JOURNAL OFCLINICALMICROBIOLOGY, Sept. 1996, p. 2163–2169 Vol. 34, No. 9 0095-1137/96/$04.0010
Copyrightq1996, American Society for Microbiology
Epidemiological Study of an Outbreak Due to Multidrug-Resistant
Enterobacter aerogenes in a Medical Intensive Care Unit
C. ARPIN,1* C. COZE,1A. M. ROGUES,2J. P. GACHIE,2C. BEBEAR,3ANDC. QUENTIN1
Laboratoire de Microbiologie, Universite´ de Bordeaux II,1and Laboratoire d’Hygie`ne Hospitalie`re2and
Laboratoire de Bacte´riologie,3Hoˆpital Pellegrin, 33076 Bordeaux Cedex, France
Received 15 November 1995/Returned for modification 4 January 1996/Accepted 6 June 1996
In 1993, 63 isolates ofEnterobacter aerogeneswere collected from 41 patients in a medical intensive care unit
(ICU). During the same period, only 46 isolates from 32 patients were collected in the rest of the hospital. All isolates were analyzed by antibiotic resistance phenotype, and 77 representative isolates were differentiated by
plasmid restriction analysis, ribotyping, and arbitrarily primed (AP)-PCR. The extended-spectrumb
-lacta-mases produced by 22 strains were characterized by determination of their isoelectric points and by hybrid-ization of plasmid DNA with specific probes. The isolates were divided into 25 antibiotic resistance phenotypes, either susceptible (group I) or resistant (group II) to aminoglycosides, and exhibited three phenotypes of
resistance tob-lactams: chromosomally derepressed cephalosporinase alone or associated with either
extend-ed-spectrumb-lactamases (mainly of the SHV-4 type) or imipenem resistance. The results of the tests divided
the 77 representative isolates (group I,n521; group II,n556) into 15 plasmid profiles, 14 ribotypes, and
15 AP-PCR patterns. Although the resistant isolates (group II) exhibited different plasmid profiles, ribotyping and AP-PCR analysis demonstrated an identical chromosomal pattern, indicating an epidemiological relat-edness. They were mainly found in the medical ICU and occasionally in other units. The susceptible strains (group I) had various and distinct markers and were mainly isolated in units other than the medical ICU. In conclusion, the presence of a nosocomial outbreak in an ICU and the spread of a multidrug-resistant epidemic strain throughout the hospital was confirmed. Ribotyping and AP-PCR represent discriminatory tools for the
investigation of nosocomial outbreaks caused byE. aerogenes.
Members of the genus Enterobacter, like most members of the family Enterobacteriaceae, can be responsible for opportu-nistic infections in debilitated patients. Enterobacter cloacae is the species that is the most frequently isolated from clini-cal specimens (14), but Enterobacter aerogenes has recently emerged as an important hospital pathogen (10, 13, 29, 30). The prevalence of Enterobacter spp., like that of otherb -lac-tamase-inducible gram-negative bacilli, has greatly increased since the introduction of extended-spectrum cephalosporins into clinical use (22). In these species, stably derepressed mu-tants overproducing their chromosomal enzyme can readily be selected during therapy and can spread within the hospital environment. Other mechanisms of b-lactam resistance have been described in Enterobacter spp., such as plasmid-mediated
b-lactamases and, recently, the diminution of membrane per-meability due to a lack of porin combined with high-level constitutiveb-lactamase production, leading to imipenem re-sistance (11, 39).
Traditional techniques used for typing Enterobacter spp. have been based on phenotypic characteristics, including anti-biotic resistance phenotype analysis (18), bacteriocin typing, serotyping, and phage typing (15). These techniques are often unsufficiently sensitive for typing all strains and discriminating between isolates. The use of genotypic markers may provide useful data for demonstrating differences between strains. Var-ious genotypic assays have been found to be useful and dis-criminating for analyzing strains of E. cloacae, a species that is known to be naturally heterogeneous; they include plasmid profile analysis (27), small-fragment restriction endonuclease
analysis (20), pulsed-field gel electrophoresis (20), ribotyping (17, 33), and, recently, arbitrarily primed (AP)-PCR analysis (17). However, very few studies have been dedicated to the genotypic investigation of nosocomial infections caused by E.
aerogenes (9, 16, 18).
Before 1993, E. aerogenes was infrequently encountered in our hospital. Between January and October 1993, 109 clinical strains of E. aerogenes were isolated, including 63 strains col-lected from a medical intensive care unit (ICU). The majority of these isolates were resistant to multiple antibiotics, in par-ticular to b-lactams, because of the chromosomally dere-pressed cephalosporinase alone or in association with either extended-spectrum b-lactamase (ESBL) or imipenem resis-tance. To determine whether they were epidemiologically re-lated, all strains were differentiated by a non-molecular biolo-gy-based method, i.e., antibiotic resistance phenotyping. Representative isolates were analyzed by three molecular typ-ing methods: plasmid restriction profile analysis, ribotyptyp-ing, and AP-PCR. In addition, the ESBLs were characterized by determination of their isoelectric points and by hybridization of plasmid DNA with specific probes.
MATERIALS AND METHODS
Hospital presentation andE. aerogenesstrains.Pellegrin Hospital (Bordeaux, France) is a 1,500-bed university-affiliated hospital with mainly surgical (;950 beds) and intensive care (;200 beds) units. The 109 isolates of E. aerogenes collected from 73 patients which were subjected to this epidemiological study included 63 isolates from 41 patients hospitalized in a medical ICU (63 beds). The 46 remaining isolates were provided by 32 patients hospitalized in 20 other units from 12 different departments during the same period. The 109 clinical isolates were isolated essentially from urine samples (40.4%), respiratory sam-ples (29.4%), and pus (15.6%). In contrast, 41.3% of the isolates collected from the medical ICU were found in respiratory samples because most of the patients admitted to this unit needed mechanical ventilation. Five strains were found in 115 environmental samples from the medical ICU (in June). They came from furniture or floor surfaces. Among the 114 isolates, 77 were selected for
addi-* Corresponding author. Mailing address: Laboratoire de Microbiol-ogie, Universite´ de Bordeaux II, 146 rue Le´o Saignat, 33076 Bordeaux Cedex, France.
2163
on May 15, 2020 by guest
http://jcm.asm.org/
tional investigations: 47 isolates from the medical ICU consisting of the first isolate from each patient for each antibiotic resistance phenotype, 25 isolates from the other units, and the 5 environmental strains. Twenty-two of the 31 ESBL-producing strains were studied for theirb-lactamase contents.
The E. aerogenes strains were cultivated in Mueller-Hinton broth (Diagnostics Pasteur) and were identified by using the API 20E system (BioMerieux).
Reference strains and plasmids.Escherichia coli HB101 harbors the pKK3535
plasmid, a pBR322 derivative containing the rrnB ribosomal operon of E. coli (4), which was used as a probe for ribotyping. Two strains, E. coli HB101 with pBR322 plasmid and E. coli MD15 with pHUC37 plasmid (31), contain theb -lactamase-specific genes blaTEM-1and blaSHV-3, respectively, which were used as probes for characterization of the ESBLs. E. coli with the R111 plasmid (TEM-1) (38), E. coli K-12 C600 with the pCF04 plasmid (TEM-3) (38), Klebsiella
pneu-moniae with the pCFF64 plasmid (SHV-4) (12), and K. pneupneu-moniae with the
pCFF74 plasmid (TEM-24) (6) were used in the isoelectric focusing analysis as pI markers.
Antibiotic resistance testing.The susceptibilities of the isolates to 23 anti-microbial agents were determined by the standard disk diffusion method (8). Commercially available disks (Diagnostics Pasteur) loaded with the following antibiotics were used: ampicillin, amoxicillin-clavulanic acid, ticarcillin, ticarcil-lin-clavulanic acid, cefalothin, cefuroxime, cefoxitin, cefotaxime, ceftazidime, latamoxef, aztreonam, imipenem, gentamicin, tobramycin, amikacin, netilmicin, tetracycline, chloramphenicol, pipemidic acid, ofloxacin, co-trimoxazole (trimeth-oprim-sulfamethoxazole), fosfomycin, and colistin. The isolates were classified as susceptible, intermediate, and resistant according to the recommendations of the Antibiogram Committee of the French Society for Microbiology (1). The pres-ence of ESBL activity was determined by placing disks containing cefotaxime, ceftazidime, cefepime, or aztreonam near a disk containing ab-lactamase inhib-itor (amoxicillin-clavulanic acid). Extension of the zone of inhibition toward the disk containing clavulanate indicated the presence of an ESBL (32).
b-Lactamase extraction and analytical isoelectric focusing.b-Lactamases were released by ultrasonic treatment (35), and their pIs were determined by isoelectric focusing in polyacrylamide gels as described by Matthew et al. (28). After each electrofocusing run, enzyme activity was detected by the starch iodine method (34) with penicillin G (0.125 g/liter), which is hydrolyzed by allb -lacta-mases, and then ceftriaxone (0.25 g/liter), which is a substrate only for the ESBLs.
Plasmid and chromosomal DNA extraction and analysis. The pKK3535, pBR322, and pHUC37 plasmids were extracted and purified by using the pro-tocol and reagents of a commercial kit (Qiagen plasmid midi kit; Coger). Plasmid DNA from 5-ml cultures of E. aerogenes was extracted by the alkaline lysis procedure (3). Total cellular DNA was isolated from 3-ml cultures grown over-night. Cells were harvested, suspended in TE buffer (Tris-HCl, 10 mM; EDTA, 1 mM [pH 8.0]), and lysed after digestion with lysozyme (1 mg/ml; 30 min at 378C) and proteinase K (25mg/ml; 1 h at 378C). The DNA was purified by phenol-chloroform extraction; this was followed by ethanol precipitation. The DNA samples were digested with the restriction enzyme HindIII (plasmid DNA) or EcoRI (total DNA) by using the buffer and reaction conditions recommended by the manufacturer (Gibco BRL). The DNA was submitted to electrophoresis in agarose gels (0.8%; wt/vol) in Tris-borate buffer (pH 8.3) as described by Sambrook et al. (37). BacteriophagelDNA digested with PstI or HindIII was used as a size marker.
Blotting and hybridization.DNA was transferred from agarose gels to Hy-bond-N nylon filters (Schleicher & Schuell) with a vacuum blotting system (Oncor-Appligene) by using a 0.4 N NaOH solution. The blotted DNA was fixed by UV irradiation at 305 nm for 5 min. A quantity of about 250 ng of purified plasmid DNA (pKK3535, pBR322, and pHUC37) was labelled with 50mCi of [a-32P]dCTP (110 TBq/mmol; ICN) with a nick-translation labelling system (Gibco BRL) according to the manufacturer’s instructions. The prehybridization, hybridization, and washing steps were performed under stringent conditions as described previously (19). The blots were exposed to Fuji RX film at2708C with intensifying screens for various lengths of time.
AP-PCR analysis.After an overnight culture at 378C on bromocresol purple lactose agar medium (Diagnostics Pasteur), one colony was suspended in 1 ml of sterile water (about 108
CFU/ml) and boiled for 15 min. The supernatant of the crude lysate was recovered after centrifugation. The amplification was performed with 2 ml of this supernatant in a final volume of 50ml containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 0.2 mg of gelatin per ml, 200mM (each) deoxynucleoside triphosphate (Boerhinger Mann-heim), 0.5mM primer, and 0.25 U of Taq polymerase (Oncor-Appligene). Each sample was overlaid with 50ml of mineral oil and was amplified in a Perkin-Elmer Cetus DNA thermal cycler programmed for a first cycle of denaturation (10 min at 948C). The 35 subsequent cycles of amplification consisted of dena-turation for 1 min at 948C, annealing for 1 min at 408C, and extension for 1 min at 728C, with a final extension step of 10 min at 728C. The primer used, AP12h (59-CGGCCCCTGT-39), was a 10-mer oligonucleotide primer (G1C content, 80%) previously evaluated for E. aerogenes by Davin-Regli et al. (9). The am-plification products were compared by electrophoresis of 10-ml samples in 1.5% agarose gels in Tris-borate buffer (90 mM Tris-HCl [pH 8.3], 90 mM boric acid, 2.5 mM EDTA).
RESULTS
Antibiotic resistance phenotypes.The 109 clinical isolates
were divided into 25 antibiotic resistance phenotypes (ATB1 to ATB25) on the basis of their resistance to b-lactams, ami-noglycosides, tetracycline, chloramphenicol, co-trimoxazole, quinolones, and fosfomycin (Table 1). Two groups could be defined: the first one (ATB1 to ATB9; group I) included 36 isolates which were susceptible to aminoglycosides; the second group consisted of 73 isolates (ATB10 to ATB25; group II) which were resistant to aminoglycosides including amikacin. The latter isolates exhibited a derepressed chromosomal cephalosporinase phenotype alone (Case phenotype; ATB10 to ATB15) or associated either with an ESBL (Case-ESBL phenotype; ATB16 to ATB22) or with resistance to imipenen (Case-IPM phenotype; ATB23 to ATB25). Among the 36 sus-ceptible group I isolates, only 7 came from the medical ICU. In contrast, among the 73 resistant isolates of group II, 56 were found in the medical ICU (Table 2). The five environmental strains were either susceptible (two strains; ATB1) or resistant (three strains; ATB10).
b-Lactamase assay.Of the 31 ESBL-producing strains, 22 (1
isolate from each patient) were studied for theirb-lactamase contents. In addition to the chromosomally derepressed cepha-losporinase, all except two isolates produced an ESBL with a pI of 7.8 which comigrated with the SHV-4 reference enzyme; the other two strains (ATB19) produced an enzyme with a pI of 6.5. The two isolates with the penicillinase phenotype (ATB4) were also tested, and theirb-lactamases comigrated with the TEM-1 reference enzyme (pI 5.4) (data not shown) (Table 2).
Plasmid hybridization analysis.The undigested plasmids of
isolates producing broad-spectrum b-lactamases or ESBLs were hybridized with the blaTEM-1and blaSHV-3probes,
respec-tively. All except four plasmids gave a positive signal with the SHV probe; the other four plasmids, including those from two isolates of ATB19 and those from two isolates with the peni-cillinase phenotype (ATB4), hybridized with the TEM probe (data not shown) (Table 2).
Restriction plasmid analysis.The undigested plasmid
pro-files were found to be nondiscriminatory: only five propro-files were obtained, including three for the resistant strains, and they had no clear correlation withb-lactam phenotypes (data not shown). Moreover, the plasmid profiles were poorly repro-ducible because of the variable presence of extraneous bands from open and linear plasmid forms.
Consequently, the plasmid DNAs of representative isolates were digested with the HindIII enzyme. On the basis of differ-ent, unrelated experiments, which were done to ensure that the patterns were reproducible and gave no partial digestion, 15 plasmid profiles (H1 to H15) were obtained and are illustrated in Fig. 1. The resistant isolates of group II (ATB10 to ATB25) were associated with profiles H1 to H12. Strains with the Case (ATB10 to ATB15) and the Case-IPM (ATB23 to ATB25) phenotypes harbored the predominant profile, profile H1 (Fig. 1, lanes 1 and 2), or related profiles, profiles H2 to H4 (Fig. 1, lanes 3 to 5, respectively). Indeed, profiles H3 and H4 ap-peared to correspond to profile H1, with additional bands being due to the presence of additional plasmid(s). Only four strains of the Case phenotype harbored plasmid profiles dif-ferent from H1: three strains of ATB10 gave profiles H5 and H6 (Fig. 1, lanes 7 and 8, respectively), and one strain of ATB11 gave profile H7 (Fig. 1, lane 9; Table 2). Strains with the Case-ESBL phenotype (ATB16 to ATB22) harbored the predominant profile, profile H8 (Fig. 1, lanes 10 to 12), which is distinct from profile H1. All strains of ATB16, ATB20, and
on May 15, 2020 by guest
http://jcm.asm.org/
ATB22 belonged to profile H8. Isolates of ATB17, ATB18, and ATB21 could be divided into profiles H9 to H11 (Fig. 1, lanes 13 to 15, respectively). It is noteworthy that profile H10 seems closely related to profile H4. The two strains of ATB19 with the TEM-type ESBL harbored profile H12 (Fig. 1, lane 16). The susceptible strains generally did not contain any plas-mid (16 strains); two isolates with the penicillinase phenotype (ATB4), however, gave profile H13 (lane 17), and three iso-lates of ATB5 and ATB9 harbored profiles H14 and H15, respectively (Fig. 1, lanes 18 and 19, respectively).
Ribotyping analysis. Ribotyping with chromosomal DNA
digested with EcoRI and plasmid pKK3535 as the probe led to 14 different patterns (ribotypes E1 to E14), which are illus-trated in Fig. 2. Patterns with at least two discordant bands were considered different (16). Each strain was tested at least twice to ensure the reproducibilities of the profiles. Ribotypes were identical, providing that they exhibited similar numbers and sizes of bands. Most of the resistant isolates of ATB10 to ATB25 tested (54 of 56 isolates) exhibited the same ribotype, ribotype E1 (Fig. 2, lane 1), which contained 14 bands ranging from 20 to 0.75 kb. Only the two resistant isolates of ATB19 which produced the TEM-type ESBL gave ribotype E10 (Fig. 2, lane 10). The susceptible isolates of group I tested (19 of 21 isolates) had a ribotype distinct from ribotype E1 (Table 2). Among the environmental strains, the two isolates of ATB1 exhibited ribotype E12, and the three isolates of ATB10 ex-hibited ribotype E1.
AP-PCR analysis.After AP-PCR analysis, the strains could
be divided into 15 profiles (profiles A1 to A15). These profiles were reproducible when the annealing step was done at a temperature of 408C. Most of the resistant isolates tested (54 of 56 isolates), those of ATB10 to ATB25, with different
plas-mid profiles and ribotype E1, exhibited the same AP-PCR profile, profile A1, containing five major bands (Fig. 3A and B, lanes 1). Only the two resistant isolates of ATB19 (ribotype E10) gave profile A10 (Fig. 3B, lane 10). The susceptible strains harbored 13 AP-PCR profiles (profiles A2 to A9 and A11 to A15) which were different from profile A1 by at least two bands. The results of AP-PCR were concordant with those of ribotyping for all except four isolates: three of the six strains of ribotype E12, including the two environmental strains (ATB1), gave profile A7 (Fig. 3B, lane 7), and another strain gave profile E15 (Fig. 3B, lane 15).
DISCUSSION
In order to examine the hypothesis that an outbreak of hospital infections was due to multidrug-resistant isolates of
E. aerogenes, all strains of this species isolated during a
[image:3.612.60.554.82.351.2]10-month period were analyzed by antibiotic resistance pheno-type, and representative strains were further characterized by plasmid restriction profiling, ribotyping, AP-PCR analysis, and ESBL typing. Our results indicate that most of the resistant isolates of group II, which came mainly from a medical ICU, belonged to 15 antibiotypes (ATB10 to ATB18 and ATB20 to ATB25), with threeb-lactam phenotypes (overproduction of the chromosomal enzyme alone or associated either with an ESBL of the SHV-4 type or imipenem resistance), and exhibited 11 plasmid restriction profiles but the same ribotype (ribotype E1) and the same AP-PCR pattern (pattern A1), demonstrating an epidemiological relatedness; only the two multidrug-resistant isolates of ATB19 produced an ESBL of the TEM type (plas-mid profile H12) and had ribotype E10 and AP-PCR pattern A10. In contrast, the susceptible isolates, mainly originating
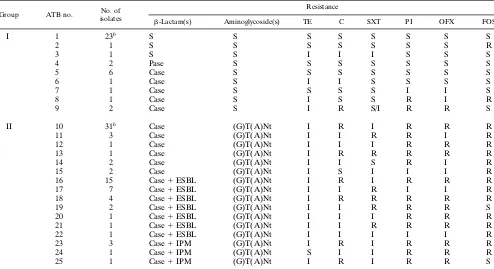
TABLE 1. Antibiotic resistance phenotypes of E. aerogenes isolatesa
Group ATB no. No. of
isolates
Resistance
b-Lactam(s) Aminoglycoside(s) TE C SXT PI OFX FOS
I 1 23b S S S S S S S S
2 1 S S S S S S S R
3 1 S S I I I S S S
4 2 Pase S S S S S S S
5 6 Case S S S S S S S
6 1 Case S I I S S S S
7 1 Case S S S S I I S
8 1 Case S I S S R I R
9 2 Case S I R S/I R R S
II 10 31b Case (G)T(A)Nt I R I R R R
11 3 Case (G)T(A)Nt I I R R I R
12 1 Case (G)T(A)Nt I I I R R R
13 1 Case (G)T(A)Nt I R R R R R
14 2 Case (G)T(A)Nt I I S R I R
15 2 Case (G)T(A)Nt I S I I I R
16 15 Case1ESBL (G)T(A)Nt I R I R R R
17 7 Case1ESBL (G)T(A)Nt I I R I I R
18 4 Case1ESBL (G)T(A)Nt I R R R R R
19 2 Case1ESBL (G)T(A)Nt I I R R R S
20 1 Case1ESBL (G)T(A)Nt I I I R R R
21 1 Case1ESBL (G)T(A)Nt I I R R R R
22 1 Case1ESBL (G)T(A)Nt I I I I I R
23 3 Case1IPM (G)T(A)Nt I R I R R R
24 1 Case1IPM (G)T(A)Nt S I I R R R
25 1 Case1IPM (G)T(A)Nt I R I R R S
aPase, penicillinase; Case, cephalosporinase; IPM, imipenem; G, gentamicin; T, tobramycin; A, amikacin; Nt, netilmicin; parentheses indicate a low level of
resistance; TE, tetracycline; C, chloramphenicol; SXT, co-trimoxazole; PI, pipemidic acid; OFX, ofloxacin; FOS, fosfomycin; S, susceptible; R, resistant; I, intermediate. bIncluding the environmental strains.
VOL. 34, 1996 EPIDEMIOLOGY OF MULTIDRUG-RESISTANT E. AEROGENES 2165
on May 15, 2020 by guest
http://jcm.asm.org/
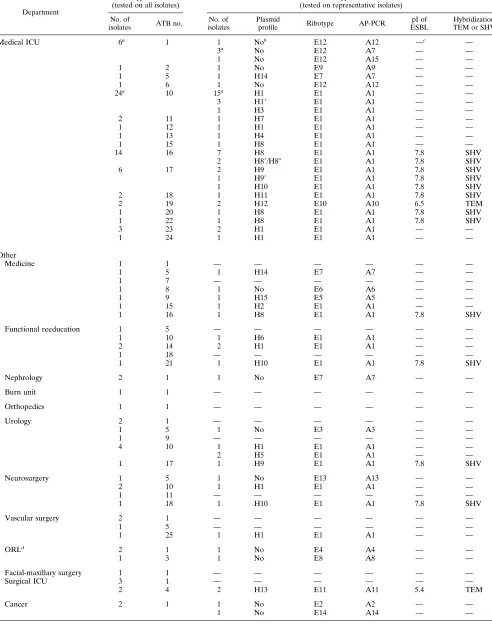
Department
Phenotypic markers (tested on all isolates)
Genotypic markers (tested on representative isolates)
No. of
isolates ATB no.
No. of isolates
Plasmid
profile Ribotype AP-PCR
pI of ESBL
Hybridization TEM or SHV
Medical ICU 6a 1 1 Nob E12 A12 —c —
3a No E12 A7 — —
1 No E12 A15 — —
1 2 1 No E9 A9 — —
1 5 1 H14 E7 A7 — —
1 6 1 No E12 A12 — —
24a 10 15a H1 E1 A1 — —
3 H19 E1 A1 — —
1 H3 E1 A1 — —
2 11 1 H7 E1 A1 — —
1 12 1 H1 E1 A1 — —
1 13 1 H4 E1 A1 — —
1 15 1 H8 E1 A1 — —
14 16 7 H8 E1 A1 7.8 SHV
2 H89/H80 E1 A1 7.8 SHV
6 17 2 H9 E1 A1 7.8 SHV
1 H99 E1 A1 7.8 SHV
1 H10 E1 A1 7.8 SHV
2 18 1 H11 E1 A1 7.8 SHV
2 19 2 H12 E10 A10 6.5 TEM
1 20 1 H8 E1 A1 7.8 SHV
1 22 1 H8 E1 A1 7.8 SHV
3 23 2 H1 E1 A1 — —
1 24 1 H1 E1 A1 — —
Other
Medicine 1 1 — — — — — —
1 5 1 H14 E7 A7 — —
1 7 — — — — — —
1 8 1 No E6 A6 — —
1 9 1 H15 E5 A5 — —
1 15 1 H2 E1 A1 — —
1 16 1 H8 E1 A1 7.8 SHV
Functional reeducation 1 5 — — — — — —
1 10 1 H6 E1 A1 — —
2 14 2 H1 E1 A1 — —
1 18 — — — — — —
1 21 1 H10 E1 A1 7.8 SHV
Nephrology 2 1 1 No E7 A7 — —
Burn unit 1 1 — — — — — —
Orthopedics 1 1 — — — — — —
Urology 2 1 — — — — — —
1 5 1 No E3 A3 — —
1 9 — — — — — —
4 10 1 H1 E1 A1 — —
2 H5 E1 A1 — —
1 17 1 H9 E1 A1 7.8 SHV
Neurosurgery 1 5 1 No E13 A13 — —
2 10 1 H1 E1 A1 — —
1 11 — — — — — —
1 18 1 H10 E1 A1 7.8 SHV
Vascular surgery 2 1 — — — — — —
1 5 — — — — — —
1 25 1 H1 E1 A1 — —
ORLd
2 1 1 No E4 A4 — —
1 3 1 No E8 A8 — —
Facial-maxillary surgery 1 1 — — — — — —
Surgical ICU 3 1 — — — — — —
2 4 2 H13 E11 A11 5.4 TEM
Cancer 2 1 1 No E2 A2 — —
1 No E14 A14 — —
aIncluding the environmental strains.
bNo, no plasmid.
c—, not determined.
dORL, otorhinolaryngology.
2166
on May 15, 2020 by guest
http://jcm.asm.org/
[image:4.612.61.553.73.697.2]from departments other than the medical ICU, belonged to nine antibiotypes (ATB1 to ATB9), generally exhibited no plasmid, but gave 12 ribotypes and 13 AP-PCR patterns. On the basis of the results of ribotyping and AP-PCR analysis, the hypothesis that the outbreak was due to a multidrug-resistant strain of E. aerogenes in the medical ICU and to the dissemi-nation of the epidemic strain throughout the hospital was con-firmed.
In our study, ribotyping was considered a good epidemio-logical method. Indeed, this method reveals stable genetic dif-ferences and shows a high degree of reproducibility. The
present results indicate that ribotyping is highly discriminatory for E. aerogenes isolates, in that as many as 14 ribotypes were delineated. These data confirm the observation of Grattard et al. (18) that E. aerogenes is a species sufficiently heterogeneous to allow for good differentiation between strains by ribotyping. An additional chromosomal marker was desirable to verify the ribotyping data. Pulsed-field gel electrophoresis, to our knowledge, has not been reported to have been used to type
E. aerogenes isolates. In contrast, the AP-PCR method has
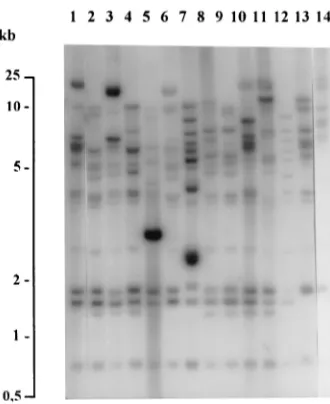
[image:5.612.318.551.70.215.2]been successfully used to document relatedness among epi-demic isolates of E. aerogenes (9, 16, 18). In our study, all the epidemic isolates of ribotype E1 harbored AP-PCR pattern A1. The results of ribotyping and AP-PCR were concordant for all strains except strains of ribotype E12: six isolates, in-cluding two environmental strains from the medical ICU, were associated with ribotype E12 (five strains of ATB1 and one of ATB6), but could be divided into three AP-PCR patterns. It is worth noting that one of the environmental strains (ATB1, E12, A7) was isolated in the immediate environment of a patient who carried an isolate with the following markers: ATB1, E12, A12. These results suggest that ribotyping exper-iments provide more conclusive evidence of strain relatedness than AP-PCR. However, as proposed elsewhere (18), ribotyp-ing and PCR can be applied usefully in combination: AP-PCR is a method that is easy to perform and that generates rapid results. This method can be used for screening, whereas ribotyping is helpful for clarifying the relatedness between nosocomial strains which give inconsistent or inconclusive re-sults by AP-PCR (18). However, in our study, the rere-sults of ribotyping and AP-PCR analysis did not strictly correlate with those of plasmid profiling and antibiotic resistance pattern analysis, and the differences deserve some explanation. Effec-tively, the plasmid profiles of epidemiologically related isolates harbored 12 plasmid restriction profiles, and the two predom-inant profiles, H1 and H8, have been found to be different. Nevertheless, the plasmid profiles of epidemiologically related isolates are not necessarily consistent over time because of either the acquisition or the loss of a plasmid(s) as a conse-quence of variable selective pressures. Conversely, genetically different host strains can acquire the same plasmid(s), either FIG. 1. Plasmid profiles after restriction of E. aerogenes isolates with HindIII.
Lane 1, profile H1; lane 2, profile H19, which differs in comparison with profile H1 by one additional band of about 25 kb; lane 3, profile H2; lane 4, profile H3; lane 5, profile H4; lane 6, profile H5; lane 7, profile H6; lane 8, profile H7; lane 9, profile H8; lanes 10 and 11, profiles H89and H80, respectively, which differ in comparison with profile H8 by one additional band of 7 and 5 kb, respectively; lane 12, profile H9; lane 13, profile H99, which differs in comparison with H9 by one additional band of 9 kb; lane 14, profile H10; lane 15, profile H11; lane 16, profile H12; lane 17, profile H13; lane 18, profile H14 and lane 19, profile H15. Lane M, marker, phagelDNA digested with HindIII (the sizes of the marker bands are indicated in kilobases).
FIG. 2. Profiles obtained by Southern blot analysis of EcoRI-digested total DNA from E. aerogenes isolates detected with thea-32P-labelled pKK3535 plas-mid as a probe. Lanes 1 to 14 correspond to profiles E1 to E14, respectively.
FIG. 3. Comparison between the AP-PCR patterns with primer AP12h of
E. aerogenes isolates. (A) AP-PCR pattern A1 obtained with isolates of group II
corresponding to different antibiotic resistance patterns and plasmid profiles. Lane 1, isolate ATB10, H1; lane 2, isolate ATB10, H3; lane 3, isolate ATB13, H4; lane 4, isolate ATB10, H5; lane 5, isolate ATB10, H6; lane 6, isolate ATB11, H7; lane 7, isolate ATB16, H8; lane 8, isolate ATB17, H9; lane 9, isolate ATB18, H11. (B) AP-PCR pattern types obtained for isolates of E. aerogenes. Lanes 1 to 15, profiles A1 to A15, respectively; lanes M, marker, phagelDNA digested with PstI (the sizes of the bands of the profile A1 are indicated in kilobases); lane C, experimental control (water).
VOL. 34, 1996 EPIDEMIOLOGY OF MULTIDRUG-RESISTANT E. AEROGENES 2167
on May 15, 2020 by guest
http://jcm.asm.org/
[image:5.612.95.260.497.699.2]randomly or specifically because of antibiotic therapy. These outbreaks are sometimes called plasmid epidemics (5). In par-ticular, plasmids encoding for the ESBLs, which are known to be large transferable plasmids that also confer resistance to aminoglycosides, can transfer among members of the family
Enterobacteriaceae (12). It is noteworthy that during the same
period (January to June 1993), another study in our laboratory (2) demonstrated the presence of an important number of SHV-4-producing strains of K. pneumoniae in the hospital (73 strains), particularly in the medical ICU (10 strains). The SHV-4-producing strains of E. aerogenes examined in the present study have often been isolated from patients also har-boring SHV-4-producing K. pneumoniae strains, sometimes in the same clinical samples. In addition, the DNA composition of antibiotic resistance plasmids may change through various transfers or because of the acquisition or loss of resistance-encoding transposons (26). These confusing problems are in-herent to the fact that plasmids are extrachromosomal ele-ments and are not part of the much more stable chromosome of the host strain. As a result, epidemiologically related isolates may exhibit different plasmid profiles which have evolved un-der antibiotic pressure. Similar variations in investigations of
E. aerogenes outbreaks (16) or outbreaks associated with other
organisms (24, 36) have been documented in longitudinal anal-yses and may limit the usefulness of this technique for the tracking of epidemic strains.
Similarly, the antibiotic resistance patterns are influenced not only by plasmid changes, as discussed above, but also by mutations to resistance which may also be selected for under antibiotic pressure. For example, overproduction of the chro-mosomal cephalosporinase is due to mutations impairing the control of inducible b-lactamase synthesis. In our study, all patients colonized by or infected with the imipenem-resistant strains of E. aerogenes had previously been treated with imi-penem. This resistance may be associated with a modification of a major outer membrane protein (7, 11, 21, 39) or a change in lipopolysaccharide (25). Other mutations altering the tar-gets and/or the permeation of antibiotics are known to lead to resistance to tetracycline, chloramphenicol, co-trimoxazole, quinolones, or fosfomycin in enterobacteria, and they could account for the minor variations observed between the resis-tant antibiotypes. The antibiotic resistance phenotype has been reported to be an undiscriminatory marker for E. aerogenes, giving very few different patterns for unrelated strains (18). The present results are in opposition to this contention, be-cause epidemiologically related strains gave different patterns. Other phenotypic markers have been proposed for use in the typing of E. aerogenes, such as biotyping (18), serotyping with
Klebsiella capsular antisera, bacteriocin typing, or phage typing
(15). However, these methods often failed to type all strains and to discriminate between isolates. On the basis of our re-sults, we came to the conclusion that the outbreak was due to a cephalosporinase-derepressed strain of E. aerogenes (ri-botype E1, AP-PCR pattern A1), which acquired various re-sistance plasmids (profiles H1 to H11) and mutations leading to several antibiotypes (ATB10 to ATB18 and ATB20 to ATB25). This evolution is probably related to the selective pressure of specific antibiotic treatments in individual patients and even in the same patient during the course of hospitaliza-tion, as demonstrated for imipenem resistance.
In the medical ICU, 54 of 63 clinical isolates were consid-ered to be the epidemic strain. The two multidrug-resistant isolates with a ribotype different from ribotype E1, which pro-duce a pI 6.5 ESBL of the TEM type (maybe TEM-24) (6), are probably imported strains, as recently described during an-other E. aerogenes outbreak (23). Three multidrug-resistant
strains of ribotype E1 were found in the environment of the medical ICU. The presence of the epidemic strain on furniture and floor surfaces suggests indirectly transmission by the hands of health care staff, as usually observed in nosocomial out-breaks caused by enterobacteria. The remaining clinical iso-lates found in this unit (7 of 63 strains) were susceptible, in particular to aminoglycosides. Among the isolates from the other departments, 29 of 46 isolates were susceptible (group I) and exhibited a ribotype different from ribotype E1 and an AP-PCR pattern different from A1. These unrelated strains had various and distinct markers. Like most members of the family Enterobacteriaceae, E. aerogenes is part of the commen-sal digestive flora of humans and can be found in the environ-ment. The bowels of colonized or infected patients are thought to be the reservoir for nosocomial infections caused by enter-obacteria. As stated above, E. aerogenes is a genetically heter-ogeneous species and may statistically be found in patients or in their environment. The remaining isolates (17 of 46 strains) correspond to the epidemic strain, ribotype E1 and AP-PCR pattern A1. They were collected from 10 patients hospitalized in nine units of five departments. The epidemiological inves-tigation revealed that six of these patients had been transferred from the medical ICU to other units during their hospitaliza-tions. These transfers explain, at least in part, the spread of the epidemic strain throughout the hospital. Hospital services (i.e., radiology, etc.) and personnel who deal with patients from many different departments are also implicated in diffusion epidemics.
Our study underlines the importance of the detection of patients colonized or infected with multidrug-resistant bacteria when they are newly admitted to a unit or to a hospital. Once established in a department, this type of strain tends to replace other more susceptible resident pathogens and to cause noso-comial outbreaks.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant (94CN34) from the Institut National de la Sante´ et de la Recherche Me´dicale (INSERM).
REFERENCES
1. Acar, J., H. Chardon, P. Choutet, P. Courvalin, H. Dabernat, H. Drugeon, L. Dubreuil, J. P. Flandrois, F. Goldstein, C. Morel, A. Philippon, B. Rouveix, J. Sirot, and A. Thabaut.1995. Statement of Antibiogram Committee of the French Society for Microbiology. Pathol. Biol. 43:1–8.
2. Bermudes, H., C. Arpin, Z. El Harrif-Heraud, F. Jude, C. Be´be´ar, and C. Quentin.1994.b-Lactamases a` spectre e´largi: caracte´risation et e´pide ´miolo-gie mole´culaire—e´tude sur un an dans un CHU, abstr. 197, p. 163. In Abstracts of the 14th Re´union Inter-disciplinaire de Chimiothe´rapie et d’Agents Infectieux.
3. Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523. 4. Brosius, J., A. Ullrich, M. A. Raker, A. Gray, T. J. Dull, R. R. Gutell, and
H. F. Noller.1981. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid 6:112–118. 5. Chabbert, Y. A., N. El Solh, M. J. Le Pors, A. Roussel, and J. L. Witchitz.
1979. Plasmid epidemics. Prog. Clin. Biol. Res. 35:27–32.
6. Chanal, C. M., D. L. Sirot, A. Petit, R. Labia, A. Morand, J. L. Sirot, and R. A. Cluzel.1989. Multiplicity of TEM-derivedb-lactamases from Klebsiella
pneumoniae strains isolated at the same hospital and relationships between
the responsible plasmids. Antimicrob. Agents Chemother. 33:1915–1920. 7. Chow, J. W., and D. M. Shlaes. 1991. Imipenem resistance associated with
the loss of a 40 kDa outer membrane protein in Enterobacter aerogenes. J. Antimicrob. Chemother. 28:499–504.
8. Courvalin, P., F. Goldstein, A. Philippon, and J. Sirot (ed.). 1985. L’anti-biogramme. mpc Vide´om, Paris.
9. Davin-Regli, A., P. Saux, C. Bollet, F. Gouin, and P. de Micco. 1996. Inves-tigation of outbreaks of Enterobacter aerogenes colonisation and infection in intensive care units by random amplification of polymorphic DNA. J. Med. Microbiol. 44:89–98.
10. de Champs, C., D. Guelon, D. Joyon, D. Sirot, M. Chanal, and J. Sirot. 1991. Treatment of a meningitis due to Enterobacter aerogenes producing a
on May 15, 2020 by guest
http://jcm.asm.org/
pressed cephalosporinase and a Klebsiella pneumoniae producing an extend-ed-spectrumb-lactamase. Infection 19:181–183.
11. de Champs, C., C. Henquell, D. Guelon, D. Sirot, N. Gazuy, and J. Sirot. 1993. Clinical and bacteriological study of nosocomial infections due to
Enterobacter aerogenes resistant to imipenem. J. Clin. Microbiol. 31:123–127.
12. de Champs, C., D. Sirot, C. Chanal, M. C. Poupart, M. P. Dumas, and J. Sirot.1991. Concomitant dissemination of three extended-spectrumb -lac-tamases among different Enterobacteriaceae isolated in a French hospital. J. Antimicrob. Chemother. 27:441–457.
13. Flynn, D. M., R. A. Weinstein, C. Nathan, M. A. Gaston, and S. A. Kabins. 1987. Patients’ endogenous flora as the source of ‘nosocomial’ Enterobacter in cardiac surgery. J. Infect. Dis. 156:363–368.
14. Gaston, M. A. 1988. Enterobacter: an emerging nosocomial pathogen. J. Hosp. Infect. 11:197–208.
15. Gaston, M. A., M. A. Strickland, B. A. Ayling-Smith, and T. L. Pitt. 1989. Epidemiological typing of Enterobacter aerogenes. J. Clin. Microbiol. 27:564– 565.
16. Georghiou, P. R., R. J. Hamill, C. E. Wright, J. Versalovic, T. Koeuth, D. A. Watson, and J. R. Lupski.1995. Molecular epidemiology of infections due to
Enterobacter aerogenes: identification of hospital outbreak-associated strains
by molecular techniques. Clin. Infect. Dis. 20:84–94.
17. Grattard, F., B. Pozzetto, P. Berthelot, I. Rayet, A. Ros, B. Lauras, and O. G. Gaudin.1994. Arbitrarily primed PCR, ribotyping, and plasmid pattern analysis applied to investigation of a nosocomial outbreak due to
Enter-obacter cloacae in a neonatal intensive care unit. J. Clin. Microbiol. 32:596–
602.
18. Grattard, F., B. Pozzetto, L. Tabard, M. Petit, A. Ros, and O. G. Gaudin. 1995. Characterization of nosocomial strains of Enterobacter aerogenes by arbitrarily primed-PCR analysis and ribotyping. Infect. Control Hosp. Epi-demiol. 16:224–230.
19. Grimont, F., and P. A. D. Grimont. 1986. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann. Inst. Pasteur Micro-biol. 137B:165–175.
20. Haertl, R., and G. Bandlow. 1993. Epidemiological fingerprinting of
Enter-obacter cloacae by small restriction endonuclease analysis and pulsed-field
gel electrophoresis of genomic restriction fragments. J. Clin. Microbiol. 31: 128–133.
21. Hopkins, J. M., and K. J. Towner. 1990. Enhancer resistance to cefotaxime and imipenem associated with outer membrane protein alterations in
Entero-bacter aerogenes. J. Antimicrob. Chemother. 25:49–55.
22. Huber, T. W., and J. S. Thomas. 1994. Detection of resistance due to inducibleb-lactamase in Enterobacter aerogenes and Enterobacter cloacae. J. Clin. Microbiol. 32:2481–2486.
23. Jarroux, E., L. Collet, O. Bellon, E. Lagier, M. Miquel, F. Grimont, A. Nguyen, P. Brunet, F. Duluc, C. Payen, P. Rousselier, M. Bietrix, Y. Assa-dourian, P. Nicolas, C. Broquet, D. Bertei, and H. Chardon.1994.
Entero-bacter aerogenes producteur de ce´phalosporinase de´re´prime´e et de be ˆtalac-tamase a` spectre e´tendu. Inte´reˆt du ce´fe´pime et du cefpirome pour la de´tection lors d’une e´pide´mie hospitalie`re en Provence, abstr. 198, p. 163. In Abstracts of the Re´union Inter-disciplinaire de Chimiothe´rapie et d’Agents Infectieux.
24. John, J. F., Jr., and J. A. Twitty. 1986. Plasmids as epidemiologic markers in
nosocomial gram-negative bacilli: experience and review of the literature. Rev. Infect. Dis. 8:693–704.
25. Leying, H., W. Cullmann, and W. Dick. 1991. Carbapenem resistance in
Enterobacter aerogenes is due to lipopolysaccharide alterations.
Chemother-apy (Basel) 37:106–113.
26. Lupski, J. R. 1987. Molecular mechanisms for transposition of drug resis-tance genes and other movable genetic elements. Rev. Infect. Dis. 9:357–368. 27. Markowitz, S. M., S. M. Smith, and D. S. Williams. 1983. Retrospective analysis of plasmid patterns in a study of burn unit outbreaks of infection due to Enterobacter cloacae. J. Infect. Dis. 148:18–23.
28. Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification ofb -lacta-mases. J. Gen. Microbiol. 88:169–178.
29. Mellencamp, M. A., J. S. Roccaforte, L. C. Preheim, C. C. Sanders, C. A. Anene, and M. J. Bittner.1990. Isolation of Enterobacter aerogenes suscep-tible to beta-lactam antibiotics despite high level beta-lactamase production. Eur. J. Clin. Microbiol. Infect. Dis. 9:827–830.
30. Meyers, H. B., E. Fontanilla, and L. Mascola. 1988. Risk factors for devel-opment of sepsis in a hospital outbreak of Enterobacter aerogenes. Am. J. Infect. Control. 16:118–122.
31. Nicolas, M. H., V. Jarlier, N. Honore, A. Philippon, and S. T. Cole. 1989. Molecular characterization of the gene encoding SHV-3b-lactamase respon-sible for transferable cefotaxime resistance in clinical isolates of Klebsiella
pneumoniae. Antimicrob. Agents Chemother. 33:2096–2100.
32. Philippon, A., R. Labia, and G. Jacoby. 1989. Extended-spectrumb -lacta-mases. Antimicrob. Agents Chemother. 33:1131–1136.
33. Poilane, I., P. Cruaud, E. Lachassinne, F. Grimont, P. A. D. Grimont, M. Collin, J. Gaudelus, J. C. Torlotin, and A. Collignon.1993. Enterobacter
cloacae cross-colonization in neonates demonstrated by ribotyping. Eur.
J. Clin. Microbiol. Infect. Dis. 12:820–826.
34. Ross, G. W., and M. G. Boulton. 1972. Improvement of the specificity of an antiserum tob-lactamase by absorption with a mutant which does not pro-duce the enzyme. J. Bacteriol. 112:1435–1437.
35. Ross, G. W., and M. G. Boulton. 1973. Purification ofb-lactamases on QAE-Sephadex. Bioch. Bioph. Acta 309:430–439.
36. Rubens, C. E., W. E. Farrar, Jr., Z. A. McGee, and W. Schaffner. 1981. Evolution of a plasmid mediating resistance to multiple antimicrobial agents during a prolonged epidemic of nosocomial infections. J. Infect. Dis. 143: 170–181.
37. Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
38. Sirot, D., J. Sirot, R. Labia, A. Morand, P. Courvalin, A. Darfeuille-Michaud, R. Perroux, and R. Cluzel.1987. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: iden-tification of CTX-1, a novelb-lactamase. J. Antimicrob. Chemother. 20:323– 334.
39. Tzouvelekis, L. S., E. Tzelepi, A. F. Mentis, A. C. Vatopoulos, and A. Tsakris. 1992. Imipenem resistance in Enterobacter aerogenes is associated with de-repression of chromosomal cephalosporinases and impaired permeability. FEMS Microbiol. Lett. 95:195–200.
VOL. 34, 1996 EPIDEMIOLOGY OF MULTIDRUG-RESISTANT E. AEROGENES 2169