metal-organic papers
Acta Cryst.(2005). E61, m1937–m1939 doi:10.1107/S1600536805027674 Shen and Yuan [Ni(C
2N3)2(C3H7NO)2]
m1937
Acta Crystallographica Section E
Structure Reports Online
ISSN 1600-5368
catena-Poly[[bis(N,N-dimethylformamide-
j
O)-nickel(II)]-di-
l
-1,5-dicyanamido-
j
N
1:
j
N
5]
Xiao-Ping Shena* and Ai-Hua Yuanb
a
School of Chemistry and Chemical Engineering, Jiangsu University, Zhenjiang 212013, People’s Republic of China, andbSchool of Materials
Science and Engineering, Jiangsu University of Science and Technology, Zhenjiang 212003, People’s Republic of China
Correspondence e-mail: xiaopingshen@163.com
Key indicators
Single-crystal X-ray study
T= 193 K
Mean(N–C) = 0.003 A˚
Rfactor = 0.023
wRfactor = 0.062
Data-to-parameter ratio = 11.8
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2005 International Union of Crystallography Printed in Great Britain – all rights reserved
In the crystal structure of the title complex, [Ni(C2N3)2 -(C3H7NO)2]nor [Ni(dca)2(DMF)2]n, where dca is dicyanamide
and DMF is N,N-dimethylformamide, each NiII atom is
six-coordinated in a distorted octahedral coordination environ-ment. Four N atoms from four dca ligands fill the equatorial positions, and two O atoms from two DMF ligands fill the axial
positions. The structure is isostructural with
[Co(dca)2(DMF)2]n but is not isostructural with
[Mn(dca)2(DMF)2]n. The Ni II
atom and the dicyanamide
bridging ligand occupy special positions of symmetry 2/mand
m, respectively. The structure consists of uniform neutral
chains where neighbouring NiIIatoms are connected through
two asymmetric end-to-end dca bridges.
Comment
Dicyanamide (dca), [N(CN)2]
, complexes have been studied extensively recently because of their fascinating topologies and interesting magnetic properties (Battenet al., 1998; Miller & Manson 2001; Jensen et al., 2000; Riggio et al., 2001). A number of nickel(II)–dca complexes have been reported (Sun,
et al., 2000; Wanget al., 2004; Konoret al., 2005). Our research interest is the construction of novel topologies of cyano complexes and studying their magnetic properties (Shenet al., 2004, 2003). In the present work, we report the crystal
struc-ture of a one-dimensional chain polymer, viz.
[image:1.610.257.406.489.670.2][Ni(dca)2(DMF)2]n, (I).
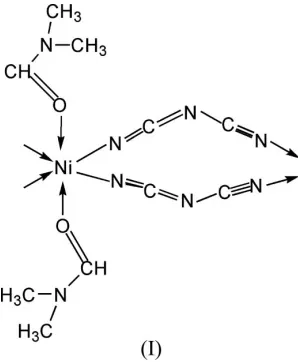
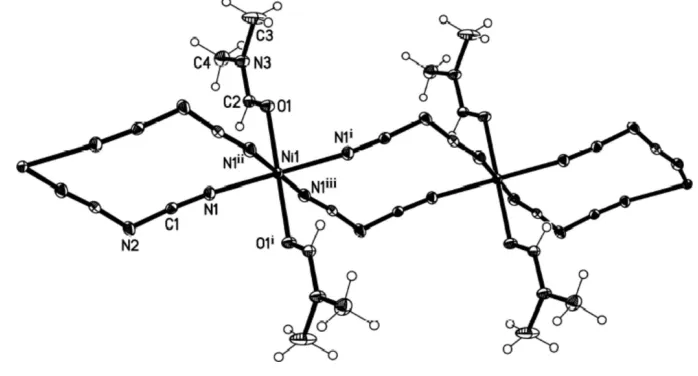
Fig. 1 shows the local coordination about the nickel(II) centre in (I). The structure of (I) is isostructural with [Co(dca)2(DMF)2]n(Tonget al., 2003) but is not isostructural
with [Mn(dca)2(DMF)2]n (Batten et al., 1999). The space
group of [Co(dca)2(DMF)2]nreported by Dong et al.(2003)
has been described incorrectly in C2; it should be C2/m, as reported by Tonget al.(2003). The structure of (I) consists of uniform neutral chains in which neighbouring nickel(II) atoms are connected through two asymmetric end-to-end dca bridges. The coordination geometry of the nickel(II) atom is distorted octahedral, being coordinated by four N atoms of four symmetry-related dca ligands in the equatorial plane and two O atoms of two symmetry-related DMF ligands at the axial positions. The N—Ni—N bond angles are in the range
87.84 (6)–92.16 (6), close to 90. The four Ni—N(dca) bond
lengths in (I) are all 2.0733 (11) A˚ , corresponding to the values reported in the dca-bridged nickel(II) complexes
[Ni(apo)-(dca)2] [2.043 (4)–2.096 (4) A˚ ; apo = 2-aminopyridine
N-oxide; Sunet al., 2000] and [Ni(tn)2(dca)](ClO4) [2.095 (4) and 2.116 (4) A˚ ; tn = trimethylenediamine; Li et al., 2002], and shorter than the Mn—N bond lengths [2.218 (2) and 2.203 (2) A˚ ] in [Mn(dca)2(DMF)2]n(Battenet al., 1999) and
the Co—N bond lengths [2.123 (2) A˚ ] in [Co(dca)2(DMF)2]n
(Tonget al., 2003); this is what one would expect from the ionic
radii (Ni2+ < Co2+ < Mn2+). The two Ni—O (DMF) bond
lengths are both 2.0670 (13) A˚ , corresponding to the values [2.0776 (19) A˚ ] in [Ni(pmbp)2(DMF)2] [Hpmbp = 1-phenyl-3-methyl-4-benzoyl-1H-pyrazol-5(4H)-one; Shen & Yuan 2004]
and shorter than the M—O bond lengths in [Mn(dca)2
-(DMF)2]n [Mn—O = 2.199 (2) A˚ ] and [Co(dca)2(DMF)2]n
[Co—O = 2.096 (2) A˚ ].
The dicyanamide (dca) ligand adopts an end-to-end coor-dination mode. Two dca ions link two nickel(II) atoms to form
a 12-membered Ni(dca)2Ni ring and neighbouring rings share
the nickel(II) atoms to form a chain of [Ni(dca)2]n. The chains
are linear, the Ni(dca)2Ni rings being in a slight chair
conformation.
The free dicyanamide (dca) ligand possessesC2vsymmetry.
The dca ligand in (I) also adopts essentiallyC2vsymmetry, with
a nitrile C N bond length of 1.1545 (18) A˚ for N1 C1,
showing the triple-bond character. The bond angle related to
the amide N atom, C1—N2—C1(x, 1y,z), is 118.61 (16),
corresponding to an amide N atom with ansp2hybrid orbital;
that related to the nitrile group, N1 C1—N2, is 174.95 (13),
corresponding to N1 and C1 with ansphybrid orbital.
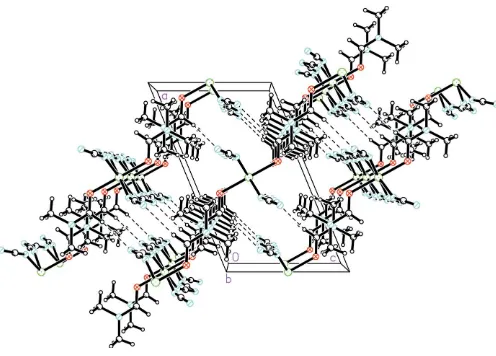
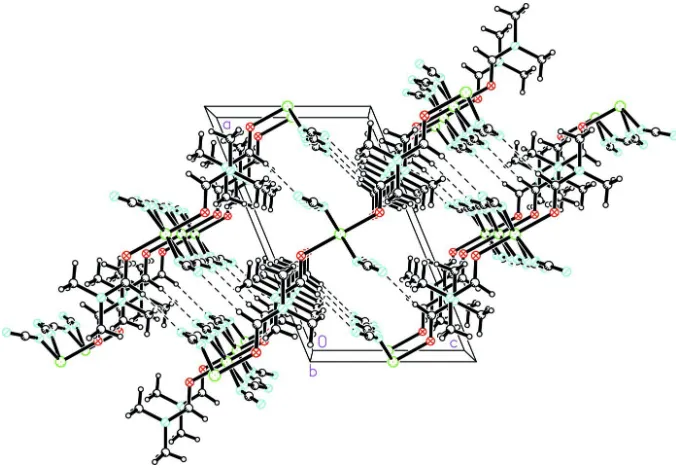
The chains propagate parallel to the crystallographicbaxis, the Ni Ni distance along the chain being equal to thebaxis length, 7.3166 (7) A˚ . The chains interdigitate such that each DMF ligand lies between two DMF ligands of an adjacent
chain, with a shortest Ni Ni interchain distance of
7.628 (2) A˚ . Adjacent chains are held together by a weak C—
H N hydrogen bond, forming layers parallel to theabplane
(Fig. 2 and Table 2).
Experimental
An aqueous solution (10 ml) of Ni(NO3)26H2O (0.146 g, 0.5 mmol)
was added to a DMF solution (10 ml) of Na[N(CN)2] (0.090 g,
1.0 mmol). Slow evaporation of the resulting mixture led to green crystals suitable for X-ray diffraction analysis. Analysis found: C 35.53, H 4.12, N 33.31%; calculated for C10H14N8NiO2: C 35.64, H
4.19, N 33.26%.
Crystal data
[Ni(C2N3)2(C3H7NO)2]
Mr= 336.98
Monoclinic,C2=m a= 13.3866 (17) A˚
b= 7.3166 (7) A˚
c= 8.0595 (10) A˚
= 112.503 (3) V= 729.28 (15) A˚3
Z= 2
Dx= 1.535 Mg m 3
MoKradiation Cell parameters from 1804
reflections
= 3.2–27.5
= 1.35 mm1
T= 193 (2) K Block, green
0.400.210.20 mm
Data collection
Rigaku Mercury CCD diffractometer
!scans
Absorption correction: multi-scan (Jacobson, 1998)
Tmin= 0.646,Tmax= 0.774
4029 measured reflections
900 independent reflections 887 reflections withI> 2(I)
Rint= 0.018
max= 27.5
h=17!15
k=8!9
l=10!10
metal-organic papers
m1938
Shen and Yuan [Ni(C2N3)2(C3H7NO)2] Acta Cryst.(2005). E61, m1937–m1939
Figure 2
[image:2.610.42.298.74.214.2] [image:2.610.46.294.275.453.2]The packing in (I), showing the C—H N hydrogen-bond interactions as dashed lines.
Figure 1
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.023
wR(F2) = 0.062
S= 1.03 900 reflections 76 parameters
H atoms treated by a mixture of independent and constrained refinement
w= 1/[2
(Fo2) + (0.0436P)2
+ 0.3824P]
whereP= (Fo2+ 2Fc2)/3
(/)max< 0.001
max= 0.15 e A˚
3
min=0.47 e A˚
[image:3.610.43.296.207.286.2]3
Table 1
Selected geometric parameters (A˚ ,).
Ni1—O1 2.0670 (13)
Ni1—N1 2.0733 (11)
O1—C2 1.243 (2)
N1—C1 1.1545 (18)
N2—C1 1.3074 (15)
N3—C2 1.319 (2)
N3—C3 1.448 (3)
N3—C4 1.458 (3)
O1—Ni1—N1 91.61 (4)
O1i—Ni1—N1 88.39 (4) N1—Ni1—N1ii
92.16 (6)
N1—Ni1—N1iii
87.84 (6) C1iv—N2—C1 118.61 (16)
N1—C1—N2 174.95 (13)
Symmetry codes: (i)xþ1;yþ1;zþ1; (ii)x;yþ1;z; (iii)xþ1;y;zþ1; (iv)x;yþ2;z.
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
C2—H2 N2v
0.95 (1) 2.51 (1) 3.453 (2) 169 (2)
Symmetry code: (v)xþ3 2;yþ
3 2;zþ1.
H atoms were found in a difference Fourier map and refined with bond-length restraints of C—H = 0.95 (1) A˚ for the methyl groups and the H H distance restrained to 1.50 (1) A˚ . One of two inde-pendent H atoms lies on the mirror plane.
Data collection: CrystalClear (Rigaku, 2000); cell refinement:
CrystalClear; data reduction: CrystalClear; method used to solve
structure: the coordinates of the Co structure of Tonget al. (2003) were used; program(s) used to refine structure:SHELXL97 (Shel-drick, 1997); molecular graphics:SHELXTL(Bruker, 1998); software used to prepare material for publication:SHELXTL.
This work was supported by the Natural Science Founda-tion of Jiangsu Province (No. BK2005056), People’s Republic of China.
References
Batten, S. R., Jensen, P., Kepert, C. J., Kurmoo, M., Moubaraki, B., Murray, K. S. & Price, D. J. (1999).J. Chem. Soc. Dalton Trans.pp. 2987–2997. Batten, S. R., Jensen, P., Moubaraki, B., Murray, K. S. & Robson, R. (1998).
Chem. Commun.pp. 439–440.
Bruker (1998). SHELXTL. Version 5.10. Bruker AXS Inc., Madison, Wisconsin, USA.
Dong, W., Liang, M., Sun, Y. Q. & Liu, Z. Q. (2003).Z. Anorg. Allg. Chem.629, 2443–2445.
Jacobson, R. (1998). Private communication to the Rigaku Corporation. Jensen, P., Batten, S. R., Moubaraki, B. & Murray, K. S. (2000). Chem.
Commun.pp. 793–740.
Konor, S., Dalai, S., Mukherjee, P. S., Drew, M. G. B., Ribas, J. & Chaudhuri, N. R. (2005).Inorg. Chim. Acta,358, 957–963.
Li, B. L., Ding, J. G., Lang, J. P., Zheng, X. & Chen, J. T. (2002).J. Mol. Struct.
616, 175–179.
Miller, J. S. & Manson, J. L. (2001).Acc. Chem. Res.34, 563–570.
Rigaku (2000). CrystalClear. Version 1.3. Rigaku Corporation, 3-9-12 Akishima, Tokyo, Japan.
Riggio, I., van Albada, G. A., Ellis, D. D., Spek, A. L. & Reedijk, J. (2001).
Inorg. Chim. Acta,313, 120–124.
Sheldrick, G. M. (1997).SHELXL97. University of Go¨ttingen, Germany. Shen, X. P., Gao, S., Yin, G., Yu, K. B. & Xu, Z. (2004).New J. Chem.28, 996–
999.
Shen, X. P., Li, B. L., Zou, J. Z., Hu, H. M. & Xu, Z. (2003).J. Mol. Struct.657, 325–331.
Shen, X. P. & Yuan, A. H. (2004).Acta Cryst.E60, m1228–m1230.
Sun, B. W., Gao, S., Ma, B. Q., Niu, D. Z. & Wang, Z. M. (2000).J. Chem. Soc. Dalton Trans.pp. 4187–4191.
Tong, M.-L., Zhou, A.-J., Hu, S., Chen, X.-M. & Ng, S. W. (2003).Acta Cryst.
E59, m405–m407.
Wang, S. W., Zhu, X., Li, B. L., Lang, J. P., Zheng, X. & Zhang, Y. (2004).Chin. J. Struct. Chem.23, 145–148.
metal-organic papers
Acta Cryst.(2005). E61, m1937–m1939 Shen and Yuan [Ni(C
supporting information
sup-1
Acta Cryst. (2005). E61, m1937–m1939
supporting information
Acta Cryst. (2005). E61, m1937–m1939 [doi:10.1107/S1600536805027674]
catena
-Poly[[bis(
N
,
N
-dimethylformamide-
κ
O
)nickel(II)]-di-
µ
-1,5-dicyanamido-κ
N
1:
κ
N
5]
Xiao-Ping Shen and Ai-Hua Yuan
S1. Comment
Dicyanamide (dca), [N(CN)2]-, complexes have been studied extensively recently because of their fascinating topologies
and interesting magnetic properties (Batten et al., 1998; Miller & Manson 2001; Jensen et al., 2000; Riggio et al., 2001).
A number of nickel(II)–dca complexes have been reported (Sun, et al., 2000; Wang et al., 2004; Konor et al., 2005). Our
research interest is construction of novel topologies of cyano complexes and studying the magnetic properties (Shen et
al., 2004, 2003). In the present work, we report the crystal structure of a one-dimensional chain polymer, viz.
[Ni(dca)2(DMF)2]n, (I).
Fig. 1 shows the local coordination about the nickel(II) center in (I). The structure of (I) is isostructural with
[Co(dca)2(DMF)2]n (Tong et al., 2003) and not isostructural with [Mn(dca)2(DMF)2]n (Batten et al., 1999). The space
group of [Co(dca)2(DMF)2]n reported by Dong et al. (2003) has been described incorrectly in C2; it should be C2/m, as
reported by Tong et al. (2003). The structure of (I) consists of uniform neutral chains where neighboring nikel(II) atoms
are connected through two asymmetric end-to-end dca bridges. The coordination geometry of the nickel(II) atom is
distorted octahedral, being coordinated by four N atoms of four symmetry-related dca ligands in the equatorial plane and
two O atoms of two symmetry-related DMF ligands at the axial positions. The N—Ni—N bond angles are in the range
87.84 (6) to 92.16 (6)°, close to 90°. The four Ni—N(dca) bond lengths in (I) are all 2.0733 (11) Å, corresponding to the
values reported in the dca-bridged nickel(II) complexes [Ni(apo)(dca)2] [2.043 (4)–2.096 (4) Å; apo = 2-aminopyridine
N-oxide; Sun et al., 2000] and [Ni(tn)2(dca)](ClO4) [2.095 (4) and 2.116 (4) Å; tn = trimethylenediamine; Li et al., 2002],
and shorter than the Mn—N bond lengths [2.218 (2) and 2.203 (2) Å] in [Mn(dca)2(DMF)2]n (Batten et al., 1999) and the
Co—N bond lengths [2.123 (2) Å] in [Co(dca)2(DMF)2]n (Tong et al., 2003); this is what one would expect from the ionic
radii (Ni2+ < Co2+ < Mn2+). The two Ni—O (DMF) bond lengths are both 2.0670 (13) Å, corresponding to the values
[2.0776 (19) Å] in [Ni(pmbp)2(DMF)2] [Hpmbp = 1-phenyl-3-methyl-4-benzoyl-1H-pyrazol-5(4H)-one; Shen & Yuan
2004] and shorter than the M—O bond lengths in [Mn(dca)2(DMF)2]n [Mn—O = 2.199 (2) Å] and in [Co(dca)2(DMF)2]n
[Co—O = 2.096 (2) Å].
The dicyanamide (dca) ligand adopts an end-to-end coordination mode. Two dca ions link two nickel(II) atoms to form
a 12-membered Ni(dca)2Ni ring and the neighboring rings share the nickel(II) atoms to form a chain of [Ni(dca)2]n. The
chains are linear, the Ni(dca)2Ni rings being in a slight chair conformation.
Free dicyanamide (dca) ligand possesses C2v symmetry. The dca ligand in (I) also adopts C2v symmetry with a nitrile C≡
N bond length of 1.1545 (18) Å for N1≡C1, showing the triple-bond character. The bond angle related to the amide N
atom, C1—N2—C1(x, 1 - y, z), is 118.61 (16)°, corresponding to an amide N atom with an sp2 hybrid orbital; tha related
supporting information
sup-2
Acta Cryst. (2005). E61, m1937–m1939
The chains propagate parallel to the crystallographic b axis, the Ni···Ni distance along the chain being equal to the b cell
length 7.3166 (7) Å. The chains interdigate such that each DMF ligand lies between two DMF ligands of an adjacent
chain, with a shortest Ni···Ni interchain distance of 7.628 (2) Å (Fig. 3). Adjacent chains are held together by a weak C—
H···N hydrogen bond, forming layers parallel to the ab plane (Fig. 2 and Table 2).
S2. Experimental
An aqueous solution (10 ml) of Ni(NO3)2·6H2O (0.146 g, 0.5 mmol) was added to a DMF solution (10 ml) of Na[N(CN)2]
(0.090 g, 1.0 mmol). Slow evaporation of the resulting mixture led to green crystals suitable for X-ray diffraction
analysis. Analysis found: C 35.53, H 4.12, N 33.31%; calculated for C10H14N8NiO2: C 35.64, H 4.19, N 33.26%.
S3. Refinement
H atoms were found in a difference Fourier map and refined with bond-length restraints of C—H = 0.95 (1) Å for the
[image:5.610.128.477.263.451.2]methyl groups and the H···H distance restrained to 1.50 (1) Å. One of two independent H atoms lies on the mirror plane.
Figure 1
The coordination geometry of the NiII atom in (I), with displacement ellipsoids drawn at the 50% probability level.
supporting information
sup-3
[image:6.610.135.473.71.308.2]Acta Cryst. (2005). E61, m1937–m1939
Figure 2
The packing in (I), showing the C—H···N hydrogen-bond interactions as dashed lines.
catena-Poly[[bis(N,N-dimethylformamide-κO)nickel(II)]-di- µ-1,5-dicyanamido-κN1:κN5]
Crystal data
[Ni(C2N3)2(C3H7NO)2]
Mr = 336.98 Monoclinic, C2/m
Hall symbol: -C 2y
a = 13.3866 (17) Å
b = 7.3166 (7) Å
c = 8.0595 (10) Å
β = 112.503 (3)°
V = 729.28 (15) Å3
Z = 2
F(000) = 348
Dx = 1.535 Mg m−3
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 1804 reflections
θ = 3.2–27.5°
µ = 1.35 mm−1
T = 193 K Block, blue
0.40 × 0.21 × 0.20 mm
Data collection
Rigaku Mercury CCD diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
ω scans
Absorption correction: multi-scan (Jacobson, 1998)
Tmin = 0.646, Tmax = 0.774
4029 measured reflections 900 independent reflections 887 reflections with I > 2σ(I)
Rint = 0.018
θmax = 27.5°, θmin = 3.2°
h = −17→15
k = −8→9
l = −10→10
Refinement
Refinement on F2 Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.023
wR(F2) = 0.062
S = 1.03 900 reflections
76 parameters 9 restraints
Primary atom site location: structure-invariant direct methods
supporting information
sup-4
Acta Cryst. (2005). E61, m1937–m1939
Hydrogen site location: inferred from neighbouring sites
H atoms treated by a mixture of independent and constrained refinement
w = 1/[σ2(F
o2) + (0.0436P)2 + 0.3824P] where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001 Δρmax = 0.15 e Å−3 Δρmin = −0.47 e Å−3
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
Ni1 0.5000 0.5000 0.5000 0.01621 (13)
O1 0.58082 (11) 0.5000 0.77586 (18) 0.0230 (3)
N1 0.59942 (9) 0.70410 (15) 0.47063 (15) 0.0231 (3)
N2 0.63339 (15) 1.0000 0.3571 (2) 0.0255 (4)
N3 0.73719 (13) 0.5000 1.0204 (2) 0.0256 (4)
C1 0.61394 (9) 0.84635 (18) 0.42201 (16) 0.0181 (3)
C2 0.68128 (15) 0.5000 0.8455 (2) 0.0210 (4)
H2 0.7242 (17) 0.5000 0.774 (3) 0.026 (6)*
C3 0.6836 (2) 0.5000 1.1460 (3) 0.0541 (8)
H3A 0.7060 (14) 0.6031 (7) 1.217 (2) 0.075 (8)*
H3B 0.6084 (9) 0.5000 1.089 (4) 0.073 (11)*
C4 0.85511 (17) 0.5000 1.0956 (3) 0.0320 (5)
H4A 0.8843 (13) 0.6028 (7) 1.1684 (16) 0.044 (6)*
H4B 0.882 (2) 0.5000 1.004 (2) 0.042 (8)*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
Ni1 0.01952 (19) 0.01301 (18) 0.01555 (18) 0.000 0.00610 (13) 0.000
O1 0.0218 (6) 0.0294 (7) 0.0161 (6) 0.000 0.0052 (5) 0.000
N1 0.0268 (6) 0.0182 (5) 0.0264 (6) −0.0022 (4) 0.0127 (5) −0.0017 (4)
N2 0.0383 (9) 0.0164 (7) 0.0318 (9) 0.000 0.0245 (8) 0.000
N3 0.0235 (8) 0.0343 (9) 0.0171 (7) 0.000 0.0057 (6) 0.000
C1 0.0173 (6) 0.0192 (6) 0.0190 (6) 0.0000 (5) 0.0084 (5) −0.0038 (5)
C2 0.0242 (9) 0.0206 (8) 0.0182 (8) 0.000 0.0082 (7) 0.000
C3 0.0369 (13) 0.108 (3) 0.0189 (10) 0.000 0.0127 (9) 0.000
supporting information
sup-5
Acta Cryst. (2005). E61, m1937–m1939
Geometric parameters (Å, º)
Ni1—O1 2.0670 (13) N2—C1 1.3074 (15)
Ni1—O1i 2.0670 (13) N3—C2 1.319 (2)
Ni1—N1 2.0733 (11) N3—C3 1.448 (3)
Ni1—N1i 2.0733 (11) N3—C4 1.458 (3)
Ni1—N1ii 2.0733 (11) C2—H2 0.954 (10)
Ni1—N1iii 2.0733 (11) C3—H3A 0.923 (8)
O1—C2 1.243 (2) C3—H3B 0.933 (10)
N1—C1 1.1545 (18) C4—H4A 0.942 (8)
N2—C1iv 1.3074 (15) C4—H4B 0.938 (9)
O1—Ni1—O1i 180.0 C1—N1—Ni1 152.49 (11)
O1—Ni1—N1 91.61 (4) C1iv—N2—C1 118.61 (16)
O1i—Ni1—N1 88.39 (4) C2—N3—C3 121.12 (18)
O1—Ni1—N1i 88.39 (4) C2—N3—C4 121.70 (17)
O1i—Ni1—N1i 91.61 (4) C3—N3—C4 117.18 (18)
N1—Ni1—N1i 180.0 N1—C1—N2 174.95 (13)
O1—Ni1—N1ii 91.61 (4) O1—C2—N3 123.76 (17)
O1i—Ni1—N1ii 88.39 (4) O1—C2—H2 121.7 (15)
N1—Ni1—N1ii 92.16 (6) N3—C2—H2 114.6 (15)
N1i—Ni1—N1ii 87.84 (6) N3—C3—H3A 107.4 (14)
O1—Ni1—N1iii 88.39 (4) N3—C3—H3B 113 (2)
O1i—Ni1—N1iii 91.61 (4) H3A—C3—H3B 109.9 (11)
N1—Ni1—N1iii 87.84 (6) N3—C4—H4A 112.6 (10)
N1i—Ni1—N1iii 92.16 (6) N3—C4—H4B 110.5 (16)
N1ii—Ni1—N1iii 180.00 (6) H4A—C4—H4B 107.4 (10)
C2—O1—Ni1 121.06 (12)
N1—Ni1—O1—C2 −46.10 (3) N1ii—Ni1—N1—C1 145.1 (2)
N1i—Ni1—O1—C2 133.90 (3) N1iii—Ni1—N1—C1 −34.9 (2)
N1ii—Ni1—O1—C2 46.10 (3) Ni1—O1—C2—N3 180.0
N1iii—Ni1—O1—C2 −133.90 (3) C3—N3—C2—O1 0.0
O1—Ni1—N1—C1 −123.3 (2) C4—N3—C2—O1 180.0
O1i—Ni1—N1—C1 56.7 (2)
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, −y+1, z; (iii) −x+1, y, −z+1; (iv) x, −y+2, z.
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
C2—H2···N2v 0.95 (1) 2.51 (1) 3.453 (2) 169 (2)