organic papers
Acta Cryst.(2006). E62, o2361–o2362 doi:10.1107/S1600536806017922 Wenet al. C
19H16Cl2N4O2
o2361
Acta Crystallographica Section E Structure Reports
Online
ISSN 1600-5368
5-Chloro-1-(4-chlorophenyl)-4-(2-methoxy-benzoylhydrazonomethyl)-3-methyl-1
H
-pyrazole
Li-Rong Wen, Li-Na Zhai, Peng Liu and Ming Li*
College of Chemistry and Molecular
Engineering, Qingdao University of Science and Technology, Qingdao 266042, People’s Republic of China
Correspondence e-mail: liming928@263.net
Key indicators
Single-crystal X-ray study
T= 293 K
Mean(C–C) = 0.003 A˚
Rfactor = 0.035
wRfactor = 0.091
Data-to-parameter ratio = 13.6
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 19 April 2006 Accepted 15 May 2006
#2006 International Union of Crystallography
All rights reserved
In the title molecule, C19H16Cl2N4O2, all bond lengths and
angles show normal values. The two benzene rings make a dihedral angle of 12.55 (2). The crystal packing is stabilized
by weak intermolecular C—H N hydrogen bonds and van der Waals forces.
Comment
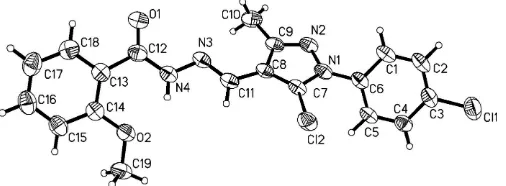
Arylpyrazoles have a widespread occurrence as substructures in a large variety of compounds with important biological activities and pharmacological properties (Nizar & James, 2002). In the course of our systematic studies aimed at the synthesis of new bioactive compounds, we have synthesized the title compound, (I) (Fig. 1).
The bond lengths and angles (Table 1) in (I) are as expected for this type of compound. The pyrazole ring is planar, the largest deviation from planarity being 0.003 (4) A˚ for atom C8, and forms a dihedral angle of 50.45 (4) with the C1–C6
benzene ring. The two benzene rings, C1–C6 and C13–C18, make a dihedral angle of 12.55 (2).
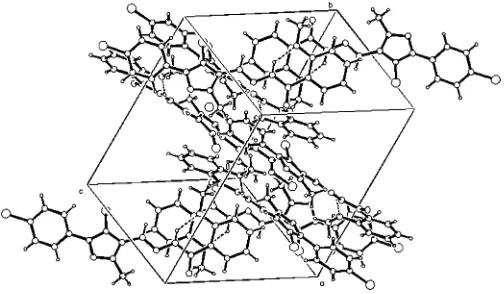
The crystal packing (Fig. 2) is stabilized by weak inter-molecular C—H N hydrogen bonds (Table 2) and van der Waals forces.
Experimental
[image:1.610.208.461.639.732.2]To anhydrous ethanol (15 ml), a mixture of 2-methyloxybenzoyl-hydrazide (3 mmol, 0.500 g) and 5-chloro-1-(4-chlorophenyl)-3-methyl-1H-4-pyrazolaldehyde (3 mmol, 0.765 g), synthesized according to Li et al.(2004), was added and refluxed for 5 h. The solvent was removed under reduced pressure and the residue was recrystallized from ethanol (m.p. 458 K).
Figure 1
Crystal data
C19H16Cl2N4O2
Mr= 403.26
Monoclinic,P21=n a= 9.3332 (10) A˚
b= 16.7115 (19) A˚
c= 12.3865 (14) A˚
= 101.839 (2) V= 1890.8 (4) A˚3
Z= 4
Dx= 1.417 Mg m
3
MoKradiation
= 0.37 mm1
T= 293 (2) K Block, colourless 0.280.220.20 mm
Data collection
Bruker APEX-II CCD area-detector diffractometer
’and!scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin= 0.885,Tmax= 0.998
(expected range = 0.824–0.929)
10116 measured reflections 3338 independent reflections 2462 reflections withI> 2(I)
Rint= 0.020
max= 25.0
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.035
wR(F2) = 0.091
S= 1.04 3338 reflections 246 parameters
H-atom parameters constrained
w= 1/[2
(Fo2) + (0.0403P)2
+ 0.4272P]
whereP= (Fo2+ 2Fc2)/3
(/)max= 0.001
max= 0.21 e A˚
3
min=0.22 e A˚
3
Table 1
Selected geometric parameters (A˚ ,).
O1—C12 1.222 (2) O2—C14 1.365 (2) N1—C7 1.354 (2) N1—N2 1.374 (2)
N2—C9 1.316 (3) N3—C11 1.278 (2) C7—C8 1.367 (3) C8—C9 1.418 (3) C7—N1—N2 109.78 (17)
C9—N2—N1 105.69 (15) N1—C7—C8 109.05 (17)
[image:2.610.45.297.71.218.2]C7—C8—C9 103.59 (18) N2—C9—C8 111.88 (18)
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
N4—H4A O2 0.86 1.98 2.617 (2) 130 C2—H2 N3i
0.93 2.54 3.421 (2) 158
Symmetry code: (i)xþ1 2;yþ
1 2;z
1 2.
All H atoms were placed in calculated positions, with C—H = 0.93 or 0.96 A˚ and N—H = 0.86 A˚, and included in the final cycles of refinement using a riding model, withUiso(H) set to 1.2Ueq(C,N) for
CH and NH groups and 1.5Ueq(C) for methyl groups.
Data collection:SMART(Bruker, 1998); cell refinement:SAINT (Bruker, 1999); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics: SHELXTL (Bruker, 1999); software used to prepare material for publication:SHELXTL.
This project was supported by the National Natural Science Foundation of China (grant No. 20572057).
References
Bruker (1998).SMART. Bruker AXS Inc., Madison, Wisconsin, USA. Bruker (1999). SAINT and SHELXTL. Bruker AXS Inc., Madison,
Wisconsin, USA.
Li, M., Wen, L. R., Jing, S. X., Zhao, G. L. & Yang, H. Z. (2004).Chin. J. Struct. Chem.23, 366–370.
Nizar, H. & James, B. (2002).Tetrahedron Lett.43, 2171–2173. Sheldrick, G. M. (1996).SADABS. University of Go¨ttingen, Germany. Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of
Go¨ttingen, Germany.
Figure 2
supporting information
sup-1 Acta Cryst. (2006). E62, o2361–o2362
supporting information
Acta Cryst. (2006). E62, o2361–o2362 [https://doi.org/10.1107/S1600536806017922]
5-Chloro-1-(4-chlorophenyl)-4-(2-methoxybenzoylhydrazonomethyl)-3-methyl-1
H
-pyrazole
Li-Rong Wen, Li-Na Zhai, Peng Liu and Ming Li
5-Chloro-1-(4-chlorophenyl)-4-(2-methoxybenzoylhydrazonomethyl)- 3-methyl-1H-pyrazole
Crystal data
C19H16Cl2N4O2 Mr = 403.26 Monoclinic, P21/n
Hall symbol: -P 2yn
a = 9.3332 (10) Å
b = 16.7115 (19) Å
c = 12.3865 (14) Å
β = 101.839 (2)°
V = 1890.8 (4) Å3
Z = 4
F(000) = 832
Dx = 1.417 Mg m−3
Mo Kα radiation, λ = 0.71073 Å
Cell parameters from 2774 reflections
θ = 2.4–22.6°
µ = 0.37 mm−1
T = 293 K
Block, colourless 0.28 × 0.22 × 0.20 mm
Data collection
Bruker APEX-II CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
φ and ω scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin = 0.885, Tmax = 0.998
10116 measured reflections 3338 independent reflections 2462 reflections with I > 2σ(I)
Rint = 0.020
θmax = 25.0°, θmin = 2.1°
h = −11→9
k = −19→19
l = −10→14
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.035 wR(F2) = 0.091
S = 1.04
3338 reflections 246 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained
w = 1/[σ2(F
o2) + (0.0403P)2 + 0.4272P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max = 0.001
Δρmax = 0.21 e Å−3
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
Cl1 1.07602 (8) 0.20432 (4) −0.36420 (6) 0.0985 (3)
Cl2 0.86797 (6) 0.08248 (3) 0.10691 (4) 0.06968 (19)
O1 0.20930 (19) 0.07607 (9) 0.31458 (14) 0.0843 (5)
O2 0.47333 (17) −0.12149 (8) 0.39147 (12) 0.0699 (4)
N1 0.67297 (18) 0.13615 (9) −0.06804 (13) 0.0565 (4)
N2 0.52624 (19) 0.15084 (9) −0.10476 (14) 0.0596 (4)
N3 0.42112 (19) 0.05855 (9) 0.19518 (14) 0.0579 (4)
N4 0.41514 (19) 0.01509 (9) 0.28839 (13) 0.0589 (4)
H4A 0.4834 −0.0187 0.3133 0.071*
C1 0.7711 (2) 0.22835 (11) −0.18525 (17) 0.0622 (6)
H1 0.7073 0.2677 −0.1705 0.075*
C2 0.8641 (3) 0.24387 (12) −0.25472 (18) 0.0676 (6)
H2 0.8638 0.2939 −0.2875 0.081*
C3 0.9582 (2) 0.18512 (13) −0.27609 (17) 0.0643 (6)
C4 0.9602 (2) 0.11053 (12) −0.22811 (17) 0.0620 (5)
H4 1.0244 0.0714 −0.2427 0.074*
C5 0.8668 (2) 0.09466 (11) −0.15877 (16) 0.0597 (5)
H5 0.8669 0.0444 −0.1264 0.072*
C6 0.7727 (2) 0.15344 (11) −0.13705 (15) 0.0539 (5)
C7 0.6954 (2) 0.10023 (11) 0.03204 (16) 0.0536 (5)
C8 0.5637 (2) 0.09034 (10) 0.06236 (15) 0.0530 (5)
C9 0.4618 (2) 0.12378 (11) −0.02696 (16) 0.0552 (5)
C10 0.2998 (2) 0.13076 (14) −0.03946 (19) 0.0742 (6)
H10A 0.2779 0.1679 0.0140 0.111*
H10B 0.2595 0.0793 −0.0281 0.111*
H10C 0.2577 0.1495 −0.1123 0.111*
C11 0.5412 (2) 0.05187 (11) 0.16170 (16) 0.0552 (5)
H11 0.6164 0.0213 0.2026 0.066*
C12 0.3033 (2) 0.02484 (12) 0.34146 (18) 0.0603 (5)
C13 0.2988 (2) −0.03115 (12) 0.43492 (16) 0.0568 (5)
C14 0.3767 (2) −0.10315 (12) 0.45685 (16) 0.0576 (5)
C15 0.3537 (3) −0.15257 (14) 0.54161 (18) 0.0687 (6)
H15 0.4060 −0.2001 0.5559 0.082*
C16 0.2552 (3) −0.13213 (17) 0.60402 (19) 0.0810 (7)
supporting information
sup-3 Acta Cryst. (2006). E62, o2361–o2362
C17 0.1790 (3) −0.06179 (18) 0.5858 (2) 0.0847 (7)
H17 0.1127 −0.0477 0.6292 0.102*
C18 0.2018 (2) −0.01192 (15) 0.50202 (18) 0.0719 (6)
H18 0.1505 0.0361 0.4903 0.086*
C19 0.5441 (3) −0.19740 (15) 0.4020 (2) 0.0890 (8)
H19A 0.6106 −0.2005 0.4722 0.134*
H19B 0.5974 −0.2038 0.3440 0.134*
H19C 0.4721 −0.2390 0.3969 0.134*
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
Cl1 0.1015 (5) 0.0989 (5) 0.0978 (5) −0.0157 (4) 0.0269 (4) 0.0265 (4)
Cl2 0.0671 (3) 0.0726 (4) 0.0614 (3) 0.0022 (3) −0.0053 (3) 0.0177 (3)
O1 0.0956 (12) 0.0603 (9) 0.1015 (13) 0.0178 (9) 0.0309 (10) 0.0124 (9)
O2 0.0809 (10) 0.0595 (9) 0.0706 (9) 0.0097 (8) 0.0182 (8) 0.0107 (7)
N1 0.0631 (11) 0.0477 (9) 0.0516 (9) 0.0012 (8) −0.0046 (8) 0.0069 (8)
N2 0.0662 (11) 0.0503 (9) 0.0551 (10) 0.0056 (8) −0.0043 (9) 0.0051 (8)
N3 0.0699 (11) 0.0424 (9) 0.0575 (10) 0.0001 (8) 0.0042 (9) 0.0019 (8)
N4 0.0629 (11) 0.0533 (9) 0.0578 (10) 0.0024 (8) 0.0059 (8) 0.0074 (8)
C1 0.0803 (15) 0.0413 (10) 0.0561 (12) −0.0023 (10) −0.0066 (11) 0.0043 (9)
C2 0.0870 (16) 0.0455 (11) 0.0606 (13) −0.0161 (11) −0.0075 (12) 0.0115 (10)
C3 0.0674 (13) 0.0620 (13) 0.0557 (12) −0.0180 (11) −0.0055 (10) 0.0105 (10)
C4 0.0651 (13) 0.0547 (12) 0.0594 (12) −0.0014 (10) −0.0030 (11) 0.0067 (10)
C5 0.0730 (14) 0.0423 (10) 0.0566 (12) −0.0035 (10) −0.0033 (11) 0.0091 (9)
C6 0.0630 (12) 0.0450 (10) 0.0459 (11) −0.0064 (9) −0.0073 (9) 0.0036 (9)
C7 0.0641 (13) 0.0404 (10) 0.0501 (11) 0.0004 (9) −0.0024 (9) 0.0046 (8)
C8 0.0665 (13) 0.0374 (9) 0.0506 (11) 0.0008 (9) 0.0012 (10) −0.0015 (8)
C9 0.0658 (13) 0.0409 (10) 0.0526 (12) 0.0039 (9) −0.0028 (10) −0.0043 (9)
C10 0.0701 (15) 0.0729 (15) 0.0715 (15) 0.0117 (11) −0.0043 (12) 0.0005 (12)
C11 0.0633 (13) 0.0427 (10) 0.0536 (12) 0.0022 (9) −0.0019 (10) −0.0009 (9)
C12 0.0667 (13) 0.0458 (11) 0.0654 (13) −0.0031 (10) 0.0068 (11) −0.0079 (10)
C13 0.0605 (12) 0.0541 (12) 0.0516 (12) −0.0087 (10) 0.0017 (10) −0.0078 (9)
C14 0.0587 (12) 0.0586 (12) 0.0507 (12) −0.0080 (10) 0.0005 (10) −0.0049 (10)
C15 0.0749 (15) 0.0701 (14) 0.0557 (13) −0.0060 (11) 0.0009 (12) 0.0062 (11)
C16 0.0877 (18) 0.096 (2) 0.0564 (14) −0.0128 (15) 0.0088 (13) 0.0089 (13)
C17 0.0833 (17) 0.113 (2) 0.0593 (14) −0.0039 (16) 0.0189 (13) −0.0057 (15)
C18 0.0730 (15) 0.0779 (15) 0.0620 (14) 0.0038 (12) 0.0073 (12) −0.0085 (12)
C19 0.0999 (19) 0.0799 (17) 0.0879 (18) 0.0344 (14) 0.0210 (15) 0.0231 (14)
Geometric parameters (Å, º)
Cl1—C3 1.731 (2) C7—C8 1.367 (3)
Cl2—C7 1.7117 (19) C8—C9 1.418 (3)
O1—C12 1.222 (2) C8—C11 1.441 (3)
O2—C14 1.365 (2) C9—C10 1.493 (3)
O2—C19 1.424 (3) C10—H10A 0.9600
N1—N2 1.374 (2) C10—H10C 0.9600
N1—C6 1.416 (3) C11—H11 0.9300
N2—C9 1.316 (3) C12—C13 1.496 (3)
N3—C11 1.278 (2) C13—C18 1.386 (3)
N3—N4 1.375 (2) C13—C14 1.404 (3)
N4—C12 1.352 (3) C14—C15 1.387 (3)
N4—H4A 0.8600 C15—C16 1.360 (3)
C1—C2 1.367 (3) C15—H15 0.9300
C1—C6 1.386 (3) C16—C17 1.368 (4)
C1—H1 0.9300 C16—H16 0.9300
C2—C3 1.379 (3) C17—C18 1.381 (3)
C2—H2 0.9300 C17—H17 0.9300
C3—C4 1.379 (3) C18—H18 0.9300
C4—C5 1.369 (3) C19—H19A 0.9600
C4—H4 0.9300 C19—H19B 0.9600
C5—C6 1.381 (3) C19—H19C 0.9600
C5—H5 0.9300
C14—O2—C19 119.75 (17) C9—C10—H10A 109.5
C7—N1—N2 109.78 (17) C9—C10—H10B 109.5
C7—N1—C6 130.21 (17) H10A—C10—H10B 109.5
N2—N1—C6 119.83 (15) C9—C10—H10C 109.5
C9—N2—N1 105.69 (15) H10A—C10—H10C 109.5
C11—N3—N4 114.58 (17) H10B—C10—H10C 109.5
C12—N4—N3 120.85 (17) N3—C11—C8 121.95 (18)
C12—N4—H4A 119.6 N3—C11—H11 119.0
N3—N4—H4A 119.6 C8—C11—H11 119.0
C2—C1—C6 119.4 (2) O1—C12—N4 122.3 (2)
C2—C1—H1 120.3 O1—C12—C13 121.1 (2)
C6—C1—H1 120.3 N4—C12—C13 116.62 (18)
C1—C2—C3 119.86 (19) C18—C13—C14 117.3 (2)
C1—C2—H2 120.1 C18—C13—C12 116.41 (19)
C3—C2—H2 120.1 C14—C13—C12 126.1 (2)
C2—C3—C4 120.9 (2) O2—C14—C15 122.66 (19)
C2—C3—Cl1 120.10 (17) O2—C14—C13 117.19 (18)
C4—C3—Cl1 119.03 (19) C15—C14—C13 120.1 (2)
C5—C4—C3 119.5 (2) C16—C15—C14 120.6 (2)
C5—C4—H4 120.3 C16—C15—H15 119.7
C3—C4—H4 120.3 C14—C15—H15 119.7
C4—C5—C6 119.79 (18) C15—C16—C17 120.7 (2)
C4—C5—H5 120.1 C15—C16—H16 119.7
C6—C5—H5 120.1 C17—C16—H16 119.7
C5—C6—C1 120.6 (2) C16—C17—C18 119.2 (2)
C5—C6—N1 119.71 (17) C16—C17—H17 120.4
C1—C6—N1 119.60 (19) C18—C17—H17 120.4
N1—C7—C8 109.05 (17) C17—C18—C13 122.1 (2)
N1—C7—Cl2 121.61 (16) C17—C18—H18 119.0
supporting information
sup-5 Acta Cryst. (2006). E62, o2361–o2362
C7—C8—C9 103.59 (18) O2—C19—H19A 109.5
C7—C8—C11 125.81 (18) O2—C19—H19B 109.5
C9—C8—C11 130.6 (2) H19A—C19—H19B 109.5
N2—C9—C8 111.88 (18) O2—C19—H19C 109.5
N2—C9—C10 120.17 (18) H19A—C19—H19C 109.5
C8—C9—C10 128.0 (2) H19B—C19—H19C 109.5
C7—N1—N2—C9 0.0 (2) C7—C8—C9—N2 0.5 (2)
C6—N1—N2—C9 175.64 (16) C11—C8—C9—N2 −177.56 (18)
C11—N3—N4—C12 170.96 (17) C7—C8—C9—C10 −179.28 (19)
C6—C1—C2—C3 0.0 (3) C11—C8—C9—C10 2.7 (3)
C1—C2—C3—C4 −0.1 (3) N4—N3—C11—C8 177.07 (16)
C1—C2—C3—Cl1 179.87 (15) C7—C8—C11—N3 165.92 (18)
C2—C3—C4—C5 0.3 (3) C9—C8—C11—N3 −16.4 (3)
Cl1—C3—C4—C5 −179.65 (15) N3—N4—C12—O1 −5.5 (3)
C3—C4—C5—C6 −0.4 (3) N3—N4—C12—C13 173.86 (16)
C4—C5—C6—C1 0.3 (3) O1—C12—C13—C18 −12.5 (3)
C4—C5—C6—N1 178.02 (17) N4—C12—C13—C18 168.20 (17)
C2—C1—C6—C5 −0.1 (3) O1—C12—C13—C14 163.60 (19)
C2—C1—C6—N1 −177.80 (17) N4—C12—C13—C14 −15.7 (3)
C7—N1—C6—C5 48.2 (3) C19—O2—C14—C15 6.3 (3)
N2—N1—C6—C5 −126.41 (19) C19—O2—C14—C13 −173.41 (19)
C7—N1—C6—C1 −134.1 (2) C18—C13—C14—O2 −179.27 (17)
N2—N1—C6—C1 51.3 (2) C12—C13—C14—O2 4.7 (3)
N2—N1—C7—C8 0.3 (2) C18—C13—C14—C15 1.1 (3)
C6—N1—C7—C8 −174.72 (17) C12—C13—C14—C15 −174.98 (18)
N2—N1—C7—Cl2 −173.62 (13) O2—C14—C15—C16 −179.40 (19)
C6—N1—C7—Cl2 11.3 (3) C13—C14—C15—C16 0.3 (3)
N1—C7—C8—C9 −0.5 (2) C14—C15—C16—C17 −1.2 (4)
Cl2—C7—C8—C9 172.88 (15) C15—C16—C17—C18 0.8 (4)
N1—C7—C8—C11 177.69 (17) C16—C17—C18—C13 0.6 (4)
Cl2—C7—C8—C11 −8.9 (3) C14—C13—C18—C17 −1.5 (3)
N1—N2—C9—C8 −0.3 (2) C12—C13—C18—C17 174.9 (2)
N1—N2—C9—C10 179.50 (16)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
N4—H4A···O2 0.86 1.98 2.617 (2) 130
C2—H2···N3i 0.93 2.54 3.421 (2) 158
C18—H18···O1 0.93 2.44 2.761 (3) 100