Yiqun Duan,
and Sharon C. A. Chen
*
Research Laboratory for Infectious Skin Diseases, Department of Dermatology, Wuhan First Hospital, Wuhan 430022, People’s Republic of China,1and Centre for Infectious Diseases and Microbiology, Westmead Hospital and the University of Sydney,
Westmead, New South Wales, Australia2
Received 3 March 2008/Returned for modification 2 April 2008/Accepted 12 May 2008
A sensitive rolling-circle amplification (RCA)-based method utilizing species-specific padlock probes tar-geted to the internal transcribed spacer 2 region of the fungal ribosomal DNA gene complex was developed. The assay was rapid (2 hours) and specific. Of 28 fungal isolates (16 ofCandida, six of Aspergillus, and six of
Scedosporiumspp.), all were all identified correctly.
Fungal pathogens cause life-threatening infections in criti-cally ill and immunosuppressed patients. Contemporary
epide-miological trends reveal a shift toward species ofCandidaand
Aspergillusother thanCandida albicansandAspergillus
fumiga-tusand a range of emerging fungi includingScedosporiumspp.
and the zygomycetes (6, 19). Given the reduced susceptibility of many of these pathogens to antifungal agents, timely iden-tification to species level is essential for clinical management. However, standard culture-based identification methods are insensitive and slow (15).
To overcome both problems, PCR-based tools have been developed. In particular, the internal transcribed spacer (ITS) regions, ITS1 and ITS2, of the fungal ribosomal DNA gene complex have shown promise as targets for species identifica-tion in a variety of formats including multiplex and/or real-time PCR assays (9, 16), DNA sequence analysis (1, 2, 12), and probe-based techniques (5, 7). The latter range from Southern blotting (5, 7) and reverse line blot (RLB) hybridization meth-ods (23) to sophisticated microarray formats (10, 11, 17).
Recently, the utility of circularizable oligonucleotide (padlock) probes has been demonstrated for the detection of target nucleic acid sequences, including nucleotide polymorphisms that differ by only a few base pairs, with high sensitivity (4, 13, 20). Such probes
comprise two sequences complementary to the 5⬘and 3⬘termini
of the target sequence joined by a linker region (Fig. 1A). Upon hybridization to the target, the probe ends are joined by DNA ligase to form a closed molecule. The intensity of the probe-specific signal is then increased exponentially by rolling-circle amplification (RCA) (13) (Fig. 1B). There are few data on the application of padlock probes in the detection of polymorphisms in fungi. We report on a sensitive, RCA-based method using real-time PCR for species identification of clinically important
Candida,Aspergillus, andScedosporiumspp.
Twenty-eight clinical isolates were studied: two ofC.
albi-cans, two ofCandida glabrata, three ofCandida krusei, three of
Candida tropicalis, three ofCandida dubliniensis, three of Can-dida guilliermondii, four of A. fumigatus, two of Aspergillus flavus, and three strains each of Scedosporium apiospermum
and Scedosporium prolificans. Species identity was confirmed by standard laboratory methods (3, 21) and ITS sequence anal-ysis (23). Isolates were stored in sterile water at 25°C until required. DNA extraction and amplification of the ITS (ITS1, 5.8S rRNA, and ITS2) region, using the primers ITS1 and ITS4 (22), in preparation for hybridization with padlock probes (see below) were performed as previously described (23).
DNA sequences spanning the ITS2 region of all major
Can-dida,Aspergillus, andScedosporiumspecies were accessed from the GenBank database and compared using the Clustal W program (BioManager, ANGIS; http://biomanager.angis.org .au/) to identify informative nucleotide polymorphisms. Ten padlock probes targeting the ITS2 region (six specific for each
Candidasp., one each forA. fumigatusandA. flavus, and one
each for S. apiospermum and S. prolificans) were then
de-signed. To optimize binding to target DNA, the melting
tem-perature of the 5⬘-end probe binding arm was higher than the
temperature used for probe ligation (62°C). To increase
spec-ificity, the 3⬘-end binding arm was designed with a melting
temperature of 50 to 57°C (i.e., below the ligation tempera-ture). Furthermore, the linker region of each probe was de-signed to minimize similarity to other closely related pathogens and to allow primer binding during RCA (Fig. 1A). Table 1 gives the sequences of the two primers used for RCA (RCA primer 1 and RCA primer 2).
Purified amplified PCR product (1011-copy-number DNA
template [DNA calculator; http://www.uri.edu/research/gsc
/resources/cndna.html]) was mixed with 2 U ofPfuDNA ligase
(Stratagene, La Jolla, CA) and 0.1M padlock probe as
pre-viously described (18) and subjected to multiple cycle ligation followed by exonucleolysis to remove unligated probe and tem-plate PCR product (18). RCA reactions of the circularized probes were then performed according to previously described experimental conditions (18, 20). Probe signals were amplified by incubation at 65°C for 30 min, and accumulation of double-stranded DNA products was monitored using a Corbett
Ro-* Corresponding author. Mailing address: Centre for Infectious Dis-eases and Microbiology, Westmead Hospital, Darcy Road, Westmead, New South Wales 2145, Australia. Phone: 61-2 9845 6255. Fax: 61-2 9893 8659. E-mail: sharon.chen@swahs.health.nsw.gov.au.
† These authors contributed equally to the work.
䌤Published ahead of print on 21 May 2008.
2423
on May 16, 2020 by guest
http://jcm.asm.org/
on May 16, 2020 by guest
http://jcm.asm.org/
torGene 6000 machine (Corbett Research, Mortlake, Austra-lia); probe signals were also visualized on a 1.5% agarose gel to verify the specificity of probe-template binding.
The universal fungal primers ITS1 and ITS4 amplified the ITS region of all study isolates. Further, individual species-specific probes, designed on the basis of species-species-specific signa-tures identified by alignment of ITS2 sequence data,
success-fully identified all strains. To assess assay sensitivity, RCA was
performed on serial dilutions of target template (1011copies).
For all isolates, a measurable signal was observed using real-time PCR (schematically shown in Fig. 1B). The sensitivity of
detection was 107copies (data not shown). The duration of the
RCA procedure was 2 h, but a positive signal was evident 15 min after commencement of the reaction.
gatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtcta 736AGATCAACACCGAGTTG720
CKR C. krusei AF246989 pa-406GCCAGCTTCGCTCCCTTT389
gatcaTGCTTCTTCGGTGCCCATtacgag gtgcggatagctacCGCGCAGACACGATAgtcta 423AAAGTCTAGTTCGCTCG407
CTR C. tropicalis AF268095 pa-365GCGTATTGCTCAACACCAAACC344
gatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtcta 383TTCTTTCAAACAAACCTA366
CDU C. dubliniensis AJ249485 pa-450CGCCTTAGCAATGTTTTTGGTTA428
gatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtcta 463CGACGCCAGAGAC451
CGU C. guilliermondii AF455495 pa-513CGGGCCAACAATACCAGAAATAT491
gatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtcta 528TGGTTGTTGTAAGGC514
AFUM A. fumigatus AY660923 pa-144CGGCTGGCGCCGGCCGGG127
gatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtcta 162AAAAATAAAGTTGGGTGT145
AFLA A. flavus EF197070 pa-176TTTGCGTTCGGCAAGCGC159
gatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtcta 193ACCTGGAAAAAGATTGA177
SAPI S. apiospermum AY228118 pa-582AACCGCGGCGGGACCGCC565
gatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtcta 598TTACTACGCAGAAGGC583
SPRO S. prolificans AY228117 pa-443GTAAATCTACTACGCAGAAGGCGC420
gatcaTGCTTCTTCGGTGCCCATtacgaggtgcggatagctacCGCGCAGACACGATAgtcta 457ACCCAATGCGAGTT444
Primers
RCA primer 1b ATGGGCACCGAAGAAGCAb
RCA primer 2c CGCGCAGACACGATAc
a
Refers to the 5⬘end of the probe; “p-” indicates 5⬘phosphorylation. The sequences of the 5⬘and 3⬘binding arms of the probes (derived from reference GenBank
sequences) are underlined; these are joined by the backbone of the probe including the nonspecific linker region where the sequence is shown in lowercase.
b
Binding site of RCA primer 1 to the padlock probe, generating a long single-stranded DNA. The sequence of RCA primer 1 is the complement of the segments, in reverse, represented by bold italic nucleotides.
c
Binding site of RCA primer 2. RCA primer 2 binds to nascent single-stranded DNAs as their binding sites become available. Its sequence is the same as that of the segments shown in nonitalic bold.
FIG. 1. (A) Typical design of a circularizable padlock probe as exemplified by theCandida albicans-specific (CAL) probe. The probe comprises (i) a 5⬘-phosphorylated end, (ii) a “backbone” containing binding sites for the RCA primers (RCA primers 1 and 2, respectively; designated by bold uppercase letters) as well as the nonspecific linker regions (designated by bold lowercase letters), and (iii) a 3⬘end. The 5⬘and 3⬘ends of the probe are complementary to the 5⬘and 3⬘termini of the target sequence in reverse, in this example to theC. albicanssequence (GenBank accession no. AF455531). Abbreviations: 5⬘-P, 5⬘-phosphorylated binding arm; 3⬘, 3⬘binding arm. (B) Pictorial representation of the RCA method. Step 1, hybridization. Hybridization of padlock probe, containing target-complementary segments, to a target DNA sequence. Step 2, ligation. The probe is circularized by DNA ligase. Step 3, RCA and primer extension I. Ligated probe and binding of RCA primer 1 for RCA. Tandem repeat sequences complementary to the circular probe are generated by RCA. The reverse primer (RCA primer 2) binds to each tandem repeat generated by the rolling circle. Step 4, RCA and primer extension II. As the original RCA strand elongates, further priming events are initiated by primer 2, generating displaced DNA strands. As a result, new priming sites for the first primer (primer 1) are generated. The two primers thus function to generate a self-propagating pattern of DNA fragment release events (20). Step 5, detection of amplified product. RCA may be monitored using real-time PCR or agarose gel electrophoresis. ssDNA, single-stranded DNA.
on May 16, 2020 by guest
http://jcm.asm.org/
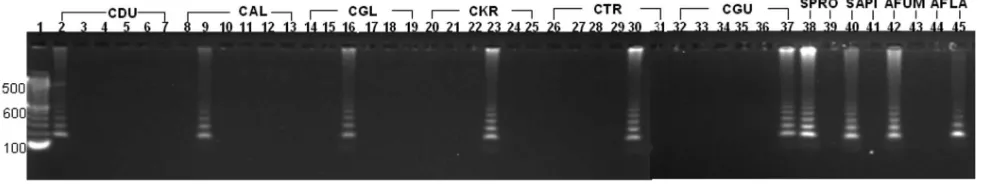
[image:3.585.46.537.80.426.2]The RCA assay was also highly specific. Amplification of probe signals was seen only with matched template-probe mix-tures; DNA from species which contained ITS2 polymor-phisms not targeted by the padlock probe in use produced no signal (Fig. 2). Species-specific probes correctly identified all six Candida species studied including C. dubliniensis and C. guilliermondii. The assay also clearly differentiated betweenA. fumigatusand A. flavusand between S. apiospermum and S. prolificans. Concordance with phenotypic identification and ITS sequencing results (23) was 100%.
The continued development of reliable diagnostic tools for the early detection and identification of fungi remains a prior-ity for improving patient outcomes. This proof-of principle study confirms the great potential of a simple, rapid (2-h), and highly specific RCA-based assay for the identification of
patho-genic fungal species, in this instanceCandida,Aspergillus, and
Scedosporium.
ITS2-targeted padlock probes were employed as the ITS2 region contains species-specific sequences well suited for iden-tification of yeasts and molds (2, 12). Signature polymorphisms
between the majorCandidaspp. were successfully targeted by
individual probes. In particular,C. albicanswas clearly
distin-guished fromC. dubliniensis, in contrast to the use of
pheno-typic methods, when separation of these two species is often unreliable (8). The RCA assay also distinguished
unambigu-ously between A. fumigatus and A. flavus and between S.
apiospermumand S. prolificans. The latter are two significant pathogens of seriously ill patients which have different epide-miological and clinical associations (14).
Among an increasing number of molecular methods estab-lished for species identification of fungal pathogens (especially
CandidaandAspergillus), ITS sequence analysis has emerged as the “gold standard.” However, sequencing is expensive, has a 2-to 3-day turnaround time, and is impractical for analyzing large numbers of isolates. Probe-based methods thus far offer a choice between broad-range detection and identification of a limited number of species, although more recently develop-ment of sensitive RLB and microarray formats has allowed the simultaneous detection of multiple pathogens in one test (10, 23). RLB approaches are best suited to “batch testing” of isolates, while microarray systems require expensive
special-ized equipment and are not readily adaptable in clinical labo-ratories.
Advantages of the RCA-based assay developed in this study include its specificity and flexibility in that probes can be cus-tom made to meet specific requirements, e.g., for a particular clinical setting. While the setup costs of the assay ($A300 per probe) are relatively high, the probes may be used up to 5,000 times and there is no need for specialized equipment. The technique has recently been used to detect single nucleotide polymorphisms within species of bacteria (18), indicating that RCA is a practicable option whereby selective targeting of single nucleotide polymorphisms can discriminate between closely related species. Further evaluation of this system using a broad range of fungal species is indicated to clarify its role in routine diagnosis.
We thank Ping Zhu for her assistance in the preparation of the figures.
REFERENCES
1.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. LaFe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA
genes. J. Clin. Microbiol.38:2302–2310.
2.Ciardo, D. E., G. Schar, E. C. Bottger, M. Altwegg, and P. P. Bosshard.2006. Internal transcribed spacer sequencing versus biochemical profiling for
iden-tification of medically important yeasts. J. Clin. Microbiol.44:77–84.
3.de Hoog, G. S., J. Guarro, J. Gene, and M. J. Figueras.2000. Atlas of clinical fungi, 2nd ed., p. 899–901. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands.
4.Demidov, V. V.2002. Rolling-circle amplification in DNA diagnostics: the
power of simplicity. Expert Rev. Mol. Diagn.2:542–548.
5.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher.
1997. Detection and identification of fungal pathogens in blood by using
molecular probes. J. Clin. Microbiol.35:1353–1360.
6.Enoch, D. A., H. A. Ludlam, and N. M. Brown.2006. Invasive fungal infec-tions: a review of epidemiology and management options. J. Med. Microbiol.
55:809–818.
7.Flahaut, M., D. Sanglard, M. Monod, J. Bille, and M. Rossier.1998. Rapid
detection ofCandida albicansin clinical samples by DNA amplification of
common regions fromC. albicans-secreted aspartic proteinase genes. J. Clin.
Microbiol.36:395–401.
8.Gutierrez, J., P. Morales, M. A. Gonzalez, and G. Quindos.2002.Candida dubliniensis, a new fungal pathogen. J. Basic Microbiol.42:207–227. 9.Hendolin, P. H., L. Paulin, P. Koukila-Kahkola, V. J. Anttila, H. Malmberg,
[image:4.585.45.541.70.162.2]M. Richardson, and J. Ylikoski.2000. Panfungal PCR and multiplex liquid
FIG. 2. Gel representation of specificity of RCA probes. Amplification of probe signals was seen only with matched template-probe mixtures (empty lanes denote absence of signals with unmatched template-probe mixtures). The species-specific probes are labeled as shown at the top of the figure (CDU,Candida dubliniensis; CAL,Candida albicans; CGL,Candida glabrata; CKR,Candida krusei; CTR,Candida tropicalis; CGU,
Candida guilliermondii; SPRO,Scedosporium prolificans; SAPI, Scedosporium apiospermum; AFUM,Aspergillus fumigatus; AFLA, Aspergillus flavus). Lane 1, 100-bp DNA Ladder MW marker (Invitrogen, Mount Waverly, Australia); lanes 2, 8, 14, 20, 26, and 32,C. dubliniensisWM 03.119; lanes 3, 9, 15, 21, 27, and 33,C. albicansWM 01.218; lanes 4, 10, 16, 22, 28, and 34,C. glabrataWM 02.58; lanes 5, 11, 17, 23, 29, and 35,C. krusei
WM 03.204; lanes 6, 12, 18, 24, 30, and 36,C. tropicalisWM 233; lanes 7, 13, 19, 25, 31, and 37,C. guilliermondiiWM 02.72; lane 38,S. prolificansWM 06.372; lane 39,S. apiospermumWM 06.466; lane 40,S. apiospermumWM 06.466; lane 41,S. prolificansWM 06.372; lane 42,A. fumigatusWM 06.96; lane 43,A. flavusWM 06.93; lane 44,A. fumigatusWM 06.96; lane 45,A. flavusWM 06.93.
on May 16, 2020 by guest
http://jcm.asm.org/
analysis of the internal transcribed spacer regions. J. Clin. Microbiol.44:
693–699.
13.Nilsson, M., F. Dahl, C. Larsson, M. Gullberg, and J. Stenberg.2006. Analyzing genes using closing and replicating circles. Trends Biotechnol.
24:83–88.
14.Panackal, A. A., and K. A. Marr.2004.Scedosporium/Pseudallescheria
infec-tions. Semin. Respir. Crit. Care Med.25:171–181.
15.Reiss, E., T. Obayashi, K. Orle, M. Yoshida, and R. M. Zancope-Oliveira.
2000. Non-culture based diagnostic tests for mycotic infections. Med. Mycol.
38(Suppl. 1):147–159.
16.Schabereiter-Gurtner, C., B. Selitsch, M. L. Rotter, A. M. Hirschl, and B. Willinger.2007. Development of novel real-time PCR assays for detection
and differentiation of eleven medically importantAspergillusandCandida
species in clinical specimens. J. Clin. Microbiol.45:906–914.
17.Spiess, B., W. Seifarth, M. Hummel, O. Frank, A. Fabarius, C. Zheng, H.
Y. Hou, and N. K. Saksena.2005. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification.
J. Clin. Microbiol.43:2339–2344.
21.Warren, N., and K. Hazen.1999.Candida,Cryptococcus, and other yeasts of
medical importance, p. 1184–1199.InR. P. Murray, E. J. Baron, M. A.
Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbi-ology. ASM Press, Washington, DC.
22.White, T., T. Bruns, S. Lee, and J. Taylor.1990. Amplification and direct
sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–336.In
M. Innis, D. Gelfand, J. Sninsky, and T. White (ed.), PCR protocols: a guide to methods and application. Academic Press, San Diego, CA.
23.Zeng, X., F. Kong, C. Halliday, S. Chen, A. Lau, G. Playford, and T. C. Sorrell. 2007. Reverse line blot hybridization assay for identification of medically important fungi from culture and clinical specimens. J. Clin.
Mi-crobiol.45:2872–2880.