A Protein Secondary Structure Prediction Method Based on BP Neural Network
Full text
Figure
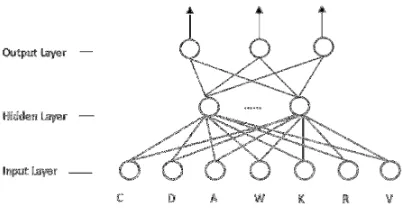
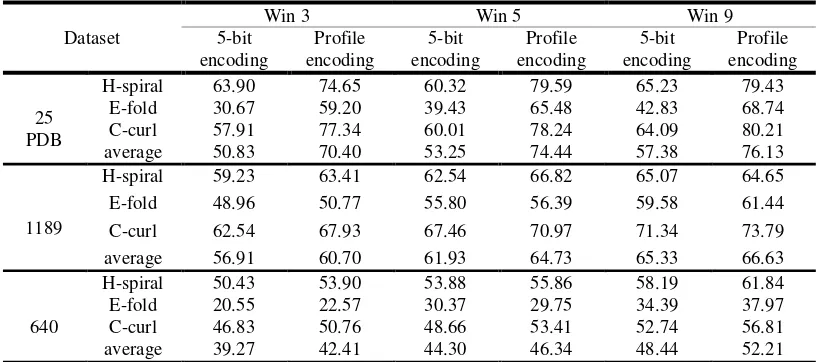
Related documents
(E) Strong MIF mRNA (purple) expression and macrophage accumulation (brown-purple) in a damaged tubules of UA groups, where some evidence for deposition of uric acid crystals
To avoid a potential collision with the ACK (as discussed earlier), the source node transmits DATA at the power level pmax, periodically, for just enough time so that
1. Particle size and surface characteristics of nanoparticles can be easily manipulated to achieve both passive and active drug targeting after
3 Department of Clinical Genetics, Karolinska University Hospital, Stockholm, Sweden.. 4 Division of Pediatric Orthopaedics, Seoul National University Children ’ s Hospital,
tography revealed a minor peak coeluting with human insulin and a major peak of proinsulin-like materials. The insulin gene of the patient was amplified by the polymerase
Three analyses were done; one analysis (analysis A) involved only patients with interventions, one analysis (analysis B) involved all patients for whom antimicrobial testing was
Ischemia resulted in a modest increase of short-chain acyl- carnitines in anterior, ischemic zones in animals that had not been treated with POCA compared with values in
Treatment of healthy rats and mice with a single intravenous injection of recombinant human tumor necrosis factor-alpha (rHuTNF-alpha) caused a dose-dependent..