organic papers
o762
Zhuet al. C28H46O6 doi:10.1107/S1600536806002418 Acta Cryst.(2006). E62, o762–o764 Acta Crystallographica Section EStructure Reports
Online
ISSN 1600-5368
A rearranged labdane diterpene glycoside
Ying Zhu, Cai-Xia Yang and Zhong-Jian Jia*
State Key Laboratory of Applied Organic Chemistry, Chemistry and Chemical Engineering College, Lanzhou University, Lanzhou 730000, People’s Republic of China
Correspondence e-mail: jiazj@lzu.edu.cn
Key indicators
Single-crystal X-ray study
T= 293 K
Mean(C–C) = 0.004 A˚
Rfactor = 0.038
wRfactor = 0.082
Data-to-parameter ratio = 10.3
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
#2006 International Union of Crystallography Printed in Great Britain – all rights reserved
The title compound, 13(R)-9 -methyl-1(10),14-dienefried-labda-13-O--l-20-acetylrhamonopyanoside or
3-methyl-5- (1,2,5,5-tetramethyl-1,2,3,4,4a,5,6,7-octahydronaphthyl)pent-1-en-3-yl 2-O-acetylrhamnopyanoside, C28H46O6, is a bicyclic
diterpenoid glycoside. The rearranged methyl group (from C10 to C9) is in aorientation and a double bond is formed between C1 and C10 in the rearrangement. Thel-rhamnose
group on the cyclohexane ring is equatorial and the configuration of the C atom to which the methylene group is attached isR. The cyclohexene, cyclohexane andl-rhamnose
rings in the molecule adopt half-chair, chair and chair conformations, respectively. In the crystal structure, molecules are linked by O—H O hydrogen bonds. The l-20
-acetyl-rhamnose hydroxy groups serve as hydrogen-bond donors, forming molecular chains along thebaxis.
Comment
Labdane diterpenoids are among the most common types of diterpenes isolated from terrestrial higher plants and sponges (Hanson, 1997, 1998, 1999, 2000, 2001; Tanaka, et al., 2001). Many of these terpenoids possess significant pharmacological properties, such as cytotoxic, antibacterial, antifungal, anti-inflammatory, analgesic, antitumor and antimutagenic (Ahsan
et al., 2003; Itokawa & Morita, 1988; Dimas et al., 1998; Kittakoop et al., 2001; Kubo et al.2003; Minami et al., 2002; Miyazawaet al., 1995). Therefore, the semisynthesis of minor components from other abundant natural products is of long-standing interest. To date, a number of semisyntheses of these biologically active labdane-type diterpenoids have been reported (Pathaket al.2005). The current interest of our group in the phytochemical study of northwest Chinese plants aims to find new natural compounds with interesting biological activities. In this connection we have studied labdane diter-penoids (Wanget al., 2002; Yang,et al., 2005).
In this paper, we report the crystal structure and relative stereochemistry of a diterpenoid glycoside, (I), with a
ranged labdane skeleton, fromAster homochlamydeus Hand-mazz. Crystal structures of typical labdane-type diterpenoids have previously been reported (Nagashima et al., 1995; Bernardinelliet al., 1988; Tavanaiepouret al., 1987; Bja˚meret al., 1968).
Compound (I) has the molecular formula C28H46O6,
established by FAB-MS, which gave [M+Na] atm/z501 and [M+Li] at 485. Its IR spectrum showed the presence of hydroxyl functionalities (3483 and 3373 cm1). Since compound (I) has five degrees of unsaturation, it must contain one glycoside, two olefin bonds and two carbocyclic rings.
As shown in Fig. 1, compound (I) is a bicyclic diterpene glycoside of a C20 skelton with a terminal double bond,
C14 C15. It is slightly different from typical labdane diter-penes. The methyl group (C20) normally located at C10 is moved to C9 and the C1—C10 bond length indicates double-bond character. The methyl group at C9 is in the-orientation. Thus compound (I) is a diterpenoid with a rearranged labdane-type skelton (Uroneset al., 1994; Feresinet al., 2003). The bond distance between C9 and C20 is 1.532 (3) A˚ . The olefin bond distance in a carbocyclic ring, between C1 and C10, is 1.328 (4) A˚ , and corresponding bond angles show the characteristics of an olefin [C2—C1—C10 = 125.4 (3) and C1—C10—C5 121.2 (3)]. The bond distance between C14 and C15 is 1.264 (4) A˚ , with C13—C14—C15 = 128.8 (5). The structure demonstrates that an equatoriall-20-acetylrhamnose
group and a methyl group are attached at C13. The bond lengths to these substituents [C13—C16 = 1.509 (4) A˚ and C13—O1 = 1.455 (3) A˚ ] are in good agreement with the standard values observed for C—C (methyl) and C—O (sugar hydroxyl) distances, respectively (Allen et al., 1987). The R
configuration at C13 is confirmed unambiguously (Bernardi-nelli et al., 1988). Furthermore, the relative configurations at C4, C5, C8 and C9 were also determined, as shown in Fig. 1.
The endocyclic torsion angles (Table 1) show that the cyclohexene ring is in the half-chair conformation (Dauxet al., 1974, 1976), which has an approximate twofold axis passing
through the mid-point of the C3—C4 bond and the mid-point of the C1—C10 bond, with an asymmetry parameterC2of
0.88. The cyclohexane ring and l-20-acetylrhamnose groups
are in approximate chair conformations (Daux et al., 1974, 1976); their conformations are similar to others in the litera-ture (Tavanaiepour et al., 1987). The molecules of (I) are associated in the crystal state by O—H O hydrogen bonds between the hydroxyl functions in the l-20-acetylrhamnose
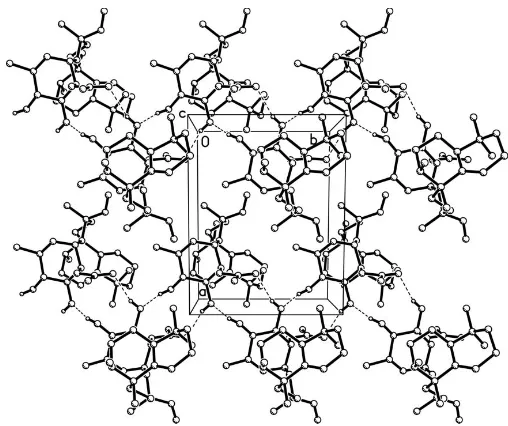
group (Table 2). As shown in Fig. 2, the molecules form columns along thebaxis.
Experimental
The extraction and isolation of compound (I) have already been described (Yang et al., 2005). Compound (I) was dissolved in chloroform and acetone (1:1v/v), and slow evaporation gave crystals suitable for X-ray diffraction. The optical rotation is []D
25 = +33.0 (c 0.4, CHCl3) and its melting point is 420 K.
Crystal data
C28H46O6
Mr= 478.65
Monoclinic,P21
a= 11.542 (2) A˚
b= 8.793 (2) A˚
c= 14.095 (2) A˚
= 102.14 (1)
V= 1398.6 (4) A˚3
Z= 2
Dx= 1.137 Mg m 3
MoKradiation Cell parameters from 42
reflections
= 2.7–14.3 = 0.08 mm1
T= 293 (2) K Prism, colourless 0.580.400.16 mm
Data collection
Siemens P4 diffractometer
!scans
Absorption correction: none 3583 measured reflections 3289 independent reflections 1841 reflections withI> 2(I)
Rint= 0.020
max= 27.1
h= 0!14
k= 0!11
l=18!17 3 standard reflections
every 97 reflections intensity decay: 3.0%
organic papers
[image:2.610.54.291.67.261.2]Acta Cryst.(2006). E62, o762–o764 Zhuet al. C28H46O6
o763
Figure 2
The molecular packing of (I), viewed along the c axis. Dashed lines indicate hydrogen bonds. H atoms not involved in hydrogen bonding have been omitted.
Figure 1
[image:2.610.311.565.68.281.2]Refinement
Refinement onF2 R[F2> 2(F2)] = 0.038
wR(F2) = 0.082
S= 0.81 3289 reflections 320 parameters
H-atom parameters constrained
w= 1/[2
(Fo2) + (0.04P)2]
whereP= (Fo2+ 2Fc2)/3
(/)max< 0.001 max= 0.12 e A˚
3
min=0.13 e A˚ 3
Extinction correction:SHELXL97
Extinction coefficient: 0.0125 (13)
Table 1
Selected torsion angles ().
C10—C1—C2—C3 15.5 (4) C1—C2—C3—C4 44.8 (4) C2—C3—C4—C5 60.4 (3) C3—C4—C5—C10 45.6 (3) C10—C5—C6—C7 55.5 (3) C5—C6—C7—C8 52.5 (3) C6—C7—C8—C9 50.3 (3) C7—C8—C9—C10 51.3 (3) C2—C1—C10—C5 2.1 (4)
C4—C5—C10—C1 17.8 (3) C6—C5—C10—C9 62.0 (3) C8—C9—C10—C5 59.5 (3) C25—O2—C21—C22 59.6 (3) O2—C21—C22—C23 52.9 (3) C21—C22—C23—C24 50.6 (3) C22—C23—C24—C25 52.8 (3) C21—O2—C25—C24 60.2 (3) C23—C24—C25—O2 55.0 (3)
Table 2
Hydrogen-bond geometry (A˚ ,).
D—H A D—H H A D A D—H A
O3—H3O O4i
0.82 1.91 2.714 (3) 165 O4—H4O O6i 0.82 2.17 2.968 (3) 163 Symmetry code: (i)x;y1
2;zþ1.
All H atoms were refined as riding on their parent atoms with C— H distances of 0.93–0.98 A˚ and O—H = 0.82 A˚, and withUiso(H) = 1.2Ueq(C,O). In the absence of significant anomalous scattering, Friedel pairs were merged and the absolute configuration is arbitrary. Data collection: XSCANS (Siemens, 1994); cell refinement:
XSCANS; data reduction:SHELXTL(Sheldrick, 1994; program(s) used to solve structure: SHELXS97 (Sheldrick, 1997); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997); molecular graphics:SHELXTL; software used to prepare material for publi-cation:SHELXTL.
The authors thank the National Natural Science Foundation of China (grant No. 29972017) for financial support.
References
Ahsan, M., Islam, S. K. N., Gray, A. I. & Stimson, W. H. (2003).J. Nat. Prod.
66, 958–961.
Allen F. H., Kennard. O., Waston, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987).J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
Bernardinelli, G., Vial, C., Starkemann, S. & Naf, F. (1988).Acta Cryst.C44, 715–717.
Bja˚mer, K., Ferguson, G. & Melville, R. D. (1968).Acta Cryst.B24, 855– 865.
Daux, W. L., Weeks, C. M. & Rohrer, D. C. (1974).Topics of Stereochemistry, Vol. 8, edited by E. L. Eliel & N. L. Allinger, pp. 165–187. New York: John Wiley.
Daux, W. L., Weeks, C. M. & Rohrer, D. C. (1976).Topics of Sterochemistry, Vol. 9, edited by E. L. Eliel & N. L. Allinger, pp. 279–289. New York: John Wiley.
Dimas, K., Demetzos, C., Marsellos, M., Sotiriadou, R., Malamas, M. & Kokkinopoulos, D. (1998).Planta Med.64, 208–211.
Feresin, G. E., Tapia, A., Gimenez, A., Ravelo, A. Z., Sortino, M. & Schmeda-Hirschmann, G. (2003).J. Ethnopharmacol.89, 71–80.
Hanson, J. R. (1997).Nat. Prod. Rep.14, 245–258. Hanson, J. R. (1998).Nat. Prod. Rep.15, 93–106. Hanson, J. R. (1999).Nat. Prod. Rep.16, 209–219. Hanson, J. R. (2000).Nat. Prod. Rep.17, 165–174. Hanson, J. R. (2001).Nat. Prod. Rep.18, 88–94.
Itokawa, H. & Morita, H. (1988).Planta Med.54, 117–120.
Kittakoop, P., Wanasith, S., Watts, P., Kramyu, J., Tanticharoen, M. & Thebtaranonth, Y. (2001).J. Nat. Prod.64, 385–388.
Kubo, I., Fujita, K. I., Kubo, A., Nihei, K. I. & Lunde, C. S. (2003).J. Agric. Food. Chem.51, 3951–3957.
Minami, T., Wada, S. I., Tokuda, H., Tanabe, G., Muraoka, O. & Tanaka, R. (2002).J. Nat. Prod.65, 1921–1923.
Miyazawa, M., Shimamura, H., Nakamura, S. & Kameoka, H. (1995).J. Agric. Food. Chem.43, 3012–3015.
Nagashima, F., Tanaka, H., Takaoka, S. & Asakawa, Y. (1995).Phytochemistry,
45, 353–363.
Pathak, A., Aslaoui, J. & Morin, C. (2005).J. Org. Chem.70, 4184–4187. Sheldrick, G. M. (1994).SHELXTL. Siemens Analytical X-ray Instruments
Inc., Madison, Wisconsin, USA.
Sheldrick, G. M. (1997). SHELXS97 and SHELXL97. University of Go¨ttingen, Germany.
Siemens (1994).XSCANS.Version 2.10b. Siemens Analytical X-ray Instru-ments Inc., Madison, Wisconsin, USA.
Tanaka, J., Marriott, G. & Higa, T. (2001).J. Nat. Prod.64, 1468–1470. Tavanaiepour, I., Watson, W. H., Gao, F. & Mabrg, T. J. (1987).Acta Cryst.C43,
754–756.
Urones, J. G., Marcos, I. S., Basabe, P., Sexmero, M. J., Carrillo, H. & Melchor, M. J. (1994).Phytochemistry,37, 1359–1361.
Wang, W.-S., Li, E.-W. & Jia, Z.-J. (2002).Pharmazie,57, 343–345. Yang, C.-X., Zhang, Q. & Jia, Z.-J. (2005).Pharmazie,60, 461–463.
organic papers
supporting information
sup-1
Acta Cryst. (2006). E62, o762–o764
supporting information
Acta Cryst. (2006). E62, o762–o764 [https://doi.org/10.1107/S1600536806002418]
A rearranged labdane diterpene glycoside
Ying Zhu, Cai-Xia Yang and Zhong-Jian Jia
3-methyl-5-(1,2,5,5-tetramethyl-1,2,3,4,4a,5,6,7-octahydronaphthyl)pent- 1-en-3-yl 2-O
-acetylrhamnopyanoside
Crystal data
C28H46O6
Mr = 478.65
Monoclinic, P21
Hall symbol: P 2yb
a = 11.542 (2) Å
b = 8.793 (2) Å
c = 14.095 (2) Å
β = 102.14 (1)°
V = 1398.6 (4) Å3
Z = 2
F(000) = 524
Dx = 1.137 Mg m−3
Melting point: 420 K
Mo Kα radiation, λ = 0.71073 Å Cell parameters from 42 reflections
θ = 2.7–14.3°
µ = 0.08 mm−1
T = 293 K Prism, colourless 0.58 × 0.40 × 0.16 mm
Data collection
Siemens P4 diffractometer
Radiation source: normal-focus sealed tube Graphite monochromator
ω scans
3583 measured reflections 3289 independent reflections 1841 reflections with I > 2σ(I)
Rint = 0.020
θmax = 27.1°, θmin = 1.5°
h = 0→14
k = 0→11
l = −18→17
3 standard reflections every 97 reflections intensity decay: 3.0%
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.038
wR(F2) = 0.082
S = 0.81 3289 reflections 320 parameters 2 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained
w = 1/[σ2(F
o2) + (0.04P)2]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.12 e Å−3
Δρmin = −0.13 e Å−3
Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4
supporting information
sup-2
Acta Cryst. (2006). E62, o762–o764
Special details
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq Occ. (<1)
supporting information
sup-3
Acta Cryst. (2006). E62, o762–o764
C14 0.5238 (3) 0.8442 (5) 0.6929 (2) 0.0879 (13) H14 0.4703 0.9244 0.6880 0.105* C15 0.6204 (4) 0.8740 (7) 0.6682 (2) 0.153 (2) H15A 0.6778 0.7989 0.6713 0.183* H15B 0.6341 0.9711 0.6467 0.183* C16 0.5615 (3) 0.5616 (5) 0.7199 (2) 0.0840 (12) H16A 0.5260 0.4703 0.7380 0.101* H16B 0.6379 0.5757 0.7617 0.101* H16C 0.5704 0.5534 0.6538 0.101* C17 0.2378 (3) 0.4858 (4) 1.07661 (19) 0.0737 (10) H17A 0.1938 0.5771 1.0813 0.088* H17B 0.3074 0.4839 1.1278 0.088* H17C 0.1894 0.3989 1.0823 0.088* C18 −0.0106 (3) 0.8719 (5) 0.7529 (2) 0.0892 (12) H18A −0.0409 0.9678 0.7260 0.107* H18B −0.0668 0.8260 0.7856 0.107* H18C 0.0021 0.8061 0.7017 0.107* C19 0.1867 (3) 0.9969 (4) 0.7767 (2) 0.0823 (11) H19A 0.1974 0.9490 0.7179 0.099* H19B 0.2624 1.0097 0.8198 0.099* H19C 0.1502 1.0945 0.7617 0.099* C20 0.4563 (2) 0.6392 (4) 1.04927 (16) 0.0578 (8) H20A 0.5073 0.7193 1.0362 0.069* H20B 0.5002 0.5459 1.0604 0.069* H20C 0.4261 0.6645 1.1058 0.069* C21 0.3407 (3) 0.6435 (3) 0.57607 (18) 0.0548 (8) H21 0.3920 0.7092 0.5466 0.066* C22 0.2122 (2) 0.6891 (4) 0.53807 (16) 0.0549 (8) H22 0.1984 0.7924 0.5593 0.066* C23 0.1316 (2) 0.5784 (3) 0.57355 (18) 0.0505 (7) H23 0.1431 0.5914 0.6440 0.061* C24 0.1598 (3) 0.4157 (3) 0.55485 (18) 0.0550 (8) H24 0.1401 0.3948 0.4850 0.066* C25 0.2902 (3) 0.3835 (3) 0.5949 (2) 0.0556 (8) H25 0.3068 0.3956 0.6656 0.067* C26 0.3282 (3) 0.2264 (4) 0.5707 (3) 0.0904 (12) H26A 0.3122 0.2137 0.5015 0.109* H26B 0.2851 0.1515 0.5988 0.109* H26C 0.4116 0.2142 0.5964 0.109* C27 0.1794 (2) 0.8131 (4) 0.3839 (2) 0.0558 (7) C28 0.1572 (3) 0.7892 (4) 0.27674 (19) 0.0821 (11)
supporting information
sup-4
Acta Cryst. (2006). E62, o762–o764
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
O1 0.0516 (10) 0.0610 (12) 0.0327 (9) −0.0048 (10) 0.0052 (8) −0.0039 (9) O2 0.0761 (14) 0.0573 (13) 0.0489 (11) −0.0002 (12) 0.0209 (10) −0.0087 (10) O3 0.0878 (15) 0.0541 (14) 0.0628 (12) −0.0155 (13) 0.0174 (12) −0.0022 (11) O4 0.0597 (12) 0.0703 (15) 0.0749 (14) −0.0019 (12) −0.0037 (11) 0.0141 (12) O5 0.1016 (16) 0.0496 (12) 0.0336 (10) −0.0047 (13) 0.0044 (10) 0.0008 (10) O6 0.112 (2) 0.0550 (15) 0.0699 (14) −0.0109 (14) 0.0174 (13) 0.0052 (13) C1 0.0449 (18) 0.067 (2) 0.0507 (18) 0.0019 (17) 0.0093 (14) −0.0107 (17) C2 0.071 (2) 0.067 (2) 0.077 (2) 0.0081 (19) 0.0195 (17) −0.0248 (18) C3 0.063 (2) 0.071 (2) 0.0689 (19) 0.0255 (18) 0.0143 (16) −0.0085 (17) C4 0.0580 (19) 0.063 (2) 0.0517 (17) 0.0201 (17) 0.0089 (14) 0.0002 (15) C5 0.0426 (15) 0.0528 (17) 0.0413 (15) 0.0064 (14) 0.0090 (12) −0.0006 (12) C6 0.0419 (15) 0.068 (2) 0.0571 (17) −0.0033 (17) −0.0007 (13) 0.0011 (16) C7 0.0569 (18) 0.057 (2) 0.0642 (18) −0.0100 (16) 0.0175 (15) 0.0037 (16) C8 0.0474 (18) 0.057 (2) 0.0527 (16) 0.0059 (15) 0.0122 (14) 0.0149 (14) C9 0.0351 (13) 0.0522 (17) 0.0359 (13) 0.0009 (14) 0.0071 (11) 0.0053 (12) C10 0.0373 (14) 0.0529 (18) 0.0384 (14) −0.0008 (13) 0.0124 (12) 0.0016 (13) C11 0.0405 (14) 0.0490 (17) 0.0447 (14) 0.0090 (14) 0.0077 (11) 0.0029 (13) C12 0.0438 (14) 0.066 (2) 0.0364 (14) −0.0014 (15) 0.0102 (12) −0.0020 (14) C13 0.0440 (15) 0.075 (2) 0.0406 (14) −0.0078 (17) 0.0139 (12) −0.0028 (15) C14 0.091 (3) 0.126 (4) 0.0501 (19) −0.055 (3) 0.0228 (18) −0.007 (2) C15 0.178 (4) 0.221 (6) 0.070 (2) −0.111 (5) 0.050 (3) −0.018 (3) C16 0.059 (2) 0.135 (4) 0.062 (2) 0.023 (2) 0.0213 (16) −0.011 (2) C17 0.0625 (19) 0.102 (3) 0.0606 (18) 0.001 (2) 0.0217 (15) 0.0252 (19) C18 0.081 (2) 0.100 (3) 0.074 (2) 0.051 (2) −0.0107 (18) −0.002 (2) C19 0.109 (3) 0.066 (2) 0.077 (2) 0.028 (2) 0.032 (2) 0.0172 (19) C20 0.0500 (16) 0.077 (2) 0.0438 (15) 0.0086 (16) 0.0047 (12) 0.0054 (15) C21 0.0685 (19) 0.058 (2) 0.0384 (15) −0.0054 (17) 0.0117 (14) −0.0060 (14) C22 0.077 (2) 0.0517 (18) 0.0313 (14) −0.0003 (18) 0.0016 (13) −0.0042 (14) C23 0.0582 (17) 0.0508 (19) 0.0378 (14) 0.0008 (16) −0.0009 (13) 0.0004 (14) C24 0.076 (2) 0.0504 (19) 0.0365 (15) −0.0062 (17) 0.0066 (14) −0.0028 (13) C25 0.074 (2) 0.0473 (19) 0.0487 (16) 0.0012 (17) 0.0202 (15) −0.0021 (14) C26 0.115 (3) 0.063 (3) 0.101 (3) 0.010 (2) 0.041 (2) −0.004 (2) C27 0.0613 (18) 0.053 (2) 0.0509 (17) −0.0127 (17) 0.0075 (14) 0.0059 (17) C28 0.117 (3) 0.078 (2) 0.0457 (18) −0.021 (2) 0.0045 (18) 0.0153 (17)
Geometric parameters (Å, º)
O1—C21 1.407 (3) C12—H12B 0.970
O1—C13 1.455 (3) C13—C16 1.509 (4)
O2—C21 1.403 (3) C13—C14 1.512 (5)
O2—C25 1.444 (3) C14—C15 1.264 (4)
O3—C24 1.437 (3) C14—H14 0.930
O3—H3O 0.820 C15—H15A 0.930
O4—C23 1.429 (3) C15—H15B 0.930
supporting information
sup-5
Acta Cryst. (2006). E62, o762–o764
O5—C27 1.335 (4) C16—H16B 0.960
O5—C22 1.454 (3) C16—H16C 0.960
O6—C27 1.198 (4) C17—H17A 0.960
C1—C10 1.328 (4) C17—H17B 0.960
C1—C2 1.493 (4) C17—H17C 0.960
C1—H1 0.943 (10) C18—H18A 0.960
C2—C3 1.526 (4) C18—H18B 0.960
C2—H2A 0.970 C18—H18C 0.960
C2—H2B 0.970 C19—H19A 0.960
C3—C4 1.532 (4) C19—H19B 0.960
C3—H3A 0.970 C19—H19C 0.960
C3—H3B 0.970 C20—H20A 0.960
C4—C19 1.533 (4) C20—H20B 0.960
C4—C18 1.538 (4) C20—H20C 0.960
C4—C5 1.542 (4) C21—C22 1.521 (4)
C5—C10 1.517 (3) C21—H21 0.980
C5—C6 1.526 (4) C22—C23 1.503 (4)
C5—H5 0.980 C22—H22 0.980
C6—C7 1.526 (4) C23—C24 1.503 (4)
C6—H6A 0.970 C23—H23 0.980
C6—H6B 0.970 C24—C25 1.519 (4)
C7—C8 1.537 (4) C24—H24 0.980
C7—H7A 0.970 C25—C26 1.510 (4)
C7—H7B 0.970 C25—H25 0.980
C8—C17 1.534 (3) C26—H26A 0.960
C8—C9 1.561 (4) C26—H26B 0.960
C8—H8 0.980 C26—H26C 0.960
C9—C10 1.521 (4) C27—C28 1.492 (4)
C9—C20 1.532 (3) C28—H28A 0.960
C9—C11 1.550 (3) C28—H28B 0.960
C11—C12 1.525 (4) C28—H28C 0.960
C11—H11A 0.970 C28—H28D 0.960
C11—H11B 0.970 C28—H28E 0.960
C12—C13 1.538 (3) C28—H28F 0.960
C12—H12A 0.970
C21—O1—C13 119.29 (19) H15A—C15—H15B 120.0 C21—O2—C25 114.0 (2) C13—C16—H16A 109.5 C24—O3—H3O 109.5 C13—C16—H16B 109.5 C23—O4—H4O 109.5 H16A—C16—H16B 109.5 C27—O5—C22 117.9 (2) C13—C16—H16C 109.5 C10—C1—C2 125.4 (3) H16A—C16—H16C 109.5 C10—C1—H1 117.6 (17) H16B—C16—H16C 109.5 C2—C1—H1 117.0 (17) C8—C17—H17A 109.5 C1—C2—C3 111.5 (3) C8—C17—H17B 109.5 C1—C2—H2A 109.3 H17A—C17—H17B 109.5
C3—C2—H2A 109.3 C8—C17—H17C 109.5
supporting information
sup-6
Acta Cryst. (2006). E62, o762–o764
C3—C2—H2B 109.3 H17B—C17—H17C 109.5 H2A—C2—H2B 108.0 C4—C18—H18A 109.5 C2—C3—C4 112.0 (2) C4—C18—H18B 109.5 C2—C3—H3A 109.2 H18A—C18—H18B 109.5
C4—C3—H3A 109.2 C4—C18—H18C 109.5
C2—C3—H3B 109.2 H18A—C18—H18C 109.5 C4—C3—H3B 109.2 H18B—C18—H18C 109.5 H3A—C3—H3B 107.9 C4—C19—H19A 109.5 C3—C4—C19 109.7 (3) C4—C19—H19B 109.5 C3—C4—C18 109.8 (2) H19A—C19—H19B 109.5 C19—C4—C18 108.2 (3) C4—C19—H19C 109.5 C3—C4—C5 109.0 (2) H19A—C19—H19C 109.5 C19—C4—C5 108.8 (2) H19B—C19—H19C 109.5 C18—C4—C5 111.2 (3) C9—C20—H20A 109.5 C10—C5—C6 107.7 (2) C9—C20—H20B 109.5 C10—C5—C4 113.3 (2) H20A—C20—H20B 109.5 C6—C5—C4 116.2 (2) C9—C20—H20C 109.5 C10—C5—H5 106.3 H20A—C20—H20C 109.5 C6—C5—H5 106.3 H20B—C20—H20C 109.5
C4—C5—H5 106.3 O2—C21—O1 112.7 (2)
C5—C6—C7 111.7 (2) O2—C21—C22 111.7 (2) C5—C6—H6A 109.3 O1—C21—C22 105.1 (2)
C7—C6—H6A 109.3 O2—C21—H21 109.1
C5—C6—H6B 109.3 O1—C21—H21 109.1
C7—C6—H6B 109.3 C22—C21—H21 109.1
H6A—C6—H6B 107.9 O5—C22—C23 108.7 (2) C6—C7—C8 113.9 (2) O5—C22—C21 107.5 (2) C6—C7—H7A 108.8 C23—C22—C21 109.8 (2)
C8—C7—H7A 108.8 O5—C22—H22 110.3
C6—C7—H7B 108.8 C23—C22—H22 110.3
C8—C7—H7B 108.8 C21—C22—H22 110.3
H7A—C7—H7B 107.7 O4—C23—C24 112.2 (2) C17—C8—C7 109.7 (2) O4—C23—C22 109.2 (2) C17—C8—C9 113.0 (2) C24—C23—C22 112.7 (2) C7—C8—C9 112.1 (2) O4—C23—H23 107.5
C17—C8—H8 107.3 C24—C23—H23 107.5
C7—C8—H8 107.3 C22—C23—H23 107.5
supporting information
sup-7
Acta Cryst. (2006). E62, o762–o764
C12—C11—H11A 108.4 C24—C25—H25 108.9 C9—C11—H11A 108.4 C25—C26—H26A 109.5 C12—C11—H11B 108.4 C25—C26—H26B 109.5 C9—C11—H11B 108.4 H26A—C26—H26B 109.5 H11A—C11—H11B 107.5 C25—C26—H26C 109.5 C11—C12—C13 114.0 (2) H26A—C26—H26C 109.5 C11—C12—H12A 108.8 H26B—C26—H26C 109.5 C13—C12—H12A 108.8 O6—C27—O5 123.1 (3) C11—C12—H12B 108.8 O6—C27—C28 124.7 (3) C13—C12—H12B 108.8 O5—C27—C28 112.2 (3) H12A—C12—H12B 107.7 C27—C28—H28A 109.5 O1—C13—C16 111.7 (3) C27—C28—H28B 109.5 O1—C13—C14 106.4 (2) H28A—C28—H28B 109.5 C16—C13—C14 114.3 (3) C27—C28—H28C 109.5 O1—C13—C12 102.89 (18) H28A—C28—H28C 109.5 C16—C13—C12 111.7 (2) H28B—C28—H28C 109.5 C14—C13—C12 109.3 (3) C27—C28—H28D 109.5 C15—C14—C13 128.8 (5) C27—C28—H28E 109.5 C15—C14—H14 115.6 H28D—C28—H28E 109.5 C13—C14—H14 115.6 C27—C28—H28F 109.5 C14—C15—H15A 120.0 H28D—C28—H28F 109.5 C14—C15—H15B 120.0 H28E—C28—H28F 109.5
supporting information
sup-8
Acta Cryst. (2006). E62, o762–o764
C4—C5—C10—C1 17.8 (3) C22—C23—C24—O3 172.2 (2) C6—C5—C10—C9 62.0 (3) O4—C23—C24—C25 176.6 (2) C4—C5—C10—C9 −168.0 (2) C22—C23—C24—C25 52.8 (3) C20—C9—C10—C1 −7.7 (4) C21—O2—C25—C26 −176.4 (2) C11—C9—C10—C1 −127.9 (3) C21—O2—C25—C24 60.2 (3) C8—C9—C10—C1 114.5 (3) O3—C24—C25—O2 −172.3 (2) C20—C9—C10—C5 178.3 (2) C23—C24—C25—O2 −55.0 (3) C11—C9—C10—C5 58.1 (3) O3—C24—C25—C26 68.1 (3) C8—C9—C10—C5 −59.5 (3) C23—C24—C25—C26 −174.5 (2) C10—C9—C11—C12 51.7 (3) C22—O5—C27—O6 −0.6 (4) C20—C9—C11—C12 −72.4 (3) C22—O5—C27—C28 179.6 (2) C8—C9—C11—C12 169.2 (2)
Hydrogen-bond geometry (Å, º)
D—H···A D—H H···A D···A D—H···A
O3—H3O···O4i 0.82 1.91 2.714 (3) 165
O4—H4O···O6i 0.82 2.17 2.968 (3) 163