Inhibitory interactions of Coenzyme Q10 with selected key enzymes of glucose metabolism: An in silico approach
Full text
Figure

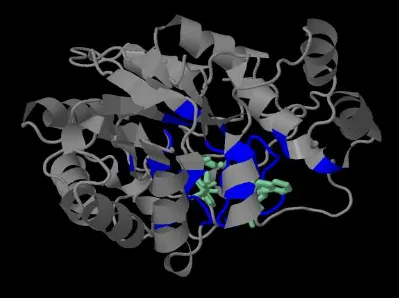
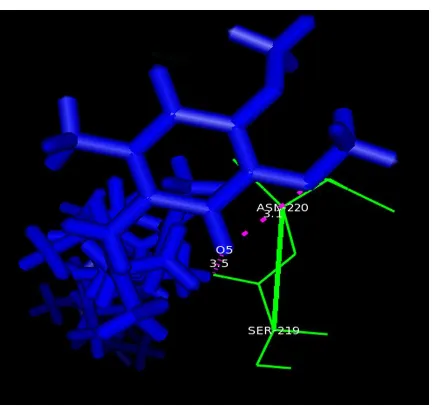
Related documents
The gene expression level is presented as a Delta Ct (Cycle threshold) using the HPRT expression as a reference value. Although there were slight differences in the levels of
• Protein Ligand Binding Site Pair Residue (PLBSP Residue) is a RDFized graph database of ligand-binding residues. • This database contains the protein–ligand interaction
[r]
● In case of round draw part with central hole, and with Progressive piercing option checked, you can change the Piercing diameter at the selected stage, then the Stamping height
Purpose: The aim of this study was to investigate whether Chikungunya virus infection (CVI) was an independent risk factor for 2-year mortality in Afro-Caribbean subjects aged 65
Ligand 4A did not form a hydrogen bond with the amino acid residues in the active side of NS5B polymerase enzyme.. Fig 11: Interacting residues of ligand 5A
As an alternative education program, we also track completion of learning modules in a learning period (persistence), credit completion for long-term LW students (credit
It would equally be a a legal and logical nonsense to make provision for use of works whose authors are not identified, while would-be users can remove identification from any