Development 139, 3838-3848 (2012) doi:10.1242/dev.080994 © 2012. Published by The Company of Biologists Ltd
INTRODUCTION
Context-dependent regulation of gene expression is key to the generation of cell-type diversity during development. RE1-silencing transcription factor [Rest; also known as Neuron-restrictive silencing factor (Nrsf)] is thought to play a central role in the transcriptional repression necessary to achieve neural-specific gene expression (Chong et al., 1995; Schoenherr and Anderson, 1995). The ~23 bp repressor element 1/neuron-restrictive silencing element (RE1/NRSE) is the binding site for Rest and is associated with hundreds of neural genes in mammals (Bruce et al., 2004; Lunyak et al., 2002). Analysis of other vertebrate genomes, including zebrafish, has revealed that many possess rest homologs and comparable numbers of RE1 sites (Mortazavi et al., 2006).
Rest alters chromatin structure by mediating the assembly of multiple chromatin-modifying complexes. The N-terminal repressor domain of Rest recruits Sin3, which dynamically interacts with a wide range of repressor proteins including the methyl-DNA-binding protein MeCP2 and histone deacetylases (HDACs) 1/2 (Grzenda et al., 2009; Naruse et al., 1999). The Rest C-terminal
domain interacts with CoREST (Ballas et al., 2001; Grimes et al., 2000) to assemble complexes that include HDACs 1/2 and histone H3 K9 methyltransferases (Lunyak et al., 2002; Roopra et al., 2004; Shi et al., 2003). The full repertoire of genes under the control of Rest is not regulated in unison and the context-specific requirements for Rest-mediated repression are unknown.
Initially, the chief function of Rest was considered to be the repression of neural genes in non-neural tissues. Recently, a more complex understanding of Rest has emerged. Rest also modulates neural gene expression within developing neurons and embryonic stem (ES) cells (Ballas et al., 2005; Singh et al., 2008). In these contexts, repression is transient, and Rest is proposed to delay target gene expression until the proper time. Consistent with this proposal, the transition from ES cell to neural progenitor to differentiated neuron is accompanied by reduction in Rest levels (Ballas et al., 2005; Guardavaccaro et al., 2008; Westbrook et al., 2008). However, the function of Rest in cultured stem and progenitor cells is controversial. Studies have reached opposing conclusions on the requirement for Rest as a regulator of stem cell pluripotency. Some have concluded that Rest is a key regulator of pluripotency or of the emergence of the neural lineage from stem cells (Singh et al., 2008; Sun et al., 2008; Gupta et al., 2009), whereas other studies differ in this view (Jørgensen et al., 2009a; Jørgensen et al., 2009b; Singh et al., 2008; Westbrook et al., 2008). Because a variety of factors influence the ability of cells to divide and differentiate, the outcomes of such experiments vary depending on the cell line and culture conditions employed.
Rest loss-of-function analysis has yet to provide a clear view of its role during embryogenesis. In the mouse Rest knockout, widespread cell death accompanies lethality by E11.5 (Chen et al., 1998). Expression of a few neural genes was observed in non-neural tissues, but the early lethality precluded further analysis; 1Department of Neurobiology and Behavior, Stony Brook University, Stony Brook,
NY 11794, USA. 2Graduate Program in Molecular and Cellular Biology, Stony Brook
University, Stony Brook, NY 11794, USA. 3Department of Organismal Biology and
Anatomy, University of Chicago, Chicago, IL 60637, USA. 4Department of Cellular
Biology and Anatomy, Georgia Health Sciences University, Augusta, GA 30912, USA.
5Genetics Graduate Program, Stony Brook University, Stony Brook, NY 11794, USA. 6Committee on Development, Regeneration and Stem Cell Biology, The University of
Chicago, Chicago, IL 60637, USA.
*Author for correspondence (hsirotkin@sunysb.edu)
Accepted 18 July 2012 SUMMARY
The transcriptional repressor Rest (Nrsf) recruits chromatin-modifying complexes to RE1 ‘silencer elements’, which are associated with hundreds of neural genes. However, the requirement for Rest-mediated transcriptional regulation of embryonic development and cell fate is poorly understood. Conflicting views of the role of Rest in controlling cell fate have emerged from recent studies. To address these controversies, we examined the developmental requirement for Rest in zebrafish using zinc-finger nuclease-mediated gene targeting. We discovered that germ layer specification progresses normally in restmutants despite derepression of target genes during embryogenesis. This analysis provides the first evidence that maternal restis essential for repression of target genes during blastula stages. Surprisingly, neurogenesis proceeds largely normally in restmutants, although abnormalities are observed within the nervous system, including defects in oligodendrocyte precursor cell development and a partial loss of facial branchiomotor neuron migration. Mutants progress normally through embryogenesis but many die as larvae (after 12 days). However, some homozygotes reach adulthood and are viable. We utilized an RE1/NRSE transgenic reporter system to dynamically monitor Rest activity. This analysis revealed that Rest is required to repress gene expression in mesodermal derivatives including muscle and notochord, as well as within the nervous system. Finally, we demonstrated that Rest is required for long-term repression of target genes in non-neural tissues in adult zebrafish. Our results point to a broad role for Rest in fine-tuning neural gene expression, rather than as a widespread regulator of neurogenesis or cell fate.
KEY WORDS: Neurogenesis, REST (NRSF), RE1/NRSE
Zebrafish
rest
regulates developmental gene expression but
not neurogenesis
Fatma O. Kok1,2, Andrew Taibi1, Sarah J. Wanner3, Xiayang Xie4, Cara E. Moravec1,5, Crystal E. Love3,6, Victoria E. Prince3,6, Jeff S. Mumm4and Howard I. Sirotkin1,2,5,*
D
E
V
E
LO
P
M
E
N
specific molecular and cellular defects underlying the lethality remain unknown. In chick, inhibiting Rest with a dominant-negative construct resulted in ectopic expression of a few neural genes in non-neural tissues, but not in alterations of fate (Chen et al., 1998). Blocking Rest activity in Xenopusresulted in ectodermal patterning defects and the derepression of target genes (Olguín et al., 2006a). Previously, we knocked down zebrafish Rest using morpholinos and observed alterations of Hedgehog signaling (Gates et al., 2010).
To circumvent caveats associated with knockdowns and dominant-negative constructs, we employed zinc-finger nucleases to disrupt zebrafish Rest. Our in-depth analysis of the rest mutant established that expression of RE1-containing genes is enhanced in the mutant, and that blastula stage repression of some target genes requires maternal rest. Unexpectedly, neurogenesis progresses largely normally in rest mutants, although abnormalities were observed in oligodendrocyte precursor cells and in facial branchiomotor neuron migration. Analysis of an RE1/NRSE transgenic reporter system revealed that Rest represses gene expression in mesodermal derivatives including muscle and notochord, as well as within the developing nervous system. Although most rest homozygous zebrafish mutants die during larval stages, some survive to maturity. In these fish, some RE1-containing genes are inappropriately expressed in adult organs, demonstrating a long-term role for Rest in the repression of target genes in non-neural tissues. Together, these results establish Rest as a repressor of gene expression during embryogenesis and in adult non-neural tissue, but not as a broad regulator of cell fate or differentiation.
MATERIALS AND METHODS Zinc-finger nuclease gene targeting
ZiFiT (http://www.zincfingers.org/software-tools.htm) was used to identify zinc-finger nuclease (ZFN) target sites that exclusively comprise GXX sequences. Two zinc-finger arrays were designed against each ‘half-target’ site (supplementary material Table S1). Conventional cloning strategies were employed to assemble arrays from clones in the Addgene ZF Kit. Each ZF array was cloned into KpnI/BamHI sites of the FokI-RR/FokI-DD vectors (Addgene). ZFN mRNA was synthesized using the mMESSAGE mMACHINE Kit (Ambion) and 10-25 pg of mRNA was injected into blastomeres of one-cell embryos. At 24 hours postfertilization (hpf), embryos were screened by PCR using primers 181 (5⬘CTGA -GGGGAAGCAGATGATG-3⬘) and 184 (5⬘TGTCCATGCTGTATCT -CACGA-3⬘).
Quantitative PCR
Total RNA was extracted from tissues or pools of five embryos using Trizol (Invitrogen). SuperScript II reverse transcriptase (Invitrogen) was used to synthesize cDNA from 0.2-0.5 g of total RNA. Quantitative (q) PCR was carried out with a LightCycler 480 (Roche) using 2⫻FastStart SYBR Green Master (Roche) or Quanta SYBR Green (Quanta Biosciences). Total RNA from each sample was normalized to -actin. In each experiment, three pools of three to five embryos or one adult organ were run in duplicate. Primer pairs are listed in supplementary material Table S3.
TSA treatments
Pronased embryos were placed in 40 nM Trichostatin A (TSA) or DMSO at the eight-cell stage and collected at 4 hpf for mRNA isolation.
Whole-mount in situ hybridization and immunostaining
Whole-mount in situ hybridization was performed as described by Thisse et al. (Thisse et al., 1993). Rest immunostaining was performed as described (Mapp et al., 2011).
RESULTS
Targeted restdisruption with zinc-finger nucleases
We disrupted zebrafish rest(Fig. 1; supplementary material Tables S1, S2) using zinc-finger nucleases (ZFNs) (Doyon et al., 2008; Kim et al., 1996; Meng et al., 2008) and recovered ten mutations in the first exon of rest. The majority of our studies utilized the restsbu29 allele (a 7 bp deletion), but we also examined viability of the restsbu34 allele (a 4 bp insertion). Both mutations produce similar frameshifts upstream of the DNA-binding domain and are likely null alleles.
[image:2.612.326.548.57.332.2]Unlike the mouse Restknockout, restsbu29/sbu29and restsbu34/sbu34 embryos appear grossly normal throughout embryogenesis and early larval stages [10 days postfertilization (dpf)]. However, adult mutants were not recovered at the expected Mendelian ratios. Of adult offspring of restsbu29/+ intercrosses, only 17/214 (7.9%) were homozygous mutants. Likewise, only 3/237 (1.3%) of adult offspring from restsbu34/+intercrosses were homozygous mutants. A subset of the restsbu29/sbu29adults had posterior defects, while all three of the restsbu34/sbu34mutants were scrawny, had a severely curved axis and died by 3 months. Some surviving restsbu29/sbu29fish were fertile. Although we cannot exclude the possibility that the Restsbu34protein has antimorphic activity, it is likely that the phenotypic variations stem from genetic background variations within the wild-type strain that was used for targeting. The surviving mutants provided a unique opportunity to study the requirement for Rest in larvae and adults. Fig. 1. Zinc-finger nuclease-mediated targeting of zebrafish rest. (A)Sequence chromatograms from wild-type siblings (left) and rest
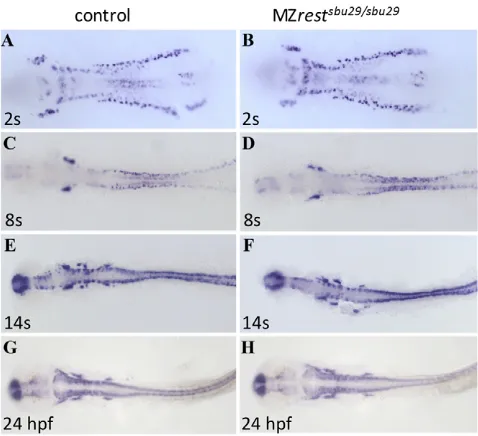
mutants (right). The 7 bp deletion (7) in SBU29 and 4 bp insertion (+4) in SBU34 are illustrated beneath. (B)Domain structure of wild-type Rest and truncated proteins produced by SBU29 and SBU34 mutations. (C)Animal pole views of 6-hpf wild-type and MZrestsbu29/sbu29embryos. Rest immunoreactivity, TOPRO-3 staining of nuclei, and merged channels are shown. Abundant cytoplasmic Rest protein is detected in wild-type embryos, but not in MZrestsbu29/sbu29mutants. Confocal images are single 1-m stacks taken at 40⫻magnification.
D
E
V
E
LO
P
M
E
N
RT-PCR analysis of restsbu29/sbu29 mutants revealed that both wild-type and restsbu29 messages are present at 6 hpf (early gastrula), but not at later stages (data not shown). The presence of wild-type restmRNA in restsbu29/sbu29gastrulae reflects maternal restexpression and is consistent with our previous analysis (Gates et al., 2010). The lack of morphological defects in restmutant embryos might be due to compensation by maternal restmRNA. To eliminate maternal Rest activity, we generated embryos devoid of both maternal and zygotic rest(MZrestsbu29/sbu29) by crossing homozygous restsbu29/sbu29females and males. To our surprise, these mutants also lacked overt morphological defects, demonstrating that Rest is dispensable for embryogenesis in zebrafish.
The nature of the restlesions (indels) and their position within the first exon make it unlikely that either readthrough or alternative splicing can produce functional protein, as the first exon encodes zinc fingers that constitute part of the DNA-binding domain. To confirm the absence of Rest protein, we performed immunohistochemistry, which revealed abundant Rest protein in wild-type 6-hpf embryos but not in MZrestsbu29/sbu29mutants (Fig. 1C). This supports the conclusion that SBU29 is a null allele.
Rest represses target gene expression during embryogenesis
The zebrafish genome has ~1000 canonical RE1 sites (Mortazavi et al., 2006) (our unpublished results) and mammalian Rest binds teleost RE1 elements (Johnson et al., 2009; Tan et al., 2010). To
determine whether loss of Rest affects target gene expression, we selected a set of RE1-containing genes with diverse cellular functions to analyze in the mutant. We studied their expression by qRT-PCR in MZrestsbu29/sbu29and wild-type embryos between 4 hpf (late blastula) and 16 hpf (mid-somitogenesis) (Fig. 2). During this time, expression of some Rest target genes was enhanced in mutants. At 4 hpf, when the embryo comprises pluripotent blastomeres, the expression levels of most RE1-containing genes are either very low or absent in wild-type embryos (Fig. 2). At this stage, transcripts of gpr27,kcnh8and snap25bcan be detected in MZrestsbu29/sbu29mutants, but not in controls (Fig. 2A-C). Similarly, during somitogenesis (11 and 16 hpf), expression of these genes is low or absent in wild-type embryos, but much more robust in the mutant. Injection of morpholinos to impede resttranslation also enhanced snap25bexpression (data not shown).
[image:3.612.57.523.57.374.2]Expression of bdnf, snap25a, grin1a, pcad (pcdh1g2) and cacng2 was enhanced in a stage-dependent manner in MZrestsbu29/sbu29. However, we identified a group of Rest target genes (neurod, spop, gfap and bsx) that showed unchanged expression levels in MZrestsbu29/sbu29mutants, suggesting that both genomic context and cell type influence Rest activity. Consistent with our findings, previous studies have shown that Rest function is cell type dependent, and that blocking Rest does not derepress all target genes (Belyaev et al., 2004; Chen et al., 1998; Otto et al., 2007). Our findings provide the first evidence that Rest is required to suppress target gene expression in blastulae.
Fig. 2. Expression levels of RE1-containing genes during early development of restmutants.qPCR analysis showing fold differences relative to MZrestsbu29/sbu29transcript levels (defined as 1). (A-C)gpr27, kcnh8and snap25bare either not expressed or expressed at low levels in wild-type zebrafish embryos, but are expressed in MZrestsbu29/sbu29mutants. (D-F)bdnf, snap25aand grin1ashow stage-specific alterations in expression in restmutants. (G-L)Expression of pcad, cacng2,neurod,gfap,spopand bsxis not significantly altered in the restmutant. Error bars indicate s.e. ND*, not detected.
D
E
V
E
LO
P
M
E
N
Maternal Rest function is required for early gene regulation
Our initial experiments did not distinguish the contributions of zygotic versus maternal Rest to the repression of target genes. To elucidate the role of maternal Rest in the regulation of RE1-containing target genes, we compared gene expression in progeny from restsbu29/sbu29 females and wild-type males (maternal, Mrestsbu29/+embryos) to offspring of restsbu29/sbu29males and wild-type females (zygotic, Zrestsbu29/+embryos) as well as wild-type controls. Although Zrestsbu29/+fish have the same genotype as Mrestsbu29/+ fish, early Rest activity stems from maternal rest mRNA that is absent from Mrestsbu29/+embryos.
Four RE1-containing genes that are expressed at blastula stages were used for this analysis. qRT-PCR demonstrated that loss of maternal restfunction increased levels of snap25b, bdnfand pcad at mid-blastula stage (Fig. 3A-C). In the reciprocal experiment, the expression levels of the RE1-containing target genes in Zrestsbu29/+ embryos were more similar to wild-type levels (Fig. 3A-C), suggesting that maternal Rest function is both necessary and sufficient for early repression of RE1-containing genes. gpr27 expression was undetectable in all three genotypes (Fig. 3D), indicating that either maternal or zygotic rest is sufficient for repression of this locus. These findings provide the first evidence that maternal restsuppresses zygotic gene expression.
REST-mediated histone modifications in blastulae are target gene dependent
Rest recruits chromatin-modifying enzymes to RE1/NRSE sites to alter histone acetylation and methylation states (Ballas et al., 2001; Huang et al., 1999). To determine whether effects on histone acetylation are central to the function of Rest during blastula stages, we examined the effects of the HDAC inhibitor TSA on target gene expression in wild-type and MZrestsbu29/sbu29embryos (Fig. 4). The difference between the effects of TSA on wild type versus rest mutants reflects the relative contribution of HDAC-independent activities of Rest, namely histone methylation. At least three response profiles were apparent among the Rest target genes analyzed.
The expression levels of snap25aand gpr27 in TSA-treated wild-type embryos were comparable to DMSO-treated MZrestsbu29/sbu29 embryos (Fig. 4A,B). Alone, these findings suggest that HDAC recruitment might be the major component of Rest function at these loci. However, examination of transcript levels in TSA-treated MZrestsbu29/sbu29 mutants revealed more intricate effects of Rest on chromatin. At both loci the impact of HDAC inhibition was greater in the absence of Rest than in wild-type embryos. This implies that Rest-mediated effects on histone methylation act cooperatively with HDACs recruited by other transcriptional regulators to repress transcription of these loci.
snap25bexpression showed a very different response to TSA. Wild-type TSA-treated embryos showed a 3-fold increase in snap25blevels compared with DMSO-treated controls (Fig. 4C). In comparison, DMSO-treated restmutants and TSA-treated rest mutants showed similar ~57-fold increases relative to wild-type snap25blevels. These results show that, whereas blocking histone deacetylation minimally enhances the expression of snap25bin wild-type embryos at mid-blastula stage, the effects of Rest inhibition are far greater. This demonstrates that HDAC-independent Rest modifications play a major role at the snap25b locus in blastulae.
[image:4.612.48.306.59.201.2]pcad, spopand grin1awere also challenged with TSA, but their expression profiles were not statistically different in wild type and restmutants (Fig. 4D-F). Although we cannot distinguish direct from indirect actions of HDAC inhibition, taken together these Fig. 3. Maternal restsuppresses target genes in mid-blastula
embryos.qPCR analysis of (A) snap25b, (B) bdnf, (C) pcad and (D) gpr27expression in mid-blastula zebrafish embryos. Comparison of transcript levels in wild-type, Zrestsbu29/+and Mrestsbu29/+embryos reveals derepression of target genes in Mrestbut not Zrestembryos. Fold comparisons are relative to Mresttranscript levels (defined as 1). Maternal (M) and zygotic (Z) embryos were obtained by crossing female and male restsbu29/sbu29mutants, respectively, to wild types. Error bars indicate s.e. ND*, not determined.
Fig. 4. Histone deacetylase contribution to Rest-mediated inhibition of target genes is locus specific.qPCR analysis of RE1-containing gene expression at mid-blastula stage in wild-type and MZrestsbu29/sbu29zebrafish embryos treated with DMSO or the HDAC inhibitor TSA. The fold differences are relative to transcript levels in MZrestsbu29/sbu29DMSO (defined as 1). (A,B)TSA treatment enhances expression of snap25aand gpr27in both MZrestand wild-type embryos compared with DMSO controls. (C)TSA treatment of wild-type embryos produces a much smaller increase in snap25bexpression than does removing Rest function. This suggests that Rest-dependent histone methylation is central to repression of snap25bexpression. (D-F)HDAC inhibition has little effect on pcad, spopand grin1a
expression. Error bars indicate s.e.
D
E
V
E
LO
P
M
E
N
[image:4.612.310.564.60.285.2]observations reveal complex regulatory interactions at Rest target genes with histone deacetylation playing dramatically different roles at different loci. Surprisingly, the two zebrafish snap25 orthologs respond differently to TSA treatment, with repression of snap25bbeing almost entirely independent of HDAC activity.
We next asked whether key Rest co-factors were expressed in zebrafish blastulae. qRT-PCR analysis revealed that rcor1(Corest1), rcor2(Corest2), sin3aa,sin3abandsin3btranscripts are present at blastula stage (supplementary material Fig. S1). Of these markers, only the expression of rcor1 was enhanced in MZrestsbu29/sbu29 mutants (supplementary material Fig. S1). We analyzed rcor1 expression by RNA in situ hybridization and observed that many domains of expression overlap with restexpression (data not shown) and that rcor1 expression is slightly more robust in the mutant.
Rest is not required for germ layer formation in zebrafish
Studies from other organisms have not yielded a clear picture of the role for Rest during early development. In Restknockout mice, midbrain head mesenchyme and myotomal cells in the somite are disorganized (Chen et al., 1998). However, overall germ layer formation and early patterning seem to be minimally affected (Chen et al., 1998). By contrast, perturbing XenopusRest expands the neural plate and reduces neural crest (Olguín et al., 2006b). To study whether Rest function is necessary for germ layer specification in zebrafish, we analyzed the expression of early patterning genes in MZrestsbu29/sbu29mutants. During gastrulation, expression of mesodermal (chd, ntl, myod1, foxa2), endodermal (foxa2) and neural (pax3) markers was not altered in MZrestsbu29/sbu29mutants (supplementary material Fig. S2). These results suggest that germ layer specification does not require Rest.
Neurogenesis progresses normally in restmutants Rudimentary observation of restmutant locomotor behavior and gross neural anatomy did not reveal abnormalities. To further investigate the requirement for Rest in neurogenesis, the domains of proneural and pan-neural markers were assayed in MZrestsbu29/sbu29 mutants by RNA in situ hybridization. Examination of the proneural markers ascl1aand neurog1did not reveal abnormalities in MZrestsbu29/sbu29mutants (supplementary material Fig. S3). If Rest modulates the differentiation of neural progenitors, loss of Rest function might result in ectopic or premature neural differentiation. We examined expression of the pan-neural marker huC (elavl3) in MZrestsbu29/sbu29 mutants between 10.5 hpf (2 somites) and 24 hpf. To our surprise, we did not detect inappropriate huCexpression in MZrestsbu29/sbu29mutants (Fig. 5). We also examined the expression of an huC:GFP transgenic reporter in the MZrestsbu29/sbu29mutants between 1 dpf and 5 dpf, but did not observe alterations in expression (data not shown). From these results, we conclude that Rest does not play a fundamental role in zebrafish neurogenesis.
olig2+oligodendrocyte precursors are reduced in restmutants
Neural fate determination appears grossly normal in restmutants. To study subtle differences in neural subpopulations that lack functional Rest, we examined olig2+ cells in MZrestsbu29/sbu29 using an olig2:GFP transgene (Fig. 6). This transgenic line marks progenitors in the ventral spinal cord domain that give rise to motor neurons and oligodendrocyte precursor cells (OPCs). By 3 dpf, OPCs occupy distinct positions after migrating dorsally from the progenitor population (Park et al., 2002). The ventral spinal cord expression of
olig2:GFP was not altered in restmutants, but a deficit was apparent in the number of OPCs in the restmutants at 50 hpf. On average, the restmutants had only 13.4±2.4 OPCs (± s.e.; n16) compared with 37.5±7.0 (n8) OPCs in sibling controls. OPC deficiencies remain at 74 hpf (data not shown), but by 98 hpf the number of OPCs in the mutants had recovered to levels comparable to sibling controls (Fig. 6). Similar deficits were observed in OPCs following Rest knockdown using morpholinos (data not shown).
As in other species, the zebrafish OPC population is tightly regulated and compensatory mechanisms can overcome deficits (Kirby et al., 2006). The zebrafish restmutant phenotype resembles the mouse Olig1knockout, in which early deficits of OPCs are subsequently overcome (Lu et al., 2002). Recent studies have provided evidence that Rest regulates glial differentiation, including emergence of oligodendrocytes (Abrajano et al., 2009; DeWald et al., 2011; Soldati et al., 2012). Our in vivo results implicate Rest-regulated gene expression in the development of the OPC lineage in zebrafish.
Facial branchiomotor neuron migration is impaired in restmutants
[image:5.612.318.557.60.278.2]Facial branchiomotor neurons (FBMNs) are a subset of the cranial branchiomotor neurons common to all vertebrates. Zebrafish FBMNs are born in rhombomere (r) 4 of the hindbrain and migrate tangentially to r6-r7 (Chandrasekhar, 2004). Migration begins after the first neurons are born at 16 hpf and is complete by 48 hpf, when the FBMNs have reached their final destination in r6-r7. Rest is expressed in FBMNs (supplementary material Fig. S4) and inhibition of Rest function by morpholino knockdown or a Fig. 5. Neurogenesis progresses normally in restmutants. Whole-mount in situ hybridization for the pan-neural marker huCin stage-matched (A,B) 2-somite, (C,D) 8-somite, (E,F) 14-somite and (G,H) 24-hpf wild type or restheterozygotes and MZrestsbu29/sbu29mutants.All views are dorsal, anterior to the left. MZrestmutants did not show ectopic or precocious huCexpression. To control for subtle differences in developmental timing and genetic background, comparisons were undertaken in crosses of restsbu29/sbu29females to restsbu29/+males and between wild-type intercrosses and MZrestsbu29/sbu29intercrosses. Genotypes were determined by PCR.
D
E
V
E
LO
P
M
E
N
dominant-negative approach was previously shown to cause defects in FBMN migration (Mapp et al., 2011). Using an islet1:GFP transgene, we examined rest mutants for defects in FBMN migration (Fig. 6). Following an intercross of restsbu29/+ and restsbu29/+; islet1-GFP fish, we observed three phenotypic classes. Out of 59 embryos, ten had wild-type FBMN positioning (Fig. 6E). Genotype analysis revealed that nine of these were wild type for restand one was a heterozygote. A second class showed very subtle changes in FBMN migration, exhibiting minor disorganization of neurons in r6 and r7 and a slight increase in the number of FBMNs remaining in r5 at 48 hpf (Fig. 6F). All 40 embryos in this class were restsbu29/+heterozygotes. The final phenotypic class showed more severe migration defects and seven out of nine of these embryos were restsbu29/sbu29homozygous mutants. In the mutants, many neurons were unable to migrate properly into r6, remaining instead in the r4-r5 region (Fig. 6G). These phenotypes closely resemble previously described Rest morphant phenotypes, and confirm a requirement for Rest function in control of FBMN migration.
Repression of RE1-containing genes by Rest at late embryonic and larval stages
Having observed derepression of Rest target genes in the mutant prior to 13 hpf (Fig. 2), we sought to establish whether Rest continues to repress target genes at later stages. As the embryo begins to mature and neurons differentiate, the overall expression levels of Rest target genes increase in wild-type embryos. We compared expression levels of RE1-containing genes in
MZrestsbu29/sbu29mutants and wild types by qRT-PCR between 1 and 19 dpf. Expression levels of nearly all target genes were comparable between mutants and wild types (supplementary material Fig. S5). Notable exceptions were gpr27, which showed elevated expression in restmutants at all stages, and kcnh8, bdnf and pcad, which were elevated at 24 hpf in the mutants. We also employed more sensitive approaches to assay snap25aexpression. However, neither in situ hybridization nor a new snap25a-GFP transgenic reporter line (data not shown) revealed evidence of ectopic expression.
Monitoring Rest activity with a transgenic reporter system
Although Rest-mediated repression of target genes in vitro is well studied, little is known about the spatial or temporal requirements for Rest in intact animals. Analysis is complicated by the nature of Rest activity: epigenetic chromatin modifications mediated by Rest may endure once Rest protein is degraded. To dynamically map tissues in the developing embryo where Rest activity is required, we crossed the restsbu29allele into four transgenic enhancer trap lines [Et(NRSE-cfos:KalTA4), hereafter NRCK (X.X. and J.S.M, unpublished)] that each contain an RE1/NRSE element adjacent to a minimal c-fospromoter upstream of KalTA4, an optimized Gal4-VP16 driver (Distel et al., 2009).
Activity of these lines was monitored by crossing them to a 14xUAS:NfsB-mCherrytransgenic reporter line that expresses a fusion protein between nitroreductase (Ntr) and a monomeric red fluorescent reporter, mCherry [Tg(UAS-E1b:NfsB-mCherry)c264, hereafter 14xNtrCh] (Davison et al., 2007). The UAS-linked mCherry reporter is dependent on KalTA4 activation. Therefore, the spatial and temporal expression of mCherry depends on the genomic context of the KalTA4 enhancer trap insertions. Owing to the influence of the RE1/NRSE element, NRCK lines express KalTA4 primarily in the nervous system, which in turn restricts expression of mCherry to the nervous system in double transgenics. Two of the lines, NRCKgmc606and NRCKgmc607, drive expression principally in the nervous system and weakly in the mesoderm. Two additional lines, NRCKgmc632 and NRCKgmc641, display expression patterns that are almost exclusively restricted to the nervous system. In all cases, expression of mCherry reporters was altered in restmutants.
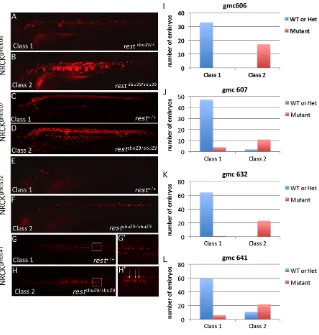
The first line, NRCKgmc606, drives mCherry expression in the spinal cord, a few notochord cells and in an occasional muscle fiber (Fig. 7A). The progeny from crosses between restsbu29/+; NRCKgmc606; 14xNtrChc264/+ and restsbu29/+ fish fell into two phenotypic classes. The first class expressed mCherry in the expected predominately neural NRCKgmc606pattern (n33/50) and comprised only wild-type and restheterozygous embryos (Fig. 7A). By contrast, the remaining embryos (n17/50) had significantly expanded mCherry expression, with many notochord cells and numerous muscle fibers expressing mCherry, and were all restmutants (class 2, Fig. 7B).
[image:6.612.52.297.60.258.2]The NRCKgmc607line expresses mCherry within the spinal cord, but also in a few muscle fibers and enteric neurons (Fig. 7C). Among progeny of restsbu29/+; NRCKgmc607; 14xNtrChc264/+and restsbu29/+ crosses, a phenotype (class 2) emerged that showed expanded neural expression, widespread expression in muscle of the trunk and head and in notochord (Fig. 7D). Genotyping revealed that 11/13 (84.6%) class 2 embryos were restmutants (Fig. 7J). Conversely, only 4/51 (7.8%) of class 1 embryos, which displayed the expected expression pattern, were restmutants. In summary, analysis of the NRCKgmc606 and NRCKgmc607 lines
Fig. 6. Rest function is crucial for the regulation of
oligodendrocyte precursor cells in the dorsal spinal cord and for facial branchiomotor migration.(A-D)Lateral views of live 50-hpf and 98-hpf Tg(olig2:GFP)(A,C) and Tg(olig2:GFP); MZrestsbu29/sbu29 mutants (B,D). The number of migrating olig2+oligodendrocyte precursors (arrows) is significantly reduced in restmutants at 50 hpf (B) compared with controls (A). The number of migrating OPCs (arrows) in 98-hpf MZrestsbu29/sbu29mutants (D) is similar to controls (C).
(E-G)Dorsal views of the hindbrain of 48-hpf Tg(islet1:GFP)transgenic zebrafish embryos. Representative embryos from a restsbu29/+,restsbu29/+;
islet1:GPF intercross. (E)rest+/+embryo showing normal FBMN migration into r6-r7. (F)FBMNs in restsbu29/+embryos have only subtle migration defects (arrows). (G)In restsbu29/sbu29embryos, a significant proportion of FBMNs remain in r4-r5 (bracket).
D
E
V
E
LO
P
M
E
N
demonstrates that Rest acts on RE1/NRSE elements to repress expression of neighboring genes in mesodermal derivatives including muscle and notochord between 3 and 6 dpf. To our knowledge, this is the first evidence for Rest activity in the notochord.
Two additional lines, NRCKgmc632 and NRCKgmc641, are expressed almost exclusively within the nervous system. The NRCKgmc632 line labels sparse cells within the spinal cord and brain. At 4 dpf, the progeny of restsbu29/+; NRCKgmc632/+; 14xNtrChc264/+and restsbu29/+crosses again fell into two phenotypic classes (Fig. 7E,F). The majority of embryos (65/85) displayed the expected pattern of expression (class 1). The second class had broader expression within the spinal cord, including numerous interneurons (Fig. 7F). All 23 class 2 embryos that had broader expression were restmutants (Fig. 7K). Class 1 comprised 64 wild-type and heterozygous embryos and a single restmutant.
The neural-specific NRCKgmc641 line expresses primarily in secondary motor neurons. The progeny of restsbu29/+; NRCKgmc641/+; 14xNtrChc264/+and restsbu29/+crosses fell into three phenotypic classes at 3 dpf. In class 1, mCherry expression was observed in two parallel rows of cells along the spinal cord (Fig. 7G,G⬘). Class 2 (n33) had similar expression except that mCherry-positive cells were frequently observed in the midline (Fig. 7H,H⬘). Genotypic analysis of class 2 revealed that mutants were predominant, representing 66.7% (22 embryos), whereas only 3% (1 embryo) was wild type (Fig. 7L). Conversely, mutants comprised only 9.2% (6/65 embryos) of class 1 (Fig. 7L). In class 3 (n31), fewer cells expressed mCherry and it was not possible to assess the midline of these individuals. Genotype analysis of this class revealed no enrichments of any genotype.
The NRCKgmc641 results suggest that either mutant neurons prematurely express the RE1/NRSE transgene or that newly differentiated mutant neurons fail to migrate properly. By 5 dpf we could no longer distinguish between class 1 and class 2 specimens, as both had many mCherry-positive midline cells (data not shown). Although we did not observe alterations in the subset of Rest target genes that we studied at larval stages, analysis of the reporter lines verified that Rest suppresses the expression of target genes between 3 and 6 dpf in muscle and notochord. In addition, analysis of these lines demonstrated that Rest is also required within the developing nervous system to repress gene expression. Taken together, these data suggest that Rest-mediated repression is highly context dependent.
Rest is required to maintain the repression of target genes in adult tissues
To determine whether Rest mediates long-term repression, we isolated brain, muscle, liver, heart, spleen and pancreas from adult wild-type and restsbu29/sbu29fish and compared snap25b,grp27, grin1a,bdnfand neurodtranscript levels (Table 1). In wild-type animals, each of these markers showed variable expression between animals. These differences might stem from unique environmental challenges faced by each animal over many months. Although this variability might have obscured some effects, clear trends emerged nonetheless. In restmutants, expression of snap25b was derepressed in pancreas, spleen and liver, whereas grin1awas derepressed in pancreas and muscle (Table 1). Expression of bdnf was enhanced in rest mutant spleens, but changes in neurod expression were not apparent in any of the organs tested. It has been suggested that Rest functions as a transcriptional activator in
Fig. 7. RE1/NRSE reporter transgenic lines are ectopically expressed in restmutants.
(A-H⬘) Lateral (A-F) or dorsal (G,H) views of live RE1/NRSE transgenic reporter lines in wild-type (A,C,E,G) or restsbu29/sbu29(B,D,F,H) backgrounds. Expression of each reporter was altered in the mutants. The boxed region in G and H is shown at higher magnification in G⬘and H⬘. Arrows mark midline cells, which are not as numerous in the wild-type background. (I-L)The proportion of embryos in each phenotypic and genotypic class for each reporter. gmc606 and gmc641 larvae are 3 dpf; gmc607, 6 dpf; gmc632, 4 dpf.
D
E
V
E
LO
P
M
E
N
[image:7.612.54.373.60.395.2]adult brain (Kuwabara et al., 2004), but no significant alterations in expression were observed in the targets that we analyzed from this organ.
Although some restsbu29/sbu29adults produce viable offspring, their fertility and fecundity often wane more rapidly than those of wild-type fish. We therefore examined the expression of RE1 target genes in ovaries and testes (Fig. 8). Strikingly, snap25b expression was near the threshold of detection in wild-type ovaries, but expression was more abundant in rest mutant ovaries. In addition, levels of gpr27,kcnh8 and pcadexpression were elevated in the mutant ovaries. To determine whether this effect was specific to female reproductive tissue, we examined all the markers in testes, but only kcnh8 expression was enhanced (Fig. 8). These observations demonstrate a long-postulated, but previously largely unproven, role for Rest in long-term repression of RE1-containing genes in non-neural tissue, and also reveal a high degree of tissue and locus specificity in the effects of Rest on target genes.
DISCUSSION
To examine the requirement for Rest in the context of an intact animal, we disrupted zebrafish rest using ZFNs. Based on previously proposed roles for Rest, we anticipated that this would cause inappropriate or failed neurogenesis. Contrary to this expectation, the mutants underwent relatively normal neurogenesis, although defects were observed in OPCs and in the migration of the FBMNs (Fig. 6). We observed inappropriate
expression of genes and reporters in the mutant (Figs 2, 3, 6 and 7), but no clear alterations of cell fate (Fig. 5; supplementary material Figs S2, S3).
Several in vitro studies have reported conflicting observations regarding the role of Rest in stem cell maintenance during development. Some reports assert that Rest is required for self-renewal and pluripotency of ES cells (Gupta et al., 2009; Singh et al., 2008), whereas others strongly contest this observation (Buckley et al., 2009; Jørgensen et al., 2009a; Jørgensen et al., 2009b; Yamada et al., 2010). Our results demonstrate that Rest is not required to maintain pluripotency or self-renewal of developing zebrafish blastomeres as germ layer specification proceeds normally in rest mutants (supplementary material Fig. S2). Although our findings do not reveal any requirement for Rest in early embryos, paradoxically, some of the largest changes in gene expression that result from Rest disruption occur during blastula stages (Fig. 2). Under some conditions, this failure to restrain inappropriate transcription might influence the proliferation or developmental potential of these cells.
Does Rest function differ in zebrafish and mouse? Mouse Restmutants undergo widespread cell death and die by E11.5 (Chen et al., 1998). By contrast, zebrafish rest mutants survive for at least 2 weeks. The disparity between the zebrafish and mouse phenotypes might stem from distinctive compensatory mechanisms present in zebrafish, or from alterations in rest function or targets between species. It is plausible that mechanisms compensate for the lack of Rest function in zebrafish and that these are less prominent in mammals. The most likely of such mechanisms would involve a second restor rest-like gene, as gene duplicates are common in zebrafish (Postlethwait et al., 2000). Functional similarities have been proposed between rest and several invertebrate genes including spr-3, spr-4, charlatanand tramtrack(Dallman et al., 2004; Lakowski et al., 2003; Tsuda et al., 2006). A zebrafish homolog of one of these factors might also compensate for loss of Rest function. We extensively searched the zebrafish genome for rest-related genes and have not detected a second rest ortholog or another rest-like gene. In addition, we found that snap25apromoter-GFP transgenic lines that contain a scrambled RE1 element generally mirror expression of the wild-Table 1. Derepression of RE1-containing genes in adult tissues
of rest mutants
Marker Muscle Pancreas Heart Brain Spleen Liver
gpr27 + ++ n.t. + +++ ++
snap25b +++ +++ – – – – – – + n.a.
bdnf + – – – – – – – – – ++ – – –
grin1a +++ ++ – – – – – – – – – – – –
neurod – – – – – – – – – – – – – – – – – –
[image:8.612.50.299.83.150.2]All markers were run on three independent tissue samples. Plus signs indicate the number of samples in which the given marker was derepressed in the mutant. n.t., not tested; n.a., no amplification.
Fig. 8. Rest target genes are depressed in germ tissue of adult restmutants.qPCR analysis of RE1-containing gene expression in wild-type and restsbu29/sbu29mutant ovary and testes (three examples of each are shown). All fold differences are relative to the average of the transcript levels in the three mutants (defined as 1). (A-D)Levels of
snap25b,kcnh8,gpr27and pcadare enhanced in the restmutant ovary. (E)Only kcnh8 expression was enhanced in restmutant testes.
D
E
V
E
LO
P
M
E
N
[image:8.612.53.434.518.738.2]type promoter (data not shown). If additional factors act on the snap25a RE1 site, mutating the site should have caused inappropriate expression of the reporter.
We favor the model that the phenotypic differences in mouse and zebrafish rest mutations stem from differences in target genes. Zebrafish have a comparable number of RE1 sites to other vertebrates (Mortazavi et al., 2006). Fish RE1/NRSE elements have been shown to bind mammalian Rest (Johnson et al., 2009) and the mammalian and zebrafish Rest DNA-binding domains exhibit a high degree of conservation (Gates et al., 2010). However, a key difference might be in the positioning of RE1/NRSE sites. Many genes have RE1/NRSE sites that are conserved between fish and mammals, but some sites are unique. Because regulatory elements are thought to be prime targets for evolutionary change (King and Wilson, 1975), such differences are expected. The cause of the lethality of the mouse Rest knockout remains unknown, but it precedes neurogenesis. The rampant cell death that is observed in the mouse knockout could potentially result from misregulation of a single locus that is not under the control of Rest in zebrafish.
While this manuscript was in preparation, a report was published showing that a neural-specific conditional Rest knockout does not alter neurogenesis, although effects were noted in cultured cells isolated from mouse (Aoki et al., 2012). Although further analysis of rest conditional knockouts is required, these findings support our conclusion that Rest is not essential for neurogenesis and highlight similarities between Rest function in zebrafish and mice.
What is the role of Rest?
Given the size and frequency of RE1 sites, strong selective pressure must maintain their presence throughout the zebrafish genome. Although we identified embryonic requirements for Rest in OPC development and in cranial nerve migration, these deficits are less severe than would be expected for a master regulator of neurogenesis. We also detected changes in the expression of RE1-containing genes during embryogenesis and early larval development. These changes were most apparent during blastula and somite stages and largely reflected precocious gene expression. The large fold differences between rest mutants and wild type during these stages reflects low or absent expression in wild type and more easily detectable expression in the mutant. Our data provide the first in vivo evidence that Rest regulates gene expression in ES cells (blastomeres). However, no alterations in cell fate were observed in the mutants. Future analysis of the mutants might reveal biases in the developmental potential of Rest-deficient blastomeres, but our analysis does not support a broad role for Rest in cell fate determination. In this context, Rest might promote the fitness of stem cells by suppressing the premature production of target transcripts.
During later stages of embryogenesis, many of the loci we analyzed appeared to be largely unaffected by loss of Rest. Our analysis of target gene expression during late embryonic and larval stages relied primarily on qPCR, and subtle alterations in gene expression might have gone undetected. However, we also examined expression of a snap25a-GFP reporter line in the mutant and failed to detect changes in expression in the absence of Rest at these stages (data not shown). Simpler reporter constructs (the NRSE KalTA4 enhancer trap lines) revealed substantial Rest activity in mesodermal derivatives including muscle and notochord, as well as within the nervous system (Fig. 7). These findings suggest that Rest-mediated repression of RE1/NRSE sites is highly
context dependent and such activity might be more apparent in the milieu of the NRSE-basal promoter constructs. From these observations, we suspect that the chief function of Rest at these stages is to fine-tune the expression of RE1-containing genes rather than as a robust silencer of expression.
Analysis of adult tissues from surviving restmutants provided the first proof that Rest is required in non-neural tissues for long-term suppression of target gene expression (Fig. 8, Table 1). These effects were most pronounced in the germ tissue. Our anecdotal observations suggest that restmutants tend to have reduced fertility compared with wild types. These observations suggest that Rest might have a role in the maintenance of the adult germline or simply contributes to overall fitness.
Rest has been proposed to function within the nervous system, but we did not observe changes in gene expression in restmutant brain. However, many Rest targets are robustly expressed in the brain and are involved in a host of essential neural processes. Discrete alteration of expression of these genes might have profound consequences for higher-order neural functions. In addition, recent studies of a conditional Restknockout in the mouse revealed a requirement for Rest in adult neurogenesis (Gao et al., 2011). Regulation of gene expression in the adult brain may be a primary Rest function. Future studies will explore the behavioral consequences of Rest depletion.
Regulation of neural migration may be a principal function of Rest. Abnormalities in FBMN migration were observed in our rest mutant. In addition, the OPC deficiencies and secondary motor neuron defects (in the NRCKgmc641reporter line) might also stem from altered migration. Adhesion molecules are abundant among Rest targets (Otto et al., 2007), and Rest has been implicated in the regulation of radial migration of mouse neocortical progenitors (Mandel et al., 2011). Together, these data suggest a key role for Rest in neuronal migration.
In conclusion, the zebrafish restmutant is a valuable tool that will help to broaden our understanding of Rest function. Our analysis reveals that although Rest represses the expression of RE1-containing target genes in several contexts, the effects often appear to modulate the timing or levels of expression during development rather than to silence vigorous expression. Although we identified discrete requirements for Rest during development, we found no evidence for a role as a fundamental regulator of neurogenesis or cell fate during development. Examination of the mutant also demonstrated that Rest-mediated repression of target genes persists in non-neural tissues in adult animals. Additional studies of the mutant will clarify the functions of Rest within the nervous system and in non-neural tissue to establish a more comprehensive view of Rest regulation of an expansive set of target genes.
Acknowledgements
We thank the many members of the community who provided reagents; Carolyn Milano and Azeez Aranmolate for experimental support; Ian Wood and Sean McCorkle for assistance identifying RE1 sites; Bernadette Holdener, Nurit Ballas and Joel Levine for comments on the manuscript; and Amanda Levine, Won Seok Kwag and Erika Wunderlich for fish care.
Funding
This work was supported by NYSTEM [C026414 to H.I.S.]; National Institutes of Health [HD066000 to H.I.S., DK064973 to V.E.P., R21 MH083614 to J.S.M.]; and March of Dimes Basil O’Connor [5-FY10-7 to J.S.M.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
D
E
V
E
LO
P
M
E
N
Supplementary material
Supplementary material available online at
http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.080994/-/DC1
References
Abrajano, J. J., Qureshi, I. A., Gokhan, S., Zheng, D., Bergman, A. and Mehler, M. F.(2009). Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS ONE4, e7665.
Aoki, H., Hara, A., Era, T., Kunisada, T. and Yamada, Y.(2012). Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development139, 667-677.
Ballas, N., Battaglioli, E., Atouf, F., Andres, M. E., Chenoweth, J., Anderson, M. E., Burger, C., Moniwa, M., Davie, J. R., Bowers, W. J. et al.(2001). Regulation of neuronal traits by a novel transcriptional complex. Neuron31, 353-365.
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C. and Mandel, G.(2005). REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell121, 645-657.
Belyaev, N. D., Wood, I. C., Bruce, A. W., Street, M., Trinh, J. B. and Buckley, N. J.(2004). Distinct RE-1 silencing transcription factor-containing complexes interact with different target genes. J. Biol. Chem. 279, 556-561.
Bruce, A. W., Donaldson, I. J., Wood, I. C., Yerbury, S. A., Sadowski, M. I., Chapman, M., Göttgens, B. and Buckley, N. J.(2004). Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA101, 10458-10463.
Buckley, N. J., Johnson, R., Sun, Y. M. and Stanton, L. W.(2009). Is REST a regulator of pluripotency? Nature457, E5-E6.
Chandrasekhar, A.(2004). Turning heads: development of vertebrate branchiomotor neurons. Dev. Dyn. 229, 143-161.
Chen, Z. F., Paquette, A. J. and Anderson, D. J.(1998). NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20, 136-142.
Chong, J. A., Tapia-Ramírez, J., Kim, S., Toledo-Aral, J. J., Zheng, Y., Boutros, M. C., Altshuller, Y. M., Frohman, M. A., Kraner, S. D. and Mandel, G.
(1995). REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell80, 949-957.
Dallman, J. E., Allopenna, J., Bassett, A., Travers, A. and Mandel, G.(2004). A conserved role but different partners for the transcriptional corepressor CoREST in fly and mammalian nervous system formation. J. Neurosci. 24, 7186-7193.
Davison, J. M., Akitake, C. M., Goll, M. G., Rhee, J. M., Gosse, N., Baier, H., Halpern, M. E., Leach, S. D. and Parsons, M. J.(2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304, 811-824.
DeWald, L. E., Rodriguez, J. P. and Levine, J. M.(2011). The RE1 binding protein REST regulates oligodendrocyte differentiation. J. Neurosci. 31, 3470-3483.
Distel, M., Wullimann, M. F. and Köster, R. W.(2009). Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. USA
106, 13365-13370.
Doyon, Y., McCammon, J. M., Miller, J. C., Faraji, F., Ngo, C., Katibah, G. E., Amora, R., Hocking, T. D., Zhang, L., Rebar, E. J. et al.(2008). Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 26, 702-708.
Gao, Z., Ure, K., Ding, P., Nashaat, M., Yuan, L., Ma, J., Hammer, R. E. and Hsieh, J.(2011). The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31, 9772-9786.
Gates, K. P., Mentzer, L., Karlstrom, R. O. and Sirotkin, H. I.(2010). The transcriptional repressor REST/NRSF modulates hedgehog signaling. Dev. Biol.
340, 293-305.
Grimes, J. A., Nielsen, S. J., Battaglioli, E., Miska, E. A., Speh, J. C., Berry, D. L., Atouf, F., Holdener, B. C., Mandel, G. and Kouzarides, T.(2000). The co-repressor mSin3A is a functional component of the REST-CoREST co-repressor complex. J. Biol. Chem. 275, 9461-9467.
Grzenda, A., Lomberk, G., Zhang, J. S. and Urrutia, R.(2009). Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta1789, 443-450.
Guardavaccaro, D., Frescas, D., Dorrello, N. V., Peschiaroli, A., Multani, A. S., Cardozo, T., Lasorella, A., Iavarone, A., Chang, S., Hernando, E. et al.
(2008). Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature452, 365-369.
Gupta, S. K., Gressens, P. and Mani, S.(2009). NRSF downregulation induces neuronal differentiation in mouse embryonic stem cells. Differentiation77, 19-28.
Huang, Y., Myers, S. J. and Dingledine, R.(1999). Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2, 867-872.
Johnson, R., Samuel, J., Ng, C. K., Jauch, R., Stanton, L. W. and Wood, I. C.
(2009). Evolution of the vertebrate gene regulatory network controlled by the transcriptional repressor REST. Mol. Biol. Evol. 26, 1491-1507.
Jørgensen, H. F., Chen, Z. F., Merkenschlager, M. and Fisher, A. G.(2009a). Is REST required for ESC pluripotency? Nature457, E4-E5.
Jørgensen, H. F., Terry, A., Beretta, C., Pereira, C. F., Leleu, M., Chen, Z. F., Kelly, C., Merkenschlager, M. and Fisher, A. G.(2009b). REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development136, 715-721.
Kim, Y. G., Cha, J. and Chandrasegaran, S.(1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA93, 1156-1160.
King, M. C. and Wilson, A. C.(1975). Evolution at two levels in humans and chimpanzees. Science188, 107-116.
Kirby, B. B., Takada, N., Latimer, A. J., Shin, J., Carney, T. J., Kelsh, R. N. and Appel, B.(2006). In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat. Neurosci. 9, 1506-1511.
Kuwabara, T., Hsieh, J., Nakashima, K., Taira, K. and Gage, F. H.(2004). A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell116, 779-793.
Lakowski, B., Eimer, S., Göbel, C., Böttcher, A., Wagler, B. and Baumeister, R.
(2003). Two suppressors of sel-12 encode C2H2 zinc-finger proteins that regulate presenilin transcription in Caenorhabditis elegans. Development130, 2117-2128.
Lu, Q. R., Sun, T., Zhu, Z., Ma, N., Garcia, M., Stiles, C. D. and Rowitch, D. H.
(2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell109, 75-86.
Lunyak, V. V., Burgess, R., Prefontaine, G. G., Nelson, C., Sze, S. H., Chenoweth, J., Schwartz, P., Pevzner, P. A., Glass, C., Mandel, G. et al.
(2002). Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science298, 1747-1752.
Mandel, G., Fiondella, C. G., Covey, M. V., Lu, D. D., Loturco, J. J. and Ballas, N.(2011). Repressor element 1 silencing transcription factor (REST) controls radial migration and temporal neuronal specification during neocortical development. Proc. Natl. Acad. Sci. USA108, 16789-16794.
Mapp, O. M., Walsh, G. S., Moens, C. B., Tada, M. and Prince, V. E.(2011). Zebrafish Prickle1b mediates facial branchiomotor neuron migration via a farnesylation-dependent nuclear activity. Development138, 2121-2132.
Meng, X., Noyes, M. B., Zhu, L. J., Lawson, N. D. and Wolfe, S. A.(2008). Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 26, 695-701.
Mortazavi, A., Leeper Thompson, E. C., Garcia, S. T., Myers, R. M. and Wold, B.(2006). Comparative genomics modeling of the NRSF/REST repressor network: from single conserved sites to genome-wide repertoire. Genome Res.
16, 1208-1221.
Naruse, Y., Aoki, T., Kojima, T. and Mori, N.(1999). Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl. Acad. Sci. USA96, 13691-13696.
Olguín, P., Armisen, R. and Kukuljan, M.(2006a). Developmental regulation of the expression of sodium currents in Xenopus primary neurons. Biol. Res. 39, 483-491.
Olguín, P., Oteíza, P., Gamboa, E., Gómez-Skármeta, J. L. and Kukuljan, M.
(2006b). RE-1 silencer of transcription/neural restrictive silencer factor modulates ectodermal patterning during Xenopus development. J. Neurosci. 26, 2820-2829.
Otto, S. J., McCorkle, S. R., Hover, J., Conaco, C., Han, J. J., Impey, S., Yochum, G. S., Dunn, J. J., Goodman, R. H. and Mandel, G.(2007). A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27, 6729-6739.
Park, H. C., Mehta, A., Richardson, J. S. and Appel, B.(2002). olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev. Biol.
248, 356-368.
Postlethwait, J. H., Woods, I. G., Ngo-Hazelett, P., Yan, Y. L., Kelly, P. D., Chu, F., Huang, H., Hill-Force, A. and Talbot, W. S.(2000). Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10, 1890-1902.
Roopra, A., Qazi, R., Schoenike, B., Daley, T. J. and Morrison, J. F.(2004). Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell14, 727-738.
Schoenherr, C. J. and Anderson, D. J.(1995). The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science
267, 1360-1363.
Shi, Y., Sawada, J., Sui, G., Affar, B., Whetstine, J. R., Lan, F., Ogawa, H., Luke, M. P., Nakatani, Y. and Shi, Y.(2003). Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature422, 735-738.
Singh, S. K., Kagalwala, M. N., Parker-Thornburg, J., Adams, H. and Majumder, S.(2008). REST maintains self-renewal and pluripotency of
embryonic stem cells. Nature453, 223-227.
D
Soldati, C., Bithell, A., Johnston, C., Wong, K. Y., Teng, S. W., Beglopoulos, V., Stanton, L. W. and Buckley, N. J.(2012). Repressor Element 1 Silencing Transcription factor couples loss of pluripotency with neural induction and neural differentiation. Stem Cells30, 425-434.
Tan, Y. P., Li, S., Jiang, X. J., Loh, W., Foo, Y. K., Loh, C. B., Xu, Q., Yuen, W. H., Jones, M., Fu, J. et al.(2010). Regulation of protocadherin gene expression by multiple neuron-restrictive silencer elements scattered in the gene cluster. Nucleic Acids Res. 38, 4985-4997.
Thisse, C., Thisse, B., Schilling, T. F. and Postlethwait, J. H.(1993). Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development119, 1203-1215.
Tsuda, L., Kaido, M., Lim, Y. M., Kato, K., Aigaki, T. and Hayashi, S.(2006). An NRSF/REST-like repressor downstream of Ebi/SMRTER/Su(H) regulates eye development in Drosophila. EMBO J. 25, 3191-3202.
Westbrook, T. F., Hu, G., Ang, X. L., Mulligan, P., Pavlova, N. N., Liang, A., Leng, Y., Maehr, R., Shi, Y., Harper, J. W. et al.(2008). SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature452, 370-374.
Yamada, Y., Aoki, H., Kunisada, T. and Hara, A.(2010). Rest promotes the early differentiation of mouse ESCs but is not required for their maintenance. Cell Stem Cell6, 10-15.