carrying the class C2 mecgene complex (21) (Fig. 2). None of the C-MRSA/NORSA strains possessed any of the three types of SCCmecwhich the majority of epidemic H-MRSA strains possess (19).
It is not clear at this moment why type IV SCCmecis
prev-FIG. 1. C-MRSA strain shows heterogeneous phenotypic expres-sion of methicillin resistance. Analysis of resistant subpopulations of the C-MRSA strain MW2, the related MSSA strain 476, and strain Mu3, with heterogeneous resistance to vancomycin, was per-formed with oxacillin (A), ceftriaxone (B), and imipenem (C) as de-scribed previously (13). Ceftriaxone was the antibiotic used in vain to treat the patient infected with MW2 (3). MW2 is an American Mid-west MRSA strain representing the major C-MRSA (see the text). Strain 476 is an MSSA strain sharing its MLST allotype with MW2 (see Table 1). Mu3 is a representative H-MRSA strain with heterogeneous resistance to vancomycin (13). It is noted that MW2 contains subpopu-lations resistant to each of the three beta-lactam antibiotics.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:3.603.46.284.62.512.2]alent in C-MRSA/NORSA strains. However, type IV SCCmec
is short (21 to 25 kb) compared to the three SCCmecs preva-lent in H-MRSA strains (34 to 67 kb) and lacks any antibiotic resistance genes other thanmecA(23) (Fig. 2). This evidently corresponds to the non-multiresistant nature of C-MRSA/ NORSA and may alleviate the fitness cost paid by H-MRSA strains carrying big SCCmecs with multiple-drug resistance de-terminants.
Although we need to explore further the reason why type IV SCCmecis prevalent in C-MRSA strains, it seems clear that we are witnessing the emergence and expansion of new MRSA clones in the community. These clones are different from any of the major H-MRSA clones in the world that we have identified by using SCCmec typing and ribotyping combinations (12, 17). In this study we realized that the antibiogram is not completely reliable in discriminating
C-TABLE 2. Primer sets used for identifying SCCmec Primer (previous name)
for detection of: Nucleotide sequence (gene[s] reactive to the primer)Expected size of product Reference
ccrgene complexa
c (2) 5⬘-ATTGCCTTGATAATAGCCITCT-3⬘ (all types ofccrB) 19
␣c 5⬘-ATCTATTTCAAAAATGAACCA-3⬘ 560 bp,c (all types ofccrA) This study
␣1 (␣2) 5⬘-AACCTATATCATCAATCAGTACGT-3⬘ 700 bp,c (type 1ccrA) 19
␣2 (␣3) 5⬘-TAAAGGCATCAATGCACAAACACT-3⬘ 1 kbp,c (type 2ccrA) 19
␣3 (␣4) 5⬘-AGCTCAAAAGCAAGCAATAGAAT-3⬘ 1.6 kbp,c (type 3ccrA) 19
mecgene complex (all types)
mecR1(MS domain)
mcR4 5⬘-GTCGTTCATTAAGATATGACG-3⬘ (mecR1—MS domain) This study
mcR3 5⬘-GTCTCCACGTTAATTCCATT-3⬘ 310 bp, mcR4 (mecR1—MS domain) 21
mecA
mA1 5⬘-TGCTATCCACCCTCAAACAGG-3⬘ (mecA) 15
mA2 5⬘-AACGTTGTAACCACCCCAAGA-3⬘ 200 bp, mA1 (mecA) 15
mecA-IS431mec
IS2 (iS-2) 5⬘-TGAGGTTATTCAGATATTTCGATGT-3⬘ 4 kbp, mA1 (IS431mec) 21
Class Cmecgene complex
mA2 5⬘-AACGTTGTAACCACCCCAAGA-3⬘ ⬍3 kbp, IS2(mecA) 21
Class Bmecgene complex
IS5 5⬘-AACGCCACTCATAACATATGGAA-3⬘ (IS1272) This study
mA6 5⬘-TATACCAAACCCGACAAC-3⬘ 2 kbp, IS5(mecA) 21
Class Amecgene complex
mecI
mI4 5⬘-CAAGTGAATTGAAACCGCCT-3⬘ (mecI) This study
mI3 5⬘-CAAAAGGACTGGACTGGAGTCCAAA-3⬘ 180 bp, mI4 (mecI) This study
mecR1(PB domain)
mcR2 5⬘-CGCTCAGAAATTTGTTGTGC-3⬘ (mecR1—PB domain) 21
mcR5 5⬘-CAGGGAATGAAAATTATTGGA-3⬘ 320 bp, mcR2 (mecR1—PB domain) This study
SCCmecsubtype IVa
4a1 5⬘-TTTGAATGCCCTCCATGAATAAAAT-3⬘ (J1 region of IVa) This study
4a2 5⬘-AGAAAAGATAGAAGTTCGAAAGA-3⬘ 450 bp, 4a1 (J1 region of IVa) This study
SCCmecsubtype IVb
4b1 5⬘-AGTACATTTTATCTTTGCGTA-3⬘ (J1 region of IVb) This study
4b2 5⬘-AGTCATCTTCAATATCGAGAAAGTA-3⬘ 1 kbp, 4b1 (J1 region of IVb) This study
accrtype is determined by PCR using primerc (the common primer for three types ofccrB) and either one of the three types ofccrA,␣1 (ccrA1),␣2 (ccrA2), and
␣3 (ccrA3). This typing actually reflects the allotype ofccrA.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:4.603.46.541.84.606.2]MRSA from H-MRSA, nor is the phenotypic expression of methicillin resistance. Even epidemiological information is not sufficient, since, for example, many C-MRSA strains have been carried in Australian hospitals (29). Therefore, no reli-able judgment can be made as to whether the strain isolat-ed in the hospital is H-MRSA or C-MRSA even basisolat-ed on the timing of isolation of the strains after admission to hospital. In this regard, SCCmec and MLST typing will be-come more important in the years to be-come for discrimina-tion of numerous C-MRSA strains prevailing in both com-munity and hospitals by reference to their ancestral MRSA clones (12).
We thank T. Naimi for providing us C-MRSA strains and for fruitful discussion. We also thank M. C. Enright for kind instruction on MLST and the BURST program and Susan Johnson and David Boxrud in Minnesota and Suwanna Trakulsomboon, Mantana Jamklang, and Fumi-hiko Takeuchi in Japan for their excellent technical assistance. We also thank Yuh Morimoto for her help in preparing the manuscript.
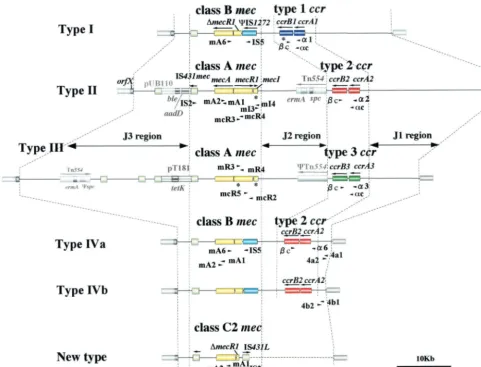
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (13226114) from The Ministry of Education, Science, Sports, Culture and Technology of Japan and the Core University System Exchange Programme under the Japan Society for the Promo-tion of Science, coordinated by the University of Tokyo Graduate School of Medicine and Mahidol University. The work was also supported by a Grant for International Health Cooperation search (11C-4) from the Ministry of Health and Welfare, by the Re-FIG. 2. Illustrative representation of various types of SCCmec.SCCmectype is defined by the combination of the type ofccrgene complex and the class ofmecgene complex. Type I SCCmecis defined by the combination of type 1ccrand class Bmec(IS1272-⌬mecR1-mecA); type II is defined by type 2ccrand class Amec(mecI-mecR1-mecA); type III is defined by type 3ccrand class Amec; and type IV is defined by type 2ccr
and class Bmec. Type IV SCCmecis further classified into subtypes (type IVa and type IVb) based on the sequence difference in the J1 region of SCCmec(J stands for “junkyard”). Positions of primers used in this study to identify and type SCCmecare shown (see Table 2 for the nucleotide sequence of each primer). The allelic class ofmecgene complex is determined by PCR detection of the presence or absence of IS1272,mecI, and
mecR1 in two domains (PB, penicillin-binding domain; and MS, membrane-spanning domain),mecA,and IS431mec. PCR amplification was
performed using 2.5 U of ExTaq(Takara Shuzo Co., Ltd., Kyoto, Japan) in 50l of reaction mixture. Thermal cycling was set at 30 cycles (30 s for denaturation at 94°C, 1 min for annealing at 50°C, and 2 min for elongation at 72°C) using the Gene Amp PCR system 9600 (Perkin-Elmer, Wellesley, Mass.). For the detection ofmecA-IS431mec, a long-range PCR method was used, set at 10 cycles (15 s for denaturation at 94°C, 30 s for annealing at 50°C, and 8 min for elongation at 68°C) followed by 20 cycles (15 s for denaturation at 94°C, 30 s for annealing at 50°C, and 12 min for elongation at 68°C). Note that this study identified a new type of SCCmecfor three C-MRSA strains that carried the class C2mecgene complex (21). The sequencing of the entire SCCmecis now ongoing.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:5.603.53.534.70.437.2]search for the Future Program of the Japan Society for the Promo-tion of Science, and by the NaPromo-tional Health and Medical Research Foundation of Australia and the Health Department of Western Australia.
REFERENCES
1. Alghaithy, A. A., N. E. Bilal, M. Gedebou, and A. H. Weily.2000. Nasal
carriage and antibiotic resistance ofStaphylococcus aureus isolates from
hospital and non-hospital personnel in Abha, Saudi Arabia. Trans. R. Soc.
Trop. Med. Hyg.94:504–507.
2. Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu.2002. Genome and virulence determinants of high virulence
community-acquired MRSA. Lancet359:1819–1827.
3. Centers for Disease Control and Prevention.1999. Four pediatric deaths
from community-acquired methicillin-resistantStaphylococcus
aureus—Min-nesota and North Dakota, 1997–1999. Morb. Mortal. Wkly. Rep.48:707–710.
4. Centers for Disease Control and Prevention. 2001. Methicillin-resistant Staphylococcus aureusskin or soft tissue infections in a state
prison—Mis-sissippi, 2000. Morb. Mortal. Wkly. Rep.50:919–922.
5. Chambers, H.2001. The changing epidemiology ofStaphylococcus aureus?
Emerg. Infect. Dis.7:178–182.
6. Enright, M. C., D. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt.2002. The evolutionary history of methicillin-resistantStaphylococcus aureus(MRSA). Proc. Natl. Acad. Sci. USA99:7687–7692.
7. Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes.
1988. CDC definitions for nosocomial infections, 1998. Am. J. Infect.
Con-trol16:128–140.
8. Gerhardt, P., R. G. E. Murray, W. A. Wood, and N. R. Krieg.1994. Methods for general and molecular bacteriology, p. 270–271. American Society for Microbiology, Washington, D.C.
9. Gosbell, I. B., J. Mercer, S. A. Neville, K. G. Chant, and R. Munro.2001.
Community-acquired, non-multiresistant oxacillin-resistantStaphylococcus
aureus(NORSA) in South Western Sydney. Pathology33:206–210. 10. Gosbell, I. B., J. Mercer, S. A. Neville, S. A. Crone, K. G. Chant, B. B.
Jalaludin, and R. Munro.2001. Non-multiresistant and multiresistant methicillin-resistant Staphylococcus aureus in community-acquired
infec-tions. Med. J. Aust.174:627–630.
11. Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum.1998.
Community-acquired methicillin-resistantStaphylococcus aureusin children with no
iden-tified predisposing risk. JAMA279:593–598.
12. Hiramatsu, K.1995. Molecular evolution of MRSA. Microbiol. Immunol.
39:531–543.
13. Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi.1997. Dissemination in Japanese hospitals of
strains ofStaphylococcus aureusheterogeneously resistant to vancomycin.
Lancet350:1670–1673.
14. Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito.2001. The emergence and
evolution of methicillin-resistantStaphylococcus aureus. Trends Microbiol.
9:486–493.
15. Hiramatsu, K., H. Kihara, and T. Yokota.1992. Analysis of
borderline-resistant strains of methicillin-borderline-resistantStaphylococcus aureususing
polymer-ase chain reaction. Microbiol. Immunol.36:445–453.
16. Hiramatsu, K., K. Okuma, X. X. Ma, M. Yamamoto, S. Hori, and M. Kapi.
2002. New trends inStaphylococcus aureusinfections: glycopeptide
resis-tance in hospital and methicillin resisresis-tance in the community. Curr. Opin.
Infect. Dis.15:407–413.
17. Hiramatsu, K., T. Ito, and H. Hanaki.1999. Mechanisms of methicillin and
vancomycin resistance in Staphylococcus aureus, p. 221–242.InR. G. Finch
and R. J. Williams (ed.), Bailliere’s clinical infectious diseases, vol. 5. Bail-liere Tindall, London, United Kingdom.
18. Igarashi, H.1993. Producibility of staphylococcal enterotoxins and toxic
shock syndrome toxin-1 on methicillin-resistant Staphylococcus aureus.
Igakunoayumi166:274–278. (In Japanese.)
19. Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiesasitorn, and K. Hiramatsu.2001. Structural comparison of three types of
staphylo-coccal cassette chromosomemecintegrated in the chromosome in
methicil-lin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:
1323–1336.
20. Johnson, A. P., H. M. Aucken, S. Cavendish, M. Ganner, M. C. Wale, M. Warner, D. M. Livermore, B. D. Cookson, and the UK EARSS Participants.
2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial
Resistance Surveillance System (EARSS). J. Antimicrob. Chemother.48:
143–144.
21. Katayama, Y., T. Ito, and K. Hiramatsu.2001. Genetic organization of the
chromosome region surroundingmecAin clinical staphylococcal strains: role
of IS431-mediatedmecIdeletion in expression of resistance in
mecA-carry-ing, low-level methicillin-resistantStaphylococcus haemolyticus. Antimicrob.
Agents Chemother.45:1955–1963.
22. Katayama, Y., T. Ito, and K. Hiramatsu.2000. A new class of genetic
element, staphylococcus cassette chromosomemec, encodes methicillin
re-sistance inStaphylococcus aureus. Antimicrob. Agents Chemother.44:1549–
1555.
23. Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu.2002. Novel type of staphylococcal
cassette chromosomemecidentified in community-acquired
methicillin-re-sistantStaphylococcus aureusstrains. Antimicrob. Agents Chemother.46:
1147–1152.
24. Naimi, T. S., K. H. LeDell, D. J. Boxrud, A. V. Groom, C. D. Steward, S. K. Johnson, J. M. Besser, C. O’Boyle, R. N. Danila, J. E. Cheek, M. T. Oster-holm, K. A. Moore, and K. E. Smith.2001. Epidemiology and clonality of
community-acquired methicillin-resistantStaphylococcus aureusin
Minne-sota, 1996–1998. Clin. Infect. Dis.33:990–996.
25. Nimmo, G. R., J. Schooneveldt, G. O’Kane, B. McCall, and A. Vickery.2000.
Community acquisition of gentamicin-sensitive methicillin-resistant
Staphy-lococcus aureusin southeast Queensland, Australia. J. Clin. Microbiol.38:
3926–3931.
26. O’Brien, F. G., J. Pearman, M. Gracey, T. V. Riley, and W. B. Grubb.1999.
Community strain of methicillin-resistantStaphylococcus aureusinvolved in
a hospital outbreak. J. Clin. Microbiol.37:2858–2862.
27. Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, P. A. Mickelsen, B. E. Murry, D. H. Persing, and B. Swaminathan.1995. Interpreting chro-mosomal DNA restriction patterns produced by pulsed-field gel
electro-phoresis: criteria for bacterial strain typing. J. Clin. Microbiol.33:2233–2239.
28. Timothy, F. J., E. K. Molly, S. S. Porter, B. Michael, and S. William.2002. An outbreak of community-acquired foodborne illness caused by
methicillin-resistantStaphylococcus aureus. Emerg. Infect. Dis.8:82–84.
29. Turnidge, J. D., and J. Bell.2000. Methicillin-resistantStaphylococcal aureus
evolution in Australia over 35 years. Microb. Drug Resist.6:223–229.