International Library for Thesis
Indexed on: October, 2014All Rights Reserved with International Library for Thesis UBN: 015-A94510112005
Photon Research Award-2014
in
Microbiology
CHARACTERIZATION OF PASTEURELLA MICROORGANISMS
ISOLATED FROM RABBITS BY USING OF RANDOM AMPLIFIED
POLYMORPHIC DNA [RAPD] MARKERS
CHARACTERIZATION OF PASTEURELLA
MICROORGANISMS ISOLATED FROM RABBITS
BY
USING OF RANDOM AMPLIFIED
POLYMORPHIC DNA
[RAPD] MARKERS
BY
EL-Sayed AL-Saied Masoud Hussein
M.V.Sc. Zagazig University [ 2000 ]Under The Supervision Of
Dr
Ashraf Mohamed Abbas
Senior Researcher of Biotechnology Unit. Vet. Serum and Vaccine Res. Inst. Abbasia, CairoA THESIS
Submitted to Zagazig University for
The Degree of Doctor of Philosophy of Veterinary Medical Science .
Dept. of Bacteriology, Mycology and Immunology [ Bacteriology ]
2004
Zagazig UniversityFaculty of Vet. Med.
Department of Bacteriology, Mycology and Immunology
Prof. Dr.
Y. Abd El- Galil Ahmed
Prof. of Microbiology Faculty of Veterinary Medicine
Zagazig University
Prof. Dr.
N.E. Amin Mohamed
Prof. of Microbiology Faculty of Veterinary Medicine
II
ACKNOWLEDGEMENT
First of all, thanks to (GOD) who made me able to accomplish this work.
I am deeply grateful to Prof. Dr. Nour EL-Din Amin, Professor of Microbiology, Fac. of Vet. Med. Zagazig University for his stimulating supervision, kind guidance, heartily encouragement, valuable help and continuous interest.
- I would like to express my thanks to Prof. Dr. Y. Abdel Galil Ahmed, Professor of Microbiology, Fac. of Vet. Med, Zagazig University for his guidance, supervision and encouragement in this work . - I would like to express my sincere gratitude for the kindness and
encouragement of Dr. Ashraf Mohamed Abbas, Senior Researcher of Biotechnology Unit. Vet. Sera and Vaccines Research Institute, Abbasia, Cairo. For his valuable guidance, help, advice, facilities throughout this work .
- I wish to express my thanks for Dr. Eman EL-Rawy, Senior Researcher of Microbiology, Vet. Sera and Vaccines Research Institute , Abbasia, Cairo.
- Great thanks to Prof. Dr. Seham Abdel-Rasheed, Head of Biotechnology Unit, Vet. Sera and Vaccines Research Institute, Abbasia, Cairo .
- Also great thanks to all Staff Members of Microbiology Department, Fac. of Vet. Med, Zagazig University for their efforts during this thesis .
III
CONTENTS
S. No
Subject
Page No.
1
Introduction
1
2
Review of literature
4
I- Incidence of Past. microorganisms in rabbits
II-Bacterial isolation and identification
III- Methods of diagnosis of Past. microorganisms
1-Biochemical methods of identification 2- Animal pathogenicity
3- Serological diagnosis
4- Random amplified polymorphic DNA
(RAPD) analysis forPast. multocida isolates
3 7 11 11 15 16 21
3
Material and Methods
28
MATERIALS I-Samples II- Media used
III- Reagents and solutions IV- Stains used
V- Standard strains of Past. multocida VI- Laboratory animals
VII- Chemicals, reagents and buffers used in biomolecular studies of Past. multocida.
VIII- Equipments and apparatuses used in biomolecular studies of Past. multocida strains.
28 28 31 32 34 34 35 35 41
IV
B- METHODS ... I- Collection of samples for bacteriological
examination ... II- Bacteriological examination ... III- Serotyping of Past. microorganisms ... IV- Random amplified polymorphic DNA (RAPD) analysis for Past. multocida isolates. ...
43
43 43 46
49
4
Results
54
1- Results of isolation and prevalence of Past. microorganisms from rabbits. ...
2- The results of physiological characters and biochemical reactions of isolated Past, microorganisms. ... 3- Results of pathogenicity test ... 4- Results of serological typing of Past. multocida
isolates... ...
5- Results of biomolecular studies on Past.
multocida strains isolated from rabbits. ...
54
57 60
62
67
5
Discussion
81
6
Summary
90
7
Conclusion
93
8
References
94
9
Vita
V
LIST OF TABLES
Title
Page
No.
1
Samples collected from apparently healthy rabbitsfrom different localities in Sharkia Governorate
29
2
Samples collected from diseased rabbits fromdifferentlocalities in Sharkia Governorate
30
3
10 - mer oligoneucleotides were used as primers in theRAPDassay
39
4
Chemicals used in one RAPD-PCR reaction40
5
Different characteristics of Past. Species45
6
Prevalence of Past. microorganisms from nasal swabs of apparently healthy rabbits in different Sharkia localities
55
7
Prevalence of Past, microorganisms from nasal swabsof diseased rabbits indifferent Sharkia localities.
56
8
Cultural and biochemical characters of isolatedPast. microorganisms ( 36 isolates ).
58
9
Fermentative ability of Past, microorganisms isolatedfromrabbits (36 isolates ).
59
10
Serological typing of Past. multocida isolated fromrabbits.
63
11
RAPD profile of Past. multocida isolates generated byprimer O!0 "TCAGAGCGCC "
VI
Title
Page
No.
12
RAPD profile of Past. multocida isolates generated byprimer DM " AGCGCCATTG "
73
13
RAPD profile of Past. multocida isolates generated byprimer C4 " CCGCATCTAC "
74
14
RAPD profile of Past. multocida isolates generated byprimer A4 " AATCGGGCTG "
75
15
RAPD profile of Past. multocida isolates generated byprimer D4 " TCTGGTGAGG "
76
16
Similarity matrix among the 14 Past. multocida isolatesVII
LIST OF FIGURES
Title Page No.
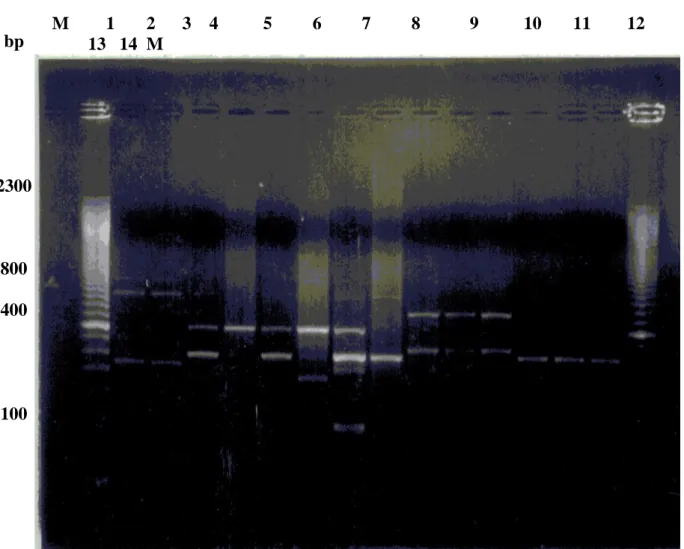
1 - Bipolarity of Past. multocida 61
2 - Genomic DNA of Past. multocida strains extracted by
CTABmethod 65
3 RAPD profiles of Past. multocida genomic DNA
isolatesamplified by primer do. 67
4 RAPD profiles of Past. multocida genomic DNA isolates
amplified by primer D11
68
5 RAPD profiles of Past. multocida genomic DNA
isolatesamplified by primer C4
69
6 RAPD profiles of Past. multocida genomic DNA
isolatesamplified by primer A4
70
7 RAPD profiles of Past. multocida genomic DNA
isolatesamplified by primer D4
71
8 UPGMA clustering dendrogram of. Past. multocida
VIII
List of Abbreviations
Word Abbreviation
Amplified Fragment Length polymorphism
AFLP
Agar gell Precipitin test
AGPT
Arbitrarly primed polymerase chain reaction AP-PCR
Amplified ribosomal DNA restriction analysis ARDRA
Base pair
Bp
Bordetella .
Bord
Hexacetyl trimethyl ammonium bromide .
CTAB
Deoxyribonucleic acid .
DNA
Enzyme linked immune - sorbent assay .
ELISA
Gell diffusion precipitation test.
GDPT
Indirect haemagglutination test.
IHAT
Mole .
M
Mycoplasma gallisepticum .
MG
Milli mole .
mM
Methyl red .
M.R.
Optical density .
OD
Pasteurella .
Past
Polymerase chain reaction .
PCR
Porcine dermatitis and nephropathy syndrome .
PDNS
Pulsed field gel electrophoresis .
PFGE
Random amplified polymorphic DNA .
IX
Restriction fragment length polymorphism .
RFLP
Subcutaneous .
S/C
Word Abbreviation
Sterile double distilled water .
S.D.D. H2O
Sodium dodecyl sulphate.
SDS
Subspecies,
SSp
Un weighted pair group method with arithmetic avarage .
UPGMA
Voges Proskauer .
INTRODUCTION
Rabbits are raised for a variety of purposes, including their use in the laboratory, meat and fur production[OKerman 1988 ].
Pasteurellosis is one of the most important bacterial diseases affecting rabbits allover the world causing severe economic losses. Affected rabbits may have signs of rhinitis [snuffles], pneumonia, otitis media or interna, abscesses and genitourinary tract infections [Sharon et al, 1996]
The polymerase chain reaction [PCR] represents a technique that is revolutionizing genetic biochemistry. This technique is used in molecular medicine, medical diagnostics, forensics, and genetic engineering [Roskoski, 1996].
DNA markers that are shown to be genetically linked to a trait of interest can be used for diagnosis of most diseases [Landegren et al., 1988]. Restriction fragment length polymorphism [RPLP] as a DNA marker is a powerful tool for the estimation of phylogenetic relationship of organisms [Botstein et al., 1980]. However detection of RPLPs by DNA hybridization technique is laborious and incompatible with high analytical throughput required for many applications
(Beckmann,1988).
It was evident that genomic comparison of Past. multocida by ribotyping and field alteration gel electrophoresis were beneficial in distinguishing isolates causing pasteurellosis, but these techniques were unable to illustrate pasteurella specific fragments or profiles [Snipes et al,l990 and Townsendet al.t 1997].
Recently a sensitive new polymerase chain reaction [PCR] based method of DNA fingerprinting, also called the randomly amplified polymorphic DNA [RAPD] or the arbitrarily primed PCR[AP-PCR] method , has been used to study heterogenicity in closely related organisms [Williams et al., 1990 and Welsh et al.,1991]. This method detects differences in the DNA sequence at sites in the genome that are defined by the single arbitrary primers [Williams et al., 1990].
Sequence variation is revealed by the number and length of amplified products which may be phylogenetically conserved [Caetatte -Anolles et al .,1991].
These primers detect polymorphism in the absence of specific information and can be used to construct genetic maps in a variety of species [Williams et al., 1990 ] .
This work was planned as an attempt to throw spot lights on the following items :
1- Isolation of pasteurella microorganisms in both apparently healthy and diseased rabbits .
2- Identification of isolated microorganisms on the basis of their biochemical activities .
3- Studying the experimental infection of isolated microorganisms . 4- Serotyping of the isolated microorganisms.
5- Identification of the genetic variability among Past. multocida isolates by using RAPD technique .
REVIEW OF LITERATURE
The available literature could be discussed under the following
categories .
I-
Incidence of Past. microorganisms in rabbits
:-Flatt and Dungworth [1971] mentioned that 20% of 8-10 weeks old rabbits showed gross lesions of an enzootic pneumonia. They stated that the most prevalent clinical signs included dyspnea and dried yellowish exudate on the external naris besides loss of condition .
Fox et al., [1971] stated that pasteurellosis in rabbits is not restricted to the respiratory form but caused otitis media, otitis interna and metritis. Moreover suppurative meningoencephalitis occurred as a result of extension of otitis interna infection. They added that out of 91 cases of otitis media, 85 [93%] were Past. multocida positive .
John and Joseph [1973] mentioned that Past. multocida caused major respiratory diseases of rabbits. Macroscopically, there were visceral congestion and haemorrhage as well as sangumineous nasal discharges. They also added that bronchopneumonia and fibrinopurulent or mucopurulent reactions were seen in the meninges, brain, middle and inner ear besides the thoracic and abdominal viscera.
Plant [1974] stated that s/c abscesses, pneumonia, otitis media, genital infection, conjunctivitis were commonly seen with cases of rabbit snuffles .
Zaher et al, [1976] isolated Past. multocida type III from 79% of diseased examined cases. They indicated that diseased rabbits showed respiratory signs, dull eyes, diarrhoea, emaciation, rough fur and conjunctival exudate. In the same time they mentioned that death was proceeded by cyanosis of the ears, eyes and slow gasping respiration.
Lu et al, [1978] stated that Past. multocida colonize the mucosal surfaces of the pharynges of rabbits in the carrier state with recurrent rhinitis .
Himi and Philip [1979] reported that all ages of rabbits were susceptible to Past. multocida infection .
Patton et al., [1980] recorded that it is quite difficult to infect rabbits with Past. multocida unless there is a high .ammonia in the air. In low ammonia environment million of Past. multocida can be sprayed intranasally with no infection resulting, but with high ammonia concentration, infection with Past. multocida occurred .
Scharf et al., [1981] revealed that bacterial culture data indicated 17% of infectivity rate was present when Pasteurella free rabbits were housed in a conventional manner for 3-6 months prior to institution of the modified barrier.
Digiacomo et al., [1983] reported that a number of clinical forms of infection can occur in pasteurellosis including upper respiratory infection (snuffles), otitis media, enzootic pneumonia, conjunctivitis, pyometra, orchitis, abscesses and septicaemia. They also reported that
23% of rabbits at 22 weeks of age had Past. multocida and in adult rabbits the percentage of infection was 72% . They also revealed that the naris of rabbits were not colonized before weaning at 8 weeks of age and detected that the earliest nasal infection was present at 12 weeks of age .
Kpodenkon [1983] stated that Past. multocida is a cause of considerable economic losses in rabbits and complication affecting the ears and brain are becoming increasingly important.
Eschborn [1985] stated that rabbits were much more sensitive to air quality (such as relative humidity, concentration of noxious gases and air movement) than to temperature. High relative humidity, linked with a high concentration of noxious gases ( especially ammonia ) and high air speed in the vicinity of rabbits, greatly favor diseases of the respiratory system particularly contagious snuffles .
Arrington and Kelley [1986] stated that snuffles may complicate to s/c abscesses , weepy eyes and wry neck .
Coudert et at., [1986] showed that most of rabbits infected with Past. multocida showed rhinitis, abscesses, sterility, otitis media and emaciation.
Cheeke [1987] stated that most respiratory diseases in rabbits were caused by Past. multocida in the presence of high ammonia level with high relative humidity in the rabbitary .
Okerman [1988] revealed that the possible complications of chronic snuffles included pneumonia, metritis, otitis media and interna,
pericarditis and s/c abscesses. He added that morbidity and mortality depends upon climatological conditions .
Digiacomo et al., [1989] detected turbinate atrophy in 77% of rabbits with Past. multpcida infection only, 71 % of rabbits with concurrent Past. multocida and Bord. bronchiseptica infections, and 6% of rabbits with Bord. bronchiseptica infection only.
Deeb et al., [1990] recorded that at weaning, about 25% of rabbits had nasal infection with Past. multocida and 75% had infection with Bord. bronchiseptica. They proved that the proportion of rabbits with both infections increased with age. They also stated that rabbits about 10 month old, 75% with Past. multocida or Past. multocida and Bord. bronchiseptica infection had upper respiratory tract.
Kawamoto et al., [1990] suggested that the Past. multocida isolates were the causal agent of rabbits rhinitis ( snuffles ) in Japan .
Rendondo et al., [1990] found that 3% of normal rabbits were carrier of Past. multocida.
Mahmoud and Abdel - Basel [1991] performed a study on 4 rabbit flocks which included 85, 65, 60 and 700 does rabbits respectively. The does were housed in individual metal hutches and were fed on commercial rabbit pellets and watered by dew drop system. They found that the mortality rate due to Past. multocida infection was high and ranged from 70 - 78 % in the investigated flocks.
Gergis et al., [1992] stated that pneumonia in domesticated rabbits regularly caused clinical manifestations and death with an incidence ranged from 51-59%.
Merrier [1992] showed that the carrier rate of Past. multocida among normal rabbits in the upper respiratory tract was in between 2-3.9 %.
Digiacomo et al., [1993] stated that naturally acquired turbinate atrophy in rabbits was associated with Past. multocida infection.
Dancla et el., [1996] stated that internal otitis, pneumonia, mastitis and metritis were commonly seen with cases of rabbit pasteurellosis.
Grossmann et al., [1998] found that the majority of isolates from rabbits ( 88% ) belonged to type P6, which was not found in strains isolates from cattle or pigs .
II-
Bacterial isolation and identification :
Hagen [1958] reported that out 857 young rabbits 35 developed respiratory infection and died . He added that Past. multocida was isolated from 54% of the cultured lungs of these dead rabbits .
Georgi et al., [1965] isolated Past. multocida from heart blood, lungs and abscesses during postmortem examination of 12 rabbits and clinical examination of 30 rabbits with respiratory involvement.
Damm and Ramisz [l967] carried out bacteriological examination of 64 cases in farm rabbits in an epidemic of pasteurellosis and isolated Past. multocida from 41 ( 64% ) cases out of them 34 ( 83% ) were septicemic while 7 ( 17% ) were pulmonary form of pasteurellosis.
Sato et al, [1968] isolated Past. multocida strains from adult domestic rabbits with respiratory diseases.
Mushin and Schoenbaum [1980] isolated Past. multocida from 4 conventional rabbit colonies . They found that amongst 51 isolates, 48 (94% ) belonged to a group of strains with similar morphological, cultural, biochemical and antigenic characteristics .
Bayoumi et al., [1984] experimentally infected 25 rabbits with
Past. multocida alone or with Diplococcus pneumoniae. E. coli or Bord. bronchiseptica. Then they noticed symptoms, performed postmortem examination and isolated microorganisms. They reported that such bacterial agents did not only play an important role in initiating rabbit pasteurellosis but also shared in the severity of infection .
Fahmy et aL., [1985] carried out a study on 120 dead and live Bauscat rabbits at Sharkia Governorate. They isolated Past. multocida from 33.3% of the investigated rabbits 40 - 60 days old .
Nakagawa et al, [1986] reported that Past. multocida was isolated from the para nasal sinuses of 53 out of 54 infected rabbits. They added that Past. multocida was also isolated from trachea, and middle and inner ear of half of the infected rabbits and occasionally from the conjunctiva ,lung and heart.
Hago et al., [1987] stated that the frequency of isolation of Past. multocida from domestic rabbit colony in Sudan was 50% from affected joints.
Rai et al., [1987] examined samples of rabbit nasal and vaginal swabs collected from 81 healthy rabbits, and they isolated Past. multocida from 56.8% and 49% of examined vaginal and nasal samples respectively .
Percy et a/., [1988] isolated Past. multocida from dead rabbits showing various pathological lesions with an incidence of 11.9 % .
Glass and Beasley [1989] recorded that Past. multocida was isolated from all ages of rabbits. They proved that sinuses were mostly colonized, followed by trachea, middle ears and lungs. They also reported that no bacteria were isolated from the liver .
Elged et al., [1990] collected 185 samples from rabbits showing signs of pneumonia, rhinitis, otitis and osteoarthritis and suspected to be suffered from pasteurellosis at different private farms of rabbits at Sharkia Governorate. They indicated that the bacteriological examination of the collected samples revealed that incidence of Past. multocida was 8.8, 11 and 5% in rabbits suffering from respiratory signs, joint abscesses and otitis respectively. More over they examined 80 nasal swabs from apparently healthy adult rabbits and they revealed that Past. multocida was the only microorganism isolated with an incidence of 3.8 % .
Olah et al., [1990] proved that 215 rabbits died in one large rabbit farm ( 1200 breeding animals ) and 20 small farms in Yugoslavia showed respiratory diseases was the cause of 38.5 - 52.1% of the
mortality. They proved that Past. multocida was isolated from 42 cases, Bord. bronchiseptica from 5 cases, Staphylococcus pyogenes from two cases and alpha - haemolytic Streptococcus from two cases .
Digiacomo et al., [1993] obtained 10 isolates of Past. multocida serotype A : 12 from adult New Zealand white rabbits with non induced atrophic rhinitis. They stated that naturally acquired turbinate atrophy in rabbits was associated with Past. multocida infection .
Ashgan and Bayoumi [1994] examined 200 diseased and dead rabbits bacteriologically at Assuit Governorate. They found that Past. multocida was isolated from 50 samples in mixed infection. The associated microorganisms were E. coli, Staphvlococcus aureus and Bordetella bronchiseptica and were isolated from heart, liver, lungs and kidneys of the affected rabbits. The incidence of associated microorganisms in mixed infection was Past. multocida + Staph. aureus + Bord. bronchiseptica 14 (28% ), Past. multocida + Staph. aureus + E . coli 12 ( 24% ), Past. multocida + Bord . bronchiseptica 9 ( 18% ), Past. multocida + Bord. bronchiseptica + E . coli 7 ( 14 % ) .
Nada [1994] examined 239 rabbits nasal swabs including 53 apparently healthy rabbits and 186 with clinical signs of respiratory disease and rhinitis . He isolated Past. multocida from 17 ( 9.1 % ) of 186 affected rabbits and 2 ( 3.8 % ) of the 53 healthy rabbits .
Dancla et al, [1996] isolated 41 strains of Past. multocida from rabbits flocks belonging to 16 breeders. Strains were generally isolated from the middle ear of breeding does suffering from internal otitis, with
some strains being obtained from other diseases such as pneumonia, mastitis or metritis .
Al-Haddawi et al., [1999J assayed forty isolates of Past. multocida from healthy (17 isolates) and diseased (23 isolates) rabbits for the presence of plasmids in seeking to determine whether any correlation exists between the presence of plasmids and health status .
III - Methods of diagnosis of pasteurella microorganisms :
I- Biochemical methods
of identification^
Khalifa [1934] grouped 49 strains of Pasteurella microorganisms isolated from a number of animals into 3 types on the chemical basis into group A, B and C .
Brigham and Rettger [1935J reported that all past. strains produced indole and fermented glucose , sucrose and mannite with the production of acid only while M.R. and V.P. tests were negative .
Merchant and Packeer [1956] proposed to classify Past, multocida by using urease activity and agglutination of pigeon red cells. They added that twenty eight of 31 strains were urease negative and sera of 14 strains agglutinated pigeon red cells .
Smith [1958] classified 250 cultures of Past. multocida on the basis of their ability to ferment maltose trehalose and dextrin with production of acid only .
Heddleston et al., [1972] observed that Past. multocida fermented maltose and xylose with production of acid only .
Daotrong et al., [1973] examined 98 strains of Past. multocida. They found that 43 isolates produced H2S and 16 isolates that did not
form indole.
EL-Ghawas [1980] reviewed the biochemical characters of 18 isolates of Past. multocida isolated from septicemic cases of domestic animals. She found that all isolates were indole positive and non motile, 14 isolates fermented arabinose but did not ferment both xylose and dulcitol.
Gilmour [1980] mentioned that Past. haemolytica could be differentiated from Past. multocida by haemolysis on 7% sheep blood agar plates whereas Past. multocida were not haemolytic. Also he found that Past. haemolytica did not produce indole and grow onto MacConkey's agar.
Biberstein [1990] reviewed that Past. multocida were positive for indole, producing no haemolysis on blood agar, not grow onto MacConkey's agar and urease utilization was negative, while Past, haemolvtica differed in reaction for indole, haemolysis on blood agar and grow on MacConkey's agar. He added that all were oxidase and nitrate positive and produced acid throughout triple sugar iron agar but no H2S .
Kawamoto et al., [1990] reported that the biochemical properties of the isolated Past. multocida from rabbits in Japan were identical except for indole production and rafinose fermentation .
Digiacomo et al., [1991] recorded that two major strains of Past. multocida were detected in the colony. One strain had mucoid colonies, fermented few carbohydrates and was serotype A:5, where the other
strain had smooth iridescent colonies, non typeable capsular antigen, type 3 somatic antigen and fermented more than twice as many carbohydrates.
EL-Battrawy et al, [1992] isolated 56 strains of Past, microorganisms from cattle and buffaloes. They found that all isolated strains were positive to indole test and negative to M.R. and V.P tests. They added that all strains fermented glucose, galactose and sorbitol.
Unchitti et al., [1992] compiled 70 isolates of Past. multocida. 17 human, 23 swine and 30 poultry. All isolates produced catalase, oxidase, indole and nitrate reduction. They failed to produce urease, gelatinase, methyl red and could not grow on MacConkey's agar, SS-agar. They found that all isolates produced acid from glucose, mannitol and mannose. They also recorded that, none of the isolates fermented lactose, maltose and dulcitol. They added that marked variation in the patterns of fermentation of arabinose and xylose were found .
Mokhtar [1997] isolated 13 strains of Past. multocida from calves. He found that all strains were indole positive and fermented glucose, xylose and sorbitol but not ferment maltose,
Schuur et al., [1997] cultured Past. multocida from the cerebrospinal fluid [ CSF ] of a - 4- month - old infant who presented with meningitis. The patient had been scratched on the head by a cat. Culture of the cat's claws also yielded Past. multocida. They found that both isolates produced acid from mannose, mannitol, sucrose, sorbitol, trehalose, xylose and glucose. Also they recorded that both isolates
produced indole, reduced nitrate and were positive for decarboxylation of ornithine and both were negative for hydrolysis of urea .
Petersen et al, [1998] characterized fifteen isolates tentatively classified as maltose positive Past. multocida in 79 biochemical tests. They found that two strains were classified as maltose positive Past. multocida ssp. multocida while six strains were classified as maltose positive Past. multocida ssp. septica. The remaining strains clustered with Past. volantium and Past. gallinarum.
Abdel-Rahman [1999]: found that all isolates of Past. multocida recovered from calves were positive to oxidase, catalase and indole tests, while were negative to urease test. On triple sugar iron agar the isolates produced H2S and fermented sucrose, glucose. He added that all the
isolates did not ferment lactose and trehalose. He noticed that the fermentation activity of the isolates were manifested by acid production without gas.
Dziva et aL, [2001] subjected eighty-one isolates presumptively identified as Past. multocida from a variety of diseases in animals in Zimbabwe to biochemical characterization. A total of 46 tests were used in the characterization of the isolates and subsequent assigning to the taxa of the genus Pasteurella. They concluded that three of the isolates were found not to be Past. multocida but they belonged to Past. canis [ one isolate ] and Past. stomatis [ two isolates ]. The majority of the isolates [65/78] could be classified into conventional taxa of the genus Pasteurella, and these were mainly from classical pasteurellosis [haemorrhagic septicaemia, pneumonia, fowl cholera ...]. Most of the isolates belonged to the taxta Past. multocida subsp. multocida and Past.
multocida subsp. septica. They added that 16/78 isolates could not be assigned to the known taxa, and these were designated " unassigned biotypes ". Also they added that dogs and cats isolates presented a greater percentage (75%) of these unassigned biotypes.
2- Animal pathogenicity:-
Carter [1967] inoculated mice subcutaneously with 0.l ml nutrient broth incubated at 37 C° for 6-8 hours. He showed that subcutaneous oedema, congestion of all parenchymatous organs and enlarged spleen in dead mice .
Fayed [1973] observed that Past. multocida recovered from apparently healthy and pneumonic lungs of cattle vary in their pathogenicity to mice, some were highly virulent and the others were of low virulence .
Sharma et al. [1979] tested 21 isolates of Past. multocida recovered from calves for their pathogenicity in mice. They observed that all mice died 24-48 hours after I/P injection of 0.5 ml of 6 hours broth culture.
Ungureanu et al, [1980] tested 176 strains of Past. multocida and 125 strains of Past. haemolytica, for their pathogenicity to mice. They observed that a total of 82% of Past. multocida. and 19% of Past. haemolytica strains were pathogenic for mice.
Magda [1998] observed that Past. multocida recovered from apparently healthy and diseased camels vary in their pathogenicity to mice, as Past. multocida B:2 were highly virulent to mice with a mean
death time 24 hours with postmortem findings of septicaemia, but the untypable strains were of less virulent, causing death of mice within 72 hours.
Abdel-Rahman [1999] proved to pathogenicity of 13 field isolates of Past. multocida recovered from calves for mice. He observed that eight isolates were highly pathogenic for mice causing its death within 18-24 hours while the other five isolates were of intermediate pathogenicity to mice and killed it within 36-72 hours.
3- Serological diagnosis
:-Cornelius [1929] grouped Past. multocida by using agglutination test into four capsular groups VIZ: (I,II,III and IV) designated by roman letters.
Ochio [1933] divided Past. multocida into four host specific serotypes VIZ:A,B,C and D. He added that type "A" was pathogenic only to chicken and was the causal agent of fowl cholera, while type "B" was host specific of domestic animals including cattle, sheep and swine.
Little and lyon [1943] divided Past. multocida into three types I, II and III by using slide agglutination test.
Robert [1947] classified Past. multocida on the bases of mouse protection test into four immunological types I,II,III and IV .
Carter and Byrne [ 1953] examined 68 strains of Past. multocida from domestic animals in Canada. They found that the serological types of the strains were A,B,C and D, which corresponded to Robert [1947] types 1,11,111 and IV respectively.
Carter [1955] used indirect haemagglutination test for serotyping of Past. multocida on the bases of capsular antigen. He found that the strains were A,B, D and E.
Carter and Rappy [1962] used the formalized erythrocytes in the indirect haemagglutination test for the serological typing of Past. multocida.
Malik [1968] used the indirect haemagglutination and passive protection test in mice to classify 14 strains of Past, organisms obtained from acute fowl cholera in different parts of Chezoslovakia. They proved that most strains [76%] belonged to group A, 8% were group D and the remaining strains could not be classified by these tests.
Heddleston et al., [1972] used gel diffusion precipitin test for serotyping Past. multocida recovered from avian species on the bases of somatic antigen . They recognized 16 serotypes .
Guo et al.,[1980] used the indirect haemagglutination test for serotyping of 10 strains of Past. multocida recovered from calves with pneumonia. They found that all isolates were belonged to Carter's capsular type " B " .
Digiacomo et ai, [1983] examined 29 isolates of Past. multocida from rabbits. They found that 93% of the isolates were somatic type 12.
Holmes et al., [1986 ] utilized an AGPT and an ELISA to detect serum antibodies against Past. multocida in nasal culture negative rabbits.
They proved that the application of an ELISA may be efficacious in identifying apparently healthy nasal cultured negative rabbits as a subclinical carrier of Past. multocida.
Rimler and Rhoades [1987] divided Past. multocida into five capsular groups designated A,B, D,E and F.
Rimler and Rhoades [1989] found that some strains of Past. multocida may represent more than one serotype .
Digiacomo et al., [1990] assayed sera of New Zealand white rabbits for class specific IgM and IgG antibodies by the ELTSA. They found that the IgM response peaked and returned to lower levels within several weeks after infection, whereas the IgG response progressively increased and remained elevated.
Okerman [1993] recorded that the capsular types among Past. multocida strains isolated from rabbits were type A and D. He added that the cause of high virulence of the D - type strain was due to production of a heat labile dermonecrotic toxin. She mentioned that most Past. multocida from rabbits related to serogroups 3, 11 or 12 [Heddleston somatic antigen ].
Kawamoto et al, [1994] evaluated the antibodies to Past. multocida in both naturally and experimentally infected rabbits. They concluded that the ELISA was superior in serodiagnosis of rabbit pasteurellosis to IHAT and GDPT.
Nada [1994] found that the most predominant serotypes of Past. multocida isolated from rabbits were 3:A [42.1%] followed by serotype 12:A with an incidence of [ 31.6%] then serotype 15:D in an incidence of 15.8% .
Borkowska et al., [1996] used an ELISA and gel diffusion precipitin test [GDPT] to determine the antibody levels in serum samples from the immunized rabbit. They concluded that the GDPT was more sensitive than the ELISA but they suggested that these methods did not detect the same antibody .
Dancla et al., [1996] studied 41 strains of Past. multocida isolated from rabbits. They found that 27 strains were serotyped (A3) , 7 (A5) , 4 (A9) and 3 were serotype (A7).
Purdy et al. [1997] examined 50 isolates of Past. multocida from calves. They found that 42 of 50 Past. multocida isolates were positive for capsule type A, while 8 were untypable. They added that five somatic type antigen profiles [ 3;3,4;3,7;3,4,7and 4] were identified among the 50 serotyped Past. multocida , while one isolate was untypable .
Abdel-Rahman [1999] used indirect - haemagglutination test for capsular serotyping and agar gel precipitation test for somatic serotyping of 13 Past. multocida isolates recovered from calves. He found that all isolates were typed as serovar [B:2] either from apparently healthy or diseased calves .
Dziva et al., [2001] subjected 78 isolates of Past. multocida recovered from a variety of diseases in animals to capsular typing. They recorded that 48 isolates were capsular group A, 6 were capsular group B, 9 were capsular group D, 10 were not capsulated, and hence not typable and 5 were not grouped at all. They added that none of the isolates belonged to either serogroup E or F.
Jonas et al., [2001] characterized nine isolates of Past. multocida recovered from field outbreaks in poultry by biotype, capsular type and somatic serotype. They revealed that three isolates were of capsular type A[A:l,3,13;A:l,3;and A:8], two isolates were of type B:2,3 and one isolate was of capsular type F. They added that the capsular group could not be identified for the remaining 3 isolates .
Huber et al., [2002] used gel diffusion precipitin test for serotyping 61 isolates of Past. multocida collected from fatal fowl cholera. They found that 7 isolates were of serotype 1, 7 were of serotype 3,21 were of serotype 3, 4, 6 were of serotype 4, leach was of serotypes 4,7 and 4, 12 and 17 were of serotype 5. They added that even after repeated testing, one isolate was untypable.
Lainson et al,[2002] characterized 66 isolates of Past. multocida from 15 pigs with porcine dermatitis and nephropathy syndrome [PDNS] and 51 pigs without PDNS by capsular and somatic antigen typing . They found that four somatic antigen patterns were present among the 15 PDNS cases. They added that the most prevalent type was 3,4,12[11 of 15] whereas types 3,3,4 and 3,12 occurred less frequently. Also they added that the frequency of somatic antigen types
among PDNS isolates was consistent with that found for non - PDNS isolates. They concluded that with the exception of a single non - PDNS isolate, all of the Past. multocida strains were capsule type A.
4- Random Amplified Polymorphic DNA C RAPD ) analysis for
Past, multocida isolates
.
Williams et al., (1990) revealed that genetic maps consisting of RAPD markers can be obtained more efficiently and with greater marker density than by RFLP or targeted PCR based methods .
Williams et al., (1992) mentioned that the presence of more than one amplification band per reaction gave the random amplified polymorphic DNA (RAPD) technique a distinct advantage over iso-enzymes or restriction fragment length polymorphism (RFLP) for differentiation among the isolates .
Dancla et al., [1996] studied 41 'strains of Past. mulrocida recovered from rabbits for epidemiological purposes by ribotyping and random amplified polymorphic DNA (RAPD) assays. They found that the results of RAPD assays were in accordance with those of ribotyping and validate the use of RAPD assays for epidemiological studies of Past. multocida strains.
Schuur et al., [1997] isolated Past. multocida from the cerebrospinal fluids ( CSF ) of a - 4 month old infant who presented with meningitis. The patient had been scratched on the head by a cat. Culture of the cat's claws also yielded Past. multocida. The isolates had identical biochemical patterns. Analysis of both strains by random amplification of polymorphic DNA (RAPD) and comparison of these strains with Past.
multocida strains isolated from other cats showed that the two strains were identical and completely different from the unrelated isolates. They concluded that RAPD analysis provided strong evidence for the causal relationship between the cat scratch and patient's meningitis. They added that application of this technique may eventually shed more light on the mechanisms by which Past. rnultocida is able to invade the CNS in humans.
Koeleman et al., (1998) compared thirty-one strains of Acinetobacter species , including type strain of the 18 genomic species and 13 clinical isolates by amplified ribosomal DNA restriction analysis (ARDRA), random amplified polymorphic DNA [RAPD] analysis and amplified fragment length polymorphism (AFLP) fingerprinting. They found that the standardized commercially available RAPD kit clearly enabled the discrimination of all Acinetobacter genomic species but showed great polymorphism between isolates of Acinetobacter baumannii.
AL-Haddawi et al., [1999] applied random amplified polymorphic DNA - polymerase chain reaction ( RAPD - PCR ) to subtype forty isolates of Past. multocida from healthy (17 isolates ) and diseased (23 isolates) rabbits. Two single primers were tested for their abilities to generate individual fingerprints by using PCR. They revealed that primer 1 grouped the isolates into 7 profiles, and primer 2 grouped them into 15. They recorded that RAPD - PCR results showed the presence of a wide heterogenicity within Past. multocida isolates. They concluded that RAPD - PCR is an efficient technique to detect the DNA polymorphism and could be used to discriminate Past. multocida of rabbit isolates together with serologic typing .
Charlton et al., [1999] used randomly amplified polymorphic DNA (RAPD) to investigate the molecular epidemiology of 26 Mycoplasma gallisepticum ( MG ) isolates obtained from turkeys. MG isolates were recovered from 5 different companies and 13 ranches. They stated that RAPD analysis of MG isolates within a ranch during an outbreak revealed only a single strain involved in each outbreak. They noticed that RAPD analysis identified an isolate from one ranch with a banding pattern identical to that of the 6/85 vaccine strain, which had been used on that particular ranch . They added that similar RAPD banding patterns of isolates from different ranches within the same company suggested horizontal spread of MG between ranches concluding that the use of two primer sets in RAPD analysis was critical to prevent misinterpretation of relationships between different isolates .
Warner and Oliver [1999] used RAPD - PCR for detecting V. vulnificus , as well as other members of the genus Vibrio. They found that RAPD method clearly differentiated between members of genus Vibrio and between isolates of V.vulnlficus. They added that each V.vulnificus strain produced a unique band pattern, indicating that the members of this species are genetically quite heterogeneous. They noticed that all of Vibrios were found to have amplification products whose sizes were within four common molecular weight ranges, while V. vulnificus strains had an additional two molecular weight range bands in common. Also they recorded that band pattern differences were observed between encapsulated and non-encapsulated isogenic morphotypes of the same strain of V.vulnificus .
Abbas et al., [2000] used random amplified polymorphic DNA (RAPD) markers, as a genetic markers to monitor genetic variability between Past. muttocida vaccinal strains. A set of single , short generic five primers of arbitrary sequence was used in this study. They revealed that a total of 111 reproducible polymorphic DNA fragments were produced by two arbitrary primers. Three common species specific amplified DNA bands were detected in all Past. multocida vaccinal strains. They found that ninety - four percent ( 94% ) and 100% of the total DNA bands, amplified by primer OP-G4 and OP-E3 respectively showed polymorphism. So these two primers can differentiate between Past. multocida vaccinal strains. They concluded that RAPD markers generated sufficient genetic data exploiting sequence differences within Past. multocida vaccinal strains.
Martinez et al., [2000] evaluate the genetic diversity of S. agalactia strains isolated from bovine milk and from asymptomatic women vaginas in Quebec, Canada, by randomly amplified polymorphic DNA ( RAPD ) analysis. They indicated that a total of 185 bovine isolates and 38 human isolates were firstly serotyped then studied by RAPD using 3 primers, designated OPS 11, OPB 17, and OPB 18. They showed that all isolates (of bovine and human origin ) shared 58% similarity. Ninety - four percent of these isolates were clustered in four groups ( I , II , III and IV ) with 70% similarity among them. Three clusters, A ( 48 isolates ), B ( 14 isolates ) and C[32 isolates], with 79 to 80 % similarity were identified within group IV, whereas the three other groups did not present any clusters. Despite some clustering of human isolates, relatively high diversity was seen among them. They recorded that high heterogenicity was observed with the RAPD profiles, not only for field strains belonging to different serotypes but also for those within a given serotype.
Ramasoota et al., [2000] used a standardized- reagents commercial kit for random amplified polymorphic DNA (RAPD) analysis for typing 58 E-coli strains that were recovered from the milk of sows, having coliform mastitis, within a single swine herd in Sweden. They stated that the RAPD analysis was fast, easily performed and required only a nanogram of DNA. They recorded that RAPD analysis is a technique that provided reproducible results for typing of E. coli strains that cause mastitis in sows. They added that all 58 strains can be differentiated by means of the RAPD technique .
Dziva et al., [2001] subjected eighty-one isolates presumptively identified as Past. multocida from a variety of diseases in animals in Zimbabwe to biochemical characterization, capsular typing and RAPD analysis. They stated that although capsular typing offers a relatively fast analysis of Past. multocida, it is shortfalls are well recognized and one of the commonly encountered obstacles is the occurrence of a capsular forms of the bacteria, which consequently become nontypeable. Such problems were not encountered when genotypic methods are employed in the typing of this organism. They showed that no relationship between RAPD pattern and capsular serogroup. They suggested that RAPD analysis is a useful tool for strain differentiation.
Huber et al., [2002] analyzed and compared 61 clinical isolates of Past. multocida recovered from turkeys that died of fowl cholera to M-9 vaccine strain. Genetic analysis of the isolates were done by random amplified polymorphic DNA[RAPD] analysis and amplified fragment length polymorphism [AFLP] fingerprinting. They found that both genetic techniques effectively identified similar subtle genomic differences. They showed that pathogenic isolates from vaccinated
turkeys were more genetically similar to the M-9 vaccine strain than isolates from non vaccinated turkeys. Statistical analysis revealed that this relationship could not have been determined by serotyping alone, demonstrating the value of RAPD and AFLP analysis in the characterization of disease causing strains.
Khoodoo et al. [2002] assessed the genetic diversity of 24 isolates of Salmonella enterica [five chicken and 19 human] by RAPD fingerprinting. They confirmed that some strains of Salmonella isolated from chicken were genetically similar to those isolated from human.
Lainson et al., [2002 ] isolated Past. multocida strains from 15 pigs with PDNS and 51 pigs without PDND. They characterized the isolates by capsule and somatic antigen typing, RAPD typing, and restriction analysis of genomic DNA using pulsed field gel electrophoresis [PFGE], They concluded that capsular, somatic and RAPD typing did not discriminate PDNS isolates. They added that all of the isolates from PDNS cases showed an identical ApaI PFGE restriction pattern. This pattern was also found in a high proportion [36%] of Past. multocida strains isolated from non PDNS cases .
Xia et al., [2002] used 94 RAPD primers of different nucleotide composition to probe the genomic differences between a highly virulent Past. mulocida strain and an attenuated vaccine strain derived from the virulent strain after culturing the latter under increasing temperature for approximately 14,400 generations. They found that the GC content of the vaccine strain was significantly lower than that of the virulent strain. They added that the frequencies of AA,TA and TT dinucleotides were
higher, and those of AT, GC and CG dinucleotides were lower in the vaccine strain than in the virulent strain .
Katsuda et al., [2003] applied RAPD method for molecular typing of 130 Past. haemolytica serotype Al isolates obtained from 13 prefectures in Japan. RAPD analysis revealed four banding patterns [types I-IV] and among 130 isolates 60.7% [79/130] of isolates were RAPD type I. They found that all of the RAPD type I isolates were grouped into clusters A-C by PFGE. They noticed that there was no relationship between molecular typing and geographic origin of these isolates. They indicated that isolates of Past. haemolytica Al strain with various profiles have already spread in Japan and may have caused sporadic infection .
MATERIALS AND METHODS
[ A ]
MATERIALS
(I)- Samples :
(a) - Samples from apparently healthy rabbits :
A total of 157 samples (nasal swabs) were aseptically collected from 157 apparently healthy rabbits of both sexes ( 16 male ) and ( 141 female ) of different age (3 months - to - 2 years) and different weight (2-5 kg) from different localities in Sharkia Governorate. From San- EL-Hagar (97 samples), Abu-Hammad (10 samples), Kafr - Sakr (16 samples), Awlad – Sakr (12 samples), Fakous (9 samples) and Abu - Kabir ( 13 samples) as it was shown in Table(l). These samples were bacteriologically examined in a trial to explore the possible existence of Past. microorganisms .
(b) Samples from diseased rabbits:
A total of 231 samples ( nasal swabs ) were aseptically collected from 231 diseased rabbits (18 male) and (213 female) from different Skarkia localities. From San-EL-Hagar (172 samples), from Abu-Hammad (8 samples ), from Kafr - Sakr (12 samples ), from Awlad -Sakr (15 samples), from Fakous (14 samples) and from Abu - Kabir (10 samples) as it was shown in Table(2). These samples were collected from rabbits and transferred without delay to the laboratory in an ice plastic bags for bacteriological examination to explore the possible existence of Past. microorganisms .
Table (1) : Nasal swab samples collected from apparently healthy rabbits from different localities in Sharkia Governorate :
Locality
No. of examined
rabbits
Male Female
1- San-EL-Hajar rabbit
projects. 97 8 89
2- Abu-Hammad farm 10 1 9
3- Kafr-Sakr farm 16 2 14
4- Awlad - Sakr farm 12 2 10
5- Fakous farm 9 1 8
6- Abu-Kabir farm 13 2 11
Total 157 16 141
Table (2) : Nasal swab samples collected from diseased rabbits from different localities in Sharkia - Governorate.
Locality
No. of examined
rabbits
Male Female Symptoms
1-San-EL-Hagar rabbit
projects
172 11 161 Rhinitis, metritis S/C abscesses, torticollis
2-
Abu-Hammad farm 8 1 7 Rhinitis, S/C abscesses 3- K;ifr-Sakr
farm 12 1 11 Rhinitis, emaciation 4- Awlad-Sakr
farm 15 2 13 Rhinitis, torticollis
5- Fakous farm 14 2 12 Rhinitis, S/C abscesses
6- Abu-Kabir
farm 10 1 9
Rhinitis, metritis, emaciation
Total 231 18 213 nasal swabs
II- Media used:
(A) Media used for isolation :
1 - Nutrient agar medium : ( Oxoid)
Basic media for the isolation of various microorganisms .
2- Blood agar medium : ( Cruickshank et al., 1975)
Nutrient agar base plus 10% defibrinated sheep blood, was used as enriched and indicator media for the isolation of Past, microorganisms and for detecting of the type of haemolysis .
3- MacConkey's agar medium ( Cruickshank et al., 1975) :
It was used as selective medium to differentiate between strains of Past. multocida and Past. haemolytica .
(B)- Media used for biochemical identification :
1- Semisolid 0.5% agar medium : (Bailey and Scott, 1974):
It was used for the demonstration of the motility of the
microorganisms and for the preservation of the isolated strains during the course of this study.
2- 2% Peptone water medium : ( Cruickshank et al., 1975).
It was used for the detection of indole production.
3-Simmon's citrate medium : ( Cruickskank et al, 1975).
It was used for the detection of citrate utilization test.
4- Urea agar medium: (Cruickshank et al., 1975)
It was used for the detection of urease activity of the isolated microorganisms.
5- Sugar media : ( Cruickshank et al., 1975)
1% peptone water containing Andrad's indicator to which 1% solution of one the following sugars were added (glucose, sucrose,
maltose, arabinose, xylose, lactose, sorbitol, mannitol, inositol and salicin) and Durham's tubes for the collection of the gas. These media were used for the studying the fermentative ability of the isolated microorganisms.
6- Nitrate reduction media : ( Cruickshank et at., 1975).
Were used for testing the reduction of nitrate into nitrite .
7- Glucose phosphate peptone water media ( Cruickshank et al., 1975).
For methyl- red ( M.R ) and Voges - Proskauer ( V.P) tests.
8- Triple sugar iron agar medium : ( Cruickshank et al., 1975)
It was used for the detection of production of hydrogen sulphide (H2S) and the detection of the fermentation of glucose, lactose and sucrose by changes in butt and slant.
C- Media used for preparation ofgenomic DNA :
1- Brain heart infusion broth medium ( Oxoid )
It was used for making heavy culture of pasteurella strains . 2- Brain heart infusion agar medium ( Oxoid ).
III- Reagents and Solutions :
1-Kovac's reagent (Cruickshank et al., 1996)
It was used in indole production test, a red colour indicated a positive reaction. It was prepared as follows :
- Isoamyl alcohol 150 ml . - P-Dimethyl - aminobenzaldehyde 10 gm . - Concentrated hydrochloric acid 50 ml .
- The aldehyde is dissolved in the alcohol with gentle heat, the acid is added after cooling the reagent and stored in a dark bottle at 4 °C .
2- Methyl red indicator solution : ( Cruickshank et al., 1975).
It was used in methyl red test, in positive case red colour developed, in negative case : yellow colour developed. It was prepared as follows :
Methyl red 0.1 gm Ethanol 300 ml
Distilled water 200 ml
3- Voges - Proskauer reagent: (Cruickshank et al., 1996) :
One ml of 40% KOH and 3 ml of 5% alpha-naphthol in absolute ethanol, used as indicator in Voges - Proskauer ( V.P ) in positive case, pink red colour developed within 2 to 5 minutes while in negative one no colour developed .
4- Oxidase reagent :
Tetramethyl-P-phenylene diamine dihydrochloride used as reagent in oxidase test ( Carpenter et al., 1947). The reagent was freshly prepared for use each time as the dye lost it's activity after 2 hours, in positive case deep pink colour rapidly developed within 5 -10 seconds finally to black. A delayed positive reaction was indicated by coloration in 10-60 seconds, while in negative one absence of coloration.
5- Reagent for nitrate reduction : (Cruickshank et al., 1996) Reagent A :
Sulphanilic acid 8.0 gm N / 5 acetic acid 1000 ml
Reagent B:
Positive reaction pink to deep red colour developed while in negative reaction no colour developed .
6- Andrad's indicator 1% of reagent:- It was added to the medium. This indicator was used in biochemical fermentation of sugar .
7- Catalase test reagent : 3% hydrogen peroxide solution was used for catalase test.
8- Physiological saline solution :
It was used for injection of experimental animals .
9- Thiomersal "1 ; 10000 " :
It was used as preservative after inactivation of serum at 56 C° for 30 minutes. Thiomersal was added to give a final concentration 1: 1 0000.
IV- Stains used :
1- Gram's stain : [ Cruickshank et. al, 1975 ]
It was used to differentiate between the isolated microorganisms into the classical Gram positive and Gram negative isolates .
2- Irishman's stain : / Cruickshank et. al., 1975 ] :
It was used to detect the bipolarity of Past. microorganisms in blood smears .
V- Standard strains of Past. multocida,
Standard strains of Past. multocida types, 1:A, 3:A, 4:A, 8:A, 9:A, 11:A, 12:A, 11:D, 1 3 :D and B were kindly obtained from National Animal Diseases Centre, Ames Iowa, USA .
VI- Laboratory animals :
1-
Mice:-A total of seventy four mice of 20 - 25 grams were used for pathogenicity test and for purification of Past, microorganisms. These mice were obtained from the laboratory animal farm of Faculty of Veterinary Medicine, Zagazig University .
2- Rabbits
:-Twenty boskat breed of rabbits weighting 2-3 kg were used for the preparation of antisera against standard strains of Past. multocida serovars [two rabbits for each strain] .
VII- Chemicals, reagents and buffers used in biomolecular studies of Past. multocida.
A- Chemicals, reagents and buffers used for DNA extraction :
- All chemicals, reagents and buffers were prepared according to
Sambrook et al. [1989], Wilson et al. [1992] and Ausubel et al., 11999]
1- Agarose molecular biology grade [ SigmaJ:
It was used for electrophoretic separation of nucleic acid molecules [DNA]. Agarose gel was prepared at concentration of 0.7% by dissolving 0.7 gm of agarose in 100 ml IX Tris Borate EDTA [ TBE ] buffer.
2-Tris-HCL:
Tris buffer [ 0.05 M] of pH ( 7.1 - 8.9 ) was prepared by mixing 50 ml of 0.1 M Tris with the indicated volume of 0.1 N HCI and the volume of mixture was then completed to 100 ml with double distilled water (dd H2O). It was used in the preparation of Tris - EDTA [TE] buffer.
3-1 mM EDTA :
0.5 M EDTA of pH 8.0 was prepared by dissolving 186.1 gm Na2
EDTA. 2H2O in 700ml d.d H2O. Adjusted pH to 8.0 with 10.0 M NaoH
[50ml], Then the mixture was completed to 1 liter by adding dd H2O .
4- TE buffer:
It was prepared as follow :
Tris - HC1 ( pH 8.0 ) 10 m M
EDTA 1.0 mM
It was used for dissolving of DNA .
5- Sodium_dodecyl sulfate (SDS) solution:
Stock solution [SDS 10%, w/v ] was prepared by dissolving 10 gm of. SDS in 100 ml double distilled water. The solution was stored at room temperature. It was used for lysis of the bacterial cells.
6- Prote_ingse ”K” soJutJoil (Sigma ):
It was used in a concentration of 20mg/ml of dd H2O, for extraction
of DNA. The solution was stored at - 20 °C.
7- 5M Nacl:
It was prepared by dissolving 292 gm of Nacl in 1 liter of dd H2O .
8-Buffered saturated phenol_:
It was used in purification of nucleic acids. It was saturated with 10 mM Tris of pH 8.0 .
9- Chloroform / isoamyl alcohol (Sigma ) :
It was used for extraction of nucleic acids. It was prepared by adding 24 ml of chloroform to 1 ml of isoamyl alcohol.
10- Phenol, Chloroform and Isoamyl alcohol :
It was used for purification of DNA. It was prepared as a mixture of the phenol, chloroform, isoamyl alcohol at a concentration of 25 : 24 : 1 .
11- CTAB / Nacl solution .
It was prepared as follow :
Hexadecyle trimethyl ammonium bromide 10 %
Nacl 0.7 M
It was used for elimination of cell wall debris, polysaccharides and denatured proteins by selective precipitation.
12- Isopropanol [ Sigma 1 :
It was used for precipitation of DNA.
13- 70 %
Ethanol:-It was used for washing of DNA. Ethanol:-It was prepared by adding 70 ml of absolute ethanol to 30 ml of dd H2O .
14- Gel loading buffer
:-Bromophenol blue 0.25 gm Xylene cyand FF 0.25 gm Ficol 400 15 gm Double distilled water up to 100 ml
15- 5X Tris Borate EDTA ( TEE) buffer (electrophoresis buffer):
Trisbase 54.0 gm Boric acid 27.5 gm 0.5 M EDTA 20.0 ml Double distilled water up to 100.0 ml
The was used as electrophoresis buffer and dissolving of agarose as working solution of IX [ pH 8.0 ] .
16- Ethidium bromide :-
It was prepared as stock of 10 mg / ml. It was used in concentration of 0.5ug/ml for staining of DNA in agarose gel. The solution was stored at room temperature.
17- DNA Marker :-
Lambda DNA digested with Hind III and phi X 174 DNA digested with Hae III. This marker covers a range <of DNA fragment size between 23130 bp and 310 bp, obtained by Agriculture Genetic Engineering Research Institute [ AGERI ], Giza, Egypt.
B- Material used in RAPD analysis
:-a- Table (3) : 10- mer oligoneucleotides were used as primers in the RAPD assay.
No. Code Primers sequence [5- - 3-]
1- O10 TCAGAGCGCC
2- D 11 AGCGCCATTG
3- C4 CCGCATCTAC
4- A4 AATCGGGCTG
5- D4 TCTGGTGAGG
b- Table (4): Chemicals used in one RAPD-PCR reaction
:-Components Master mix.
1- 10 X PCR buffer [ Promega ] 2.5 ul 2- 25 mM Mgcl2 [ Promega ] 1.5ul
3- 10 mM dNTPs [ Promega ] 0.5 ul
4- 5 uM primer 1.0 ul
5- 5 U/ ul Taq. Polymerase [ Promega ] 0.1 ul 6- 10 ng/ul template DNA 1.0 ul 7- Sterile .d.d. water 18.4ul 8- Total volume 25.0 ul
C-100 bp DNA ladder:
100 bp DNA marker [pharmacia], Catalogue number 27 - 4100 - 0 1, U.S.A. It was suitable for sizing DNA from 100 to 2300 bp.
d- Asarose eel fSisma /
Agarose ultrapure was used at a concentration of 1.5%. It was prepared by dissolving 1.5 gm of agarose in 100 ml of IX Tris- Borate EDTA [ TBE ] buffer.
VIII- Equipments and apparatuses used in biomolecular studies of Past, multocida strains :
- Sterile Guard Hood : class II type A/B3 Baker Company .
- Water jacketed incubator : 3250 forma scientific . - Cooling centrifuge : Beckman Microfuge F.
- High speed centrifuge : Beckman L-80 ultra centrifuge . - Ordinary centrifuge : Beckman GS - SR centrifuge .
- Shaker incubator : New Brunswick Scientific Company series 25 . - Pipettes : Wheaton Convenient Laboratory pipette .
- Vortex mixer : Vortex Genie 2 Scientific Industries . - Pipette aid : Willinger Bros .
- Micropipettes : Gilson pipetman .
Ranged: 0-10 ul 20-200 ul 100-1000 ul
- Tips : Fisher brand. Redi - Tip .
- Eppindorff tubes : Fischerbrand Fischer Scientific . - pH meter : Accumet model 15 - Fischer Scientific .
- Power supply : Bio- Rad model 200 / 2.0 - Bio - Rad mini sub DNA cell .
- Electronic UV Transilluminator [ ultra. Lum ] polaroid .
- MP4 Polaroid Camera : was used for photographing the DNA on agarose .
- Deep freezer : -70 C° deep freeze Reveco was used for preservation of isolates seeds .
- Perkin - Elmer Cetus DNA Thermal Cycler 480 was used for amplification of DNA-