Mycobacterium tuberculosis
Isolates to Pyrazinamide
Zhenling Cui, Jie Wang, Junmei Lu, Xiaochen Huang, Ruijuan Zheng, Zhongyi Hu
Shanghai Key Laboratory of Tuberculosis, Shanghai Pulmonary Hospital, Medical School, Tongji University, Shanghai, China
Pyrazinamide (PZA) is a first-line antituberculosis (anti-TB) drug capable of killing nonreplicating, persistentMycobacterium tubercu-losis. However, reliable testing of the susceptibility ofM. tuberculosisto PZA is challenging. Using 432 clinicalM. tuberculosisisolates, we compared the performances of five methods for the determination ofM. tuberculosissusceptibility to PZA: the MGIT 960 system, the molecular drug susceptibility test (mDST), the pyrazinamidase (PZase) activity assay, the resazurin microtiter assay (REMA), and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction test. The sensitivities of the MGIT 960 system, the PZase activity assay, the mDST, the REMA, and the MTT assay were 98.8%, 88.8%, 90.5%, 98.8%, and 98.2%, respectively. The sensitiv-ities of the PZase activity assay and the mDST were lower than those of the other three methods (P<0.05). The specificities of the MGIT 960 system, the PZase activity assay, the mDST, the REMA and the MTT assays were 99.2%, 98.9%, 90.9%, 98.5%, and 100%, respectively. The specificity of the mDST was lower than those of the other four methods (P<0.05). In conclusion, the MGIT 960 sys-tem, the MTT assay, and the REMA are superior to the PZase activity assay and the mDST in determining the susceptibility ofM. tu-berculosisto PZA. The MTT assay and the REMA might serve as alternative methods for clinical laboratories without access to the MGIT 960 system. For rapid testing in well-equipped laboratories, the mDST might be the best choice, particularly for small quantities ofM. tuberculosis. The PZase activity assay has no obvious advantage in the assessment ofM. tuberculosissusceptibility to PZA, as it is less accurate and requires larger quantities of bacteria.
P
yrazinamide (PZA) is a first-line antituberculosis (anti-TB) drug. It is often used in combination with isoniazid, ethambutol, and rifampin for the treatment of TB (1). PZA can shorten TB treatment time from the 9 to 12 months required prior to its introduction to the current standard treatment time of 6 months. This is often referred to as short-course chemotherapy (2). PZA is more effective against non-replicating, persistent Mycobacterium tuberculosis than any other conventional anti-TB drug (3). However, reliable testing of the sus-ceptibility ofM. tuberculosisto PZAin vitrois challenging since PZA has no apparent effect on actively growing TB bacilli under normal culture conditions at neutral pH. PZA is effective againstM. tubercu-losisonly at an acidic pH, and acidic media inhibit the growth ofM. tuberculosis(4,5). The Bactec 460 and MGIT 960 are the only two methods recommended by the World Health Organization (WHO) for susceptibility testing ofM. tuberculosisto PZA, due to the difficulty in standardizing its susceptibility to PZA on solid media (6). Drug susceptibility testing in liquid media is costly, especially in some re-gions that do not have enough economic capabilities (6). Several other drug susceptibility-testing methods have been developed, in-cluding the molecular drug susceptibility test (mDST) based on the detection of apncAmutation, the pyrazinamidase (PZase) activity assay, and colorimetric methods based on a minimal inhibitory con-centration (MIC) or redox indicator (7,8,9,10,11,12). In this study, we used 432 clinicalM. tuberculosisisolates to compare five methods for determination of the susceptibility ofM. tuberculosisto PZA: the MGIT 960 system, the mDST, the PZase activity assay, and two colorimetric methods [the resazurin microtiter assay (REMA) and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reduction test].MATERIALS AND METHODS
Strains.ClinicalM. tuberculosisspecimen samples were randomly
col-lected from 432 tuberculosis patients in Shanghai Pulmonary Hospital from January to December of 2011. All of the isolates were identified asM.
tuberculosisusing PCR based on the IS6110sequence and a sequence based on thepncAgene (13,14). TheM. tuberculosistype strain H37Rv (ATCC 27294) andMycobacterium bovistype strain (ATCC 19210) were gifts from the National Tuberculosis Reference Laboratory (Beijing, China). The H37Rv strain served as a PZA-susceptible control, andM. bovisserved as a PZA-resistant control. All of the strains were cultured for 3 to 4 weeks in Middlebrook 7H9 liquid medium with 10% oleic albumin dextrose catalase (OADC) supplement (Becton Dickinson, Franklin Lakes, NJ) and then used in the PZA susceptibility tests.
Testing ofM. tuberculosissusceptibility to PZA using the MGIT 960
system.MGIT 960 PZA kits were purchased from Becton Dickinson. The
concentration of PZA used was 100 mg/liter (6). Prior to inoculating the PZA set tubes, 0.8 ml Bactec MGIT 960 PZA supplement was added to the growth control and PZA tubes, and 100l PZA solution was added to the PZA tubes. Inocula were prepared in accordance with the manufac-turer’s instructions. Each colony was transferred into a sterile tube con-taining 4 ml Middlebrook 7H9 medium with 8 to 10 sterile glass beads. The suspension was vortexed for 3 min and left standing undisturbed for 20 min. The supernatant was transferred into a sterile tube and left stand-ing undisturbed for 15 min. Finally, the supernatant was transferred into a third sterile tube. The turbidity was adjusted to a 0.5 McFarland stan-dard and then diluted 1:5 with sterile saline. The MGIT PZA tubes with PZA were then inoculated with a 0.5-ml suspension. The growth control tube was inoculated with 0.5 ml of a 1:10 dilution of theM. tuberculosis suspension. All inoculated PZA sets were loaded into the Bactec MGIT 960 instrument. Using predefined algorithms, readings were automati-cally interpreted by the Epicenter data system of the MGIT 960
instru-Received5 December 2102 Returned for modification18 January 2013 Accepted4 February 2013
Published ahead of print6 February 2013
Address correspondence to Zhongyi Hu, shtblab@163.com.
Copyright © 2013, American Society for Microbiology. All Rights Reserved.
doi:10.1128/JCM.03197-12
on May 16, 2020 by guest
http://jcm.asm.org/
ment, and each colony was reported as either susceptible or resistant. The unloaded PZA set report listed growth units, time to results, and suscep-tible, resistant, or invalid results (15,16).
mDST.Genomic DNA was extracted from theM. tuberculosisstrains
as described previously (17). The entirepncAgene and 82 bp of an up-stream putative regulatory sequence were amplified by PCR with the for-ward primer 5=-CGGTTTCCTTGCCCTGCGGT-3= and the reverse primer 5=-CGATGGCCTCATGCCCCACC-3=. PrimeSTAR HS DNA polymerase (TaKaRa, Dalian, China) was used in the PCR. To determine thepncAsequence, PCR products were purified and sequenced by the BGI Company (Shenzhen, China). The primers were designed using Primer-BLAST software with reference to thepncAgene sequences ofM. tubercu-losis(GenBank accession number NC_000962.2)..The presence of any nuclear acid substitution in the promoter region of thepncAgene or any amino acid substitution in thepncAcoding region was considered indic-ative of PZA resistance. Otherwise, the strain was considered susceptible to PZA.
The C terminus of therpsAgene was amplified by PCR with the for-ward primer 5=-GCGGTGTGGACGGTCTGGTG-3= and the reverse primer 5=-GCCCGGCTGGACCACTTCAC-3=in all of the strains that were detected as strains resistant to PZA by MGIT 960 testing without mutations in the promoters of thepncAandpncAgenes or that showed positive results under the PZase activity assay (18).
PZase activity assay.The PZase activities of theM. tuberculosisstrains
were assessed using a modified Wayne’s procedure as described by Mor-lock et al. (19,20). Dubos broth (Difco Laboratories, Detroit, MI,) with 100 mg/liter PZA was used. Three individuals examined all of the tubes independently. The PZase activity assay was considered positive if a pink band was observed in the upper part of the test medium after incubation at 37°C for 10 days. Strains found positive under the PZase activity assay were considered PZA susceptible. PZA-resistantM. bovisserved as a neg-ative control, andM. tuberculosisH37Rv served as a positive control.
Colorimetric methods.Two colorimetric methods, the resazurin
mi-crotiter assay (REMA) and the MTT reduction test, were used for eachM. tuberculosisstrain simultaneously.
The REMA plate method was carried out as described by Rivoire et al. (21). The REMA was performed in a 96-well plate. Middlebrook 7H9 medium supplemented with OADC enrichment was prepared according to the manufacturer’s instructions, and the pH was adjusted to 5.9 using 1 mol/liter KH2PO4(12). PZA was diluted to final concentrations of 12.5 to 3,200 mg/liter. One hundred microliters of each standardized bacterial inoculum, with a turbidity equivalent to that of a 1 McFarland standard, was diluted 1:20 in Middlebrook 7H9 medium, the pH was adjusted to 5.9, and then the mixture was inoculated. A PZA-free 100% growth con-trol, containing the same number ofM. tuberculosiscells as the testing well with PZA, was prepared. A PZA-free 10% growth control, containing 10% of theM. tuberculosiscells of the testing well with PZA, was also prepared. The third sterile control, containing only liquid culture medium, was prepared as well (22). The plates were covered with lids, placed in a plastic bag, and incubated at 37°C. When white bacterial sediments were ob-served with the naked eye in the 10% growth control, 30l of freshly prepared 0.01% resazurin (Sigma, St. Louis, MO) solution was added to each well, and the plates were reincubated at 37°C for 24 h. The MIC was defined as the lowest PZA concentration capable of preventing the color change from blue to pink due to resazurin reduction.M. tuberculosis strains in which the MIC wasⱕ100 mg/liter were considered susceptible to PZA by the REMA; otherwise, the strains were considered resistant to PZA.
The MTT assay was carried out as described by Abate and coworkers (23). The inoculum was prepared as described above for the REMA plate method, and the same drug concentration ranges were used. Preparation of the 96-well plates was identical to that described for the REMA plate. When white bacterial sediments were observed with the naked eye in the well with the 10% growth control, 10l of the MTT solution (5 g/liter) was added to each well, and the plate was reincubated at 37°C overnight.
When a violet precipitate (formazan) appeared, 50l of SDS-DMF solu-tion (a mixture of 1:1 [vol/vol] 20% sodium dodecyl sulfate and a solusolu-tion of 50%N,N-dimethylformamide) was added to the wells, and the plate was reincubated at 37°C for 3 h. The MIC was defined as the lowest PZA concentration capable of preventing the color change from yellow to vio-let due to MTT reduction. Strains in which the MIC wasⱕ100 mg/liter were considered susceptible to PZA by the MTT assay; otherwise, the isolates were considered resistant to PZA.
Resolution of discrepancies.If there were any discrepancies in the
results for one strain among the five methods, the five methods were then repeated an additional two times. The same results in at least two of the three tests for one method were considered the final results.
Data analysis.Data analysis was carried out using MedCalc software
(MedCalc, Belgium). A comparison of the sensitivities and specificities was performed using a chi-square test.
RESULTS
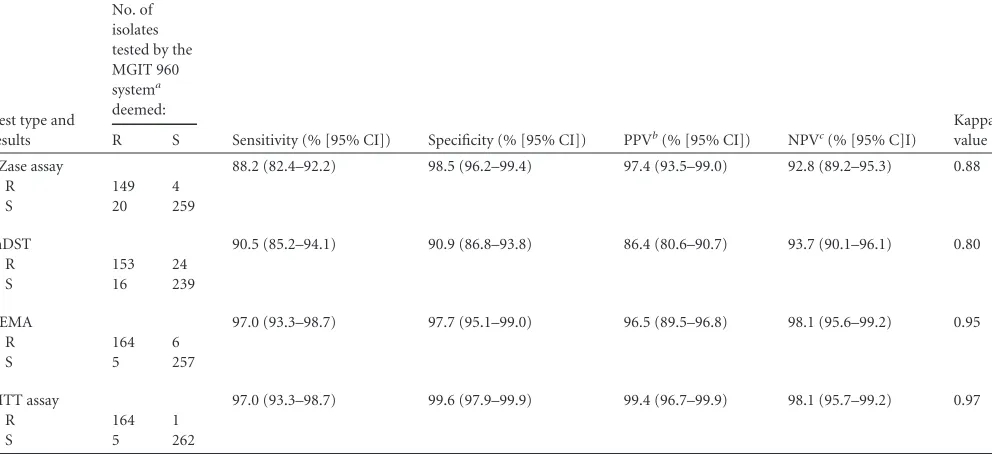
Of the 432 clinicalM. tuberculosisisolates, 169 were determined to be resistant to PZA and 263 susceptible to PZA by the MGIT 960 system, 149 strains were determined to be resistant to PZA and 259 strains susceptible to PZA by the PZase activity assay, 153 strains were determined to be resistant to PZA and 239 isolates susceptible to PZA by the mDST, 164 strains were determined to be resistant to PZA and 257 strains susceptible to PZA by the REMA, and 164 strains were determined to be resistant to PZA and 262 strains susceptible to PZA by the MTT assay (Table 1). No mutation was found in any part of the 117-amino-acid C terminus of RpsA in the 20 strains found resistant to PZA by the MGIT 960 test without mutations in the promoters and structural gene of pncAor in the 8 strains resistant to PZA by MGIT 960 testing that showed positive results by the PZase activity assay.
In comparison to the results obtained with the MGIT 960 sys-tem (as a reference of 100% sensitivity), the sensitivities of the PZase activity assay, the mDST, the REMA, and the MTT assay were 88.2%, 90.5%, 97.0%, and 97.0%, respectively. The sensitiv-ities of the REMA and the MTT assay were significantly higher than those of the mDST and the PZase activity assay (2⫽9.719, P⬍0.05, and2⫽6.144,P⬍0.05, respectively). There was no
significant difference when we compared the sensitivities of the REMA and the MTT assay and no significant difference when we compared the sensitivities of the mDST and the PZase activity assay (P⬎0.05). The specificities of the PZase activity assay, the mDST, the REMA, and the MTT assay were 98.5%, 90.9%, 97.7%, and 99.6%, respectively. The specificities of the PZase activity as-say, the REMA, and the MTT assay were significantly higher than that of the mDST (2⫽38.155,P⬍0.05). There was no
signifi-cant difference among the specificities of the PZase activity assay, the REMA, and the MTT assay (P⬎0.05).
Of the 432M. tuberculosisstrains, 374 strains showed the same drug susceptibility testing results with all of the five methods ex-amined, and 58 strains showed different results (Table 2). Among 58 discrepant results, 39 results were the same with four testing methods, while 10 results of the PZase activity assay, 26 results of the mDST, and 3 results of the REMA were different from those from the other four methods. Among 58 discrepant results, 19 results were the same for three methods but different from those for the other two methods. When the results were the same in three methods or when more than three methods yielded refer-ence results, there were 169 isolates that were resistant to PZA and 263 isolates that were susceptible to PZA. The sensitivities of the MGIT 960 system, the PZase activity, the mDST, the REMA, and
on May 16, 2020 by guest
http://jcm.asm.org/
the MTT assay were 98.8%, 88.8%, 90.5%, 98.8%, and 98.2%, respectively (Table 3). The MGIT 960 system, the REMA, and the MTT assay showed higher sensitivities than those of the PZase activity assay and the mDST (P⬍0.05). There were no significant differences among sensitivities of the MGIT 960 system, the REMA, and the MTT assay and no significant difference when comparing the sensitivities of the mDST and the PZase activity assay (P⬎0.05). The specificities of the MGIT 960 system, the PZase activity assay, the mDST, the REMA, and the MTT assay were 99.2%, 98.9%, 90.9%, 98.5%, and 100%, respectively (Table 3). The MGIT 960 system, the REMA, the MTT, and the PZase activity assay showed higher specificities than that of the mDST (2⫽60.177,P⬍0.05). There were no significant differences among the specificities of the MGIT 960 system, the REMA, the MTT, and the PZase activity assay (P⬎0.05).
Several parameters related to clinical application were also an-alyzed. The mDST was found to be the fastest of the methods examined here, needing only 1 day. The other four methods needed about 7 to 18 days once a pure isolate was obtained in order to finish the test. While the mDST required only a very small quantity of bacteria, on the order of several CFU, the PZase activ-ity assay required a much larger quantactiv-ity of bacteria (at least 3 to 5 mg). An insufficient number of bacteria in the PZase activity assay can lead to falsely negative results. The PZase activity assay, the REMA, and the MTT test do not require any special equipment other than a constant-temperature incubator; however, the MGIT 960 system and the mDST do require special equipment, such as MGIT equipment and a DNA sequencer.
DISCUSSION
Drug-resistant TB has increased patient morbidity and mortality as well as treatment-associated costs (24). Rapid and accurate drug susceptibility testing methods are important in the selection
of appropriate TB drugs. Understanding drug resistance patterns within a given community is also of enormous epidemiological significance, since it provides indicators of the existence and prev-alence of primary and acquired drug resistance, which can be use-ful in the evaluation of the efficacy of anti-TB treatment (25,26). For most anti-TB drugs, such as rifampin, isoniazid, ethambutol, and streptomycin, several WHO-recommended methods of as-sessing susceptibility testing are available. These include the Low-enstein-Jensen, Middlebrook 7H10, Middlebrook 7H11, Bactec 460, and MGIT 960 methods. However, only the Bactec 460 and MGIT 960 methods are recommended by the WHO for PZA, one of the most important early oral anti-TB agents (6). The main reason for this is that many of the other methods of testing sus-ceptibility to PZA have certain disadvantages, such as poor or slow growth and difficult standardization, that render them unsuitable for use in clinical mycobacteriology laboratories. Several other methods of testing the susceptibility ofM. tuberculosisto PZA have been developed. These include the mDST based on the de-tection ofpncAmutations, the PZase activity assay, and the color-imetric methods, which are based on MIC and redox indicators (7,
8,9,10,11,12). A few studies on the performances of these meth-ods have been reported (7,8,9,10,11,12); however, few studies have been done on the use of these methods for testing the suscep-tibility ofM. tuberculosisto PZA or comparing several methods synchronously using large quantities of clinically isolatedM. tu-berculosisstrains.
Our study was a one-to-one comparison of the performances of five methods for testing the susceptibility ofM. tuberculosisto PZA by using 432 clinicalM. tuberculosisisolates. The MGIT 960 system was used as a reference according to guidelines from the WHO (6). The sensitivity and specificity of the MTT assay were found to be the best, followed by the REMA. The sensitivities of the mDST and the PZase activity assay were similar, and the spec-TABLE 1PZA susceptibility testing of 432 clinicalM. tuberculosisisolates using the REMA, the MTT, the PZase activity assay, and the mDST
Test type and results
No. of isolates tested by the MGIT 960 systema deemed:
Sensitivity (% [95% CI]) Specificity (% [95% CI]) PPVb(% [95% CI]) NPVc(% [95% C]I)
Kappa value
R S
PZase assay 88.2 (82.4–92.2) 98.5 (96.2–99.4) 97.4 (93.5–99.0) 92.8 (89.2–95.3) 0.88
R 149 4
S 20 259
mDST 90.5 (85.2–94.1) 90.9 (86.8–93.8) 86.4 (80.6–90.7) 93.7 (90.1–96.1) 0.80
R 153 24
S 16 239
REMA 97.0 (93.3–98.7) 97.7 (95.1–99.0) 96.5 (89.5–96.8) 98.1 (95.6–99.2) 0.95
R 164 6
S 5 257
MTT assay 97.0 (93.3–98.7) 99.6 (97.9–99.9) 99.4 (96.7–99.9) 98.1 (95.7–99.2) 0.97
R 164 1
S 5 262
a
The MGIT 960 system served as the reference (100% sensitivity). R, resistance to PZA; S, susceptibility to PZA.
bPPV, positive predictive value. c
NPV, negative predictive value.
on May 16, 2020 by guest
http://jcm.asm.org/
[image:3.585.46.542.80.307.2]TABLE 2PZA susceptibility testing results of 58 strains ofM. tuberculosisshowing disagreement among the five methods tested
Strain no.
Drug susceptibilityadetermined
by: MIC (g/ml) found with: Changes in PncA amino acids MGIT 960
system
PZase activity
assay REMA MTT test Codon Amino acid substitutions 1 R S 400 200 133 Ile (ATT) to Thr (ACT) 2 R S 3,200 1,600 12 Asp (GAC) to Asn (AAC) 3 R S 200 200 151 Leu (TTG) to Ser (TCG) 4 R S 3,200 3,200 \a No substitution
5 R S 400 400 162 Gly (GGT) to Ser (AGT) 6 R S 100 50 116 Leu (CTG to Pro (CCG) 7 R S 50 50 81 Phe (TTC) to Val (GTC) 8 R S 800 400 62 Pro (CCG) to Leu (CTG) 9 R S 3,200 3,200 \ No substitution
10 R S 3,200 3,200 136 Frameshift (insert A behind 407 bp) 11 R S 1,600 1,600 \ No substitution
12 R S 200 200 \ No substitution 13 R S 3,200 3,200 76 Thr (ACT) to Pro (CCT) 14 R S 3,200 3,200 12 Asp (GAC) to Asn (AAC) 15 R S 3,200 3,200 \ No substitution 16 R S 3,200 3,200 12 Asp (GAC) to Asn (AAC) 17 R S 3,200 3,200 \ No substitution 18 R S 3,200 3,200 \ No substitution 19 R S 800 800 82 His (CAT) to Arg (CGT) 20 R S 1,600 1,600 \ No substitution 21 R R 25 50 146 Ala (GCG) to Thr (ACG) 22 R R 100 100 136 Asp(GAT) to Gly (GGT) 23 R R 50 50 171 Ala (GCG) to Val (GTG) -24 R R 3,200 3,200 \ No substitution 25 R R 1,600 1,600 \ No substitution 26 R R 3,200 3,200 \ No substitution 27 R R 1,600 1,600 \ No substitution 28 R R 800 400 \ No substitution 29 R R 800 800 \ No substitution 30 R R 3,200 3,200 \ No substitution 31 R R 800 800 \ No substitution 32 S R 12.5 12.5 136 Asp (GAT) to Gly (GGT) 33 S R 50 50 171 Ala (GCG) to Thr (ACG) 34 S R 50 50 35 Leu (CTG) to Arg (CGG) 35 S R 200 50 157 Val (GTG) to Gly (GGG) 36 S S 200 200 12 Asp(GAC) to Ala (GCC) 37 S S 200 100 \ No substitution 38 S S 200 100 \ No substitution 39 S S 200 50 \ No substitution 40 S S 200 100 5 Ile (ATC) to Thr (ACC) 41 S S 100 100 155 Val (GTG) to Ala (GCG) 42 S S 50 50 102 Ala(GCG) to Val (GTG) 43 S S 50 50 102 Ala (GCG) to Val (GTG) 44 S S 25 12.5 102 Ala (GCG) to Val (GTG) 45 S S 100 100 67–68 Insert Ser (TCG) 46 S S 100 100 11 Asn (AAC) to Asp (GAC) 47 S S 25 25 12 Asp (GAC) to Ala (GCC) 48 S S 25 25 154 Arg (AGG) to Gly (GGG)
49 S S 25 25 187 Termination codon (TGA) to Arg (CGA) 50 S S 50 50 171 Ala (GCG) to Val (GTG)
51 S S 100 100 160 Thr (ACA) to Ala (GCA) 52 S S 12.5 12.5 97 Gly (GGT) to Asp (GAT) 53 S S 100 50 116 Leu (CTG) to Pro (CCG) 54 S S 100 100 65 Ser (TCC) to Pro (CCC) 55 S S 100 100 46 Ala (GCA) to Val (GTA) 56 S S 50 25 134 Ala (GCC) to Val (GTC) 57 S S 100 50 12 Asp (GAC) to Ala (GCC) 58 S S 100 100 128 Val (GTC) to Phe (TTC)
aR, resistance to PZA; S, susceptibility to PZA. b\, no codon change.
on May 16, 2020 by guest
http://jcm.asm.org/
ificity of the PZase activity assay was found to be better than that of the mDST.
Some studies reported the potential for erroneous results indi-cating resistance when using the Bactec MGIT 960 system for test-ing the susceptibility ofMycobacterium tuberculosisto PZA (27,
28). Another study showed that the critical concentration for drug susceptibility testing ofMycobacterium tuberculosisagainst PZA should be reevaluated and revised (29). Several previous reports suggested that there were some deficiencies in the MGIT 960 sys-tem. In the current study, we used three or more methods for reference results. The performance of the MGIT 960 system showed perfect agreement with the reference results (kappa value⫽0.98). Although 0.9% (4/432) of the strains showed discrepant results between the MGIT 960 system and the reference results, no one result from the MGIT 960 system was absolutely different from those of the other four testing methods for one strain. The results using the MGIT 960 system from two strains were concordant with those from the mDST; one was concordant with the MTT assay, and one was concordant with the PZase activity assay. We speculated that the main reason for the discrepant results from the MGIT system was the difference in methodologies.
The REMA and the MTT test also showed the same perfor-mance as the MGIT 960 system for the susceptibility testing of PZA (kappa values⫽0.97 and 0.99, respectively). Although there were some discrepancies among the MGIT 960 system and the REMA and the MTT assay, the discrepancies were likely due to the difference in methodology. The MGIT 960 system records bacterial growth by oxygen consumption in liquid culture, while the REMA and the MTT test indicate bacterial growth through redox reactions. Neither of them directly reflects changes in the amount of bacteria.
The results using other methods also offered some important hints. The PZase assay and the mDST are all based on the as-sumption that PZA-susceptibleM. tuberculosisstrains produce pyrazinamidase, which hydrolyzes PZA to the antibacterial moiety pyrazinoic acid (POA) (30).pncAis a gene encoding pyrazinamidase. Previous studies have verified that the muta-tion inpncAcan inactivate PZase and confer resistance to PZA (31). The current study indicated that the assumption regard-ing the mechanism ofM. tuberculosisresistance to PZA based on pyrazinamidase did not explain all of the resistance mech-anisms ofM. tuberculosisto PZA.
In this study, 9.5% (16/169) of the PZA-resistant strains were determined to be positive to PZase activity testing, indicating that, for these strains, PZA resistance was not caused by the inactivation of PZase but by some other unknown cause(s). Other studies have also detected PZase activity in PZA-resistant strains (10,32,33). Only 1% (3/260) of the PZA-susceptible strains was negative according to the PZase activity assay, probably due to the weak activity of PZase or to the small quantity ofM. tuberculosis, which makes it difficult to observe the Wayne assay color change with the naked eye at 10 days. Recently, Zhou et al. reported a new PCR-basedin vitro-synthesized pyrazinamidase method. This method might exclude false negatives from the PZase activity assay due to the small quantity ofM. tuberculosis (34); however, this method does not solve the false susceptibil-ity of the PZase activsusceptibil-ity assay. False susceptibilsusceptibil-ity is the major limitation of the PZase activity assay and should be considered seriously in clinical settings.
[image:5.585.46.545.85.345.2]The mDST is an indirect method of assessing PZA susceptibil-ity. PZase is inferred by the assessment of changes in the sequence ofpncA. If no amino acid substitution is detected in the coding TABLE 3PZA susceptibility testing of 432 clinicalM. tuberculosisisolates using the MGIT 960 system, the REMA, the MTT test, the PZase activity assay, and the mDST
Test type and results
No. of isolates with indicated reference resultsa
Sensitivity (% [95% CI]) Specificity (% [95% CI]) PPV (% [95% CI]) NPV (% [95% CI]) Kappa value
R S
MGIT 960 98.8 (95.8–99.7) 99.2 (97.3–99.8) 98.8 (95.8–99.7) 99.24 (97.3–99.8) 0.98
R 167 2
S 2 261
PZase assay 88.8 (83.1–92.7) 98.9 (96.7–99.6) 98.0 (94.4–99.3) 93.19 (89.6–95.6) 0.89
R 150 3
S 19 260
mDST 90.5 (85.2–94.1) 90.9 (86.8–93.8) 86.4 (80.6–90.7) 93.73 (90.1–96.1) 0.81
R 153 24
S 16 239
REMA 98.8 (95.8–99.7) 98.5 (96.2–99.4) 97.7 (94.1–99.1) 99.23 (97.3–99.8) 0.97
R 167 4
S 2 259
MTT assay 98.2 (94.9–99.4) 100.0 (98.6–100.0) 100.0 (94.9–99.4) 98.87 (96.7–99.6) 0.99
R 166 0
S 3 263
aThe same results in three or more methods served as the reference (100% sensitivity). R, resistance to PZA; S, susceptibility to PZA.
on May 16, 2020 by guest
http://jcm.asm.org/
sequence and no base substitution is detected in the promoter sequence, PZase is considered active; otherwise, PZase is consid-ered inactive or absent. In this study, 9% of the strains with the wild-typepncAgene (16/169) were found to be resistant to PZA, including 8 strains that tested positive for PZase activity and 8 strains that tested negative by the PZase activity assay. These re-sults suggest that there might be other genes that regulate the expression of PZase and/or other unknown mechanisms for PZA resistance.
In this study, amino acid substitutions in thepncAgenes of PZA-susceptible strains were also detected. Most of these strains were positive for PZase activity. According to previous studies, mutations at certain amino acid sites, such as codons 8, 96, 134, and 138, at active sites, and codons 13, 49, 51, 57, and 68, coordi-nated to the Fe2⫹ion, are closely associated withM. tuberculosis resistance to PZA (35,36). Mutations at these sites can causeM. tuberculosisto lose PZase activity and render the strain resistant to PZA. In this study, most amino acid substitutions in PZA-suscep-tible strains were far from the functional sites, and almost all of the amino acid substitutions occurred in PZA-susceptible strains. For this reason, we hypothesized that these amino acid substitutions did not lead to the loss of PZase activity. Thein vitro-synthesized PZase assay can evaluate the correlation between the amino acid substitutions and PZase activity. If the amino acid substitutions far from the activity sites of PZase can be confirmed to be irrele-vant toM. tuberculosisresistance to PZA, then the specificity of the mDST can be improved. Additionally, Zhang and coworkers found that POA binds to RpsA, subsequently inhibiting trans-translation rather than canonical trans-translation (18). They found that the deletion of amino acid 438 in the C terminus of RpsA can cause resistance to PZA in a clinicalM. tuberculosisisolate lacking pncAmutations. In the current study, we did not detect the dele-tion of amino acid 438 in the C terminus of RpsA and did not find any other mutation in the C terminus of RpsA. This suggests that the mutation rate of this site in therpsAgene in clinicalM. tuber-culosisisolates is too low to be a detection target for mDST. The sensitivities of the PZase activity assay and the mDST test should be improved to further clarify what the mechanism of PZA resis-tance is.
The MGIT 960 system, used as a reference method by the WHO, is still a good PZA susceptibility testing method forM. tuberculosis. The MTT test and the REMA are suitable alternatives to the MGIT 960 system for laboratories that do not have access to the MGIT 960 system. The mDST does not take the place of the phenotype drug susceptibility test due to the low sensitivity and specificity, but it can satisfy the rapid test requirements and func-tions using smaller quantities ofM. tuberculosisthan some other types of high-end laboratory equipment. Although the specificity of the PZase activity assay is high, the PZase activity assay has been shown to be less accurate than the MGIT 960 system, the MTT test, and the REMA, owing to their low sensitivities. In addition, the PZase activity assay requires a larger quantity of bacteria and does not take significantly less time, so there is no obvious advan-tage to using the PZase activity assay for the assessment of PZA susceptibility.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant 81201323) and the Shanghai Committee of Science and Technology, China (grant 124119a5200).
We are grateful to all members of the Shanghai Key Laboratory of Tuberculosis, Shanghai Pulmonary Hospital, Medical School, Tongji University, for their knowledge, helpfulness, and willingness to share.
We declare that no conflicts of interest exist.
REFERENCES
1.World Health Organization.2010. Treatment of tuberculosis: guidelines for national programmes, 4th ed. WHO/HTM/TB/2009.420. World Health Organization, Geneva, Switzerland.
2.Mitchison DA.1985. The action of antituberculosis drugs in short-course chemotherapy. Tubercle66:219 –225.
3.Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z.2003. Mode of action of pyrazinamide: disruption ofMycobacterium tuberculosismembrane transport and energetics by pyrazinoic acid. J. Antimicrob. Chemother.
52:790 –795.
4.Tarshis MS, Jr, Weed WA.1953. Lack of significantin vitrosensitivity of Mycobacterium tuberculosisto pyrazinamide on three different solid me-dia. Am. Rev. Tuberc.67:391–395.
5.Salfinger M, Heifets LB.1988. Determination of pyrazinamide MICs for Mycobacterium tuberculosisat different pHs by the radiometric method. Antimicrob. Agents Chemother.32:1002–1004.
6.World Health Organization.2009. Guidelines for surveillance of drug resistance in tuberculosis, 4th ed. WHO/HTM/TB/2009.422. World Health Organization, Geneva, Switzerland.
7.Denkin S, Volokhov D, Chizhikov V, Zhang Y.2005. Microarray-based pncAgenotyping of pyrazinamide-resistant strains ofMycobacterium tu-berculosis. J. Med. Microbiol.54:1127–1131.
8.Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, Hooks DP, Cowan LS, Plikaytis BB, Posey JE. 2011. Molecular detection of mutations associated with first- and second-line drug resis-tance compared with conventional drug susceptibility testing of Mycobac-teriumtuberculosis. Antimicrob. Agents Chemother.55:2032–2041. 9.Miller MA, Thibert L, Desjardins F, Siddiqi SH, Dascal A.1995. Testing
of susceptibility ofMycobacterium tuberculosisto pyrazinamide: compar-ison of Bactec method with pyrazinamidase assay. J. Clin. Microbiol.33: 2468 –2470.
10. Singh P, Wesley C, Jadaun GP, Malonia SK, Das R, Upadhyay P, Faujdar J, Sharma P, Gupta P, Mishra AK, Singh K, Chauhan DS, Sharma VD, Gupta UD, Venkatesan K, Katoch VM.2007. Comparative evaluation of Löwenstein-Jensen proportion method, BacT/ALERT 3D system, and enzymatic pyrazinamidase assay for pyrazinamide suscepti-bility testing ofMycobacterium tuberculosis. J. Clin. Microbiol.45:76 – 80. 11. Leite CQ, Beretta AL, Anno IS, Telles MA. 2000. Standardization of broth microdilution method forMycobacterium tuberculosis. Mem. Inst. Oswaldo Cruz95:127–129.
12. Syre H, Øvreås K, Grewal HM.2010. Determination of the susceptibility ofMycobacterium tuberculosisto pyrazinamide in liquid and solid media assessed by a colorimetric nitrate reductase assay. J. Antimicrob. Che-mother.65:704 –712.
13. Thierry D, Cave MD, Eisenach KD, Crawford JT, Bates JH, Gicquel B, Guesdon JL.1990. IS6110, an IS-like element ofMycobacterium tubercu-losiscomplex. Nucleic Acids Res.18:188. doi:10.1093/nar/18.1.188. 14. Barouni AS, Augusto CJ, Lopes MT, Zanini MS, Salas CE.2004. A pncA
polymorphism to differentiate betweenMycobacterium bovisand Myco-bacterium tuberculosis. Mol. Cell. Probes18:167–170.
15. Kontos F, Nicolaou S, Kostopoulos C, Gitti Z, Petinaki E, Maniati M, Anagnostou S, Raftopoulou A, Papageorgiou P, Scrioubellou A, Tselen-tis I, ManiaTselen-tis AN.2003. Multicenter evaluation of the fully automated Bactec MGIT 960 system for susceptibility testing ofMycobacterium tu-berculosisto pyrazinamide: comparison with the radiometric Bactec 460TB system. J. Microbiol. Methods55:331–333.
16. Pfyffer GE, Palicova F, Rüsch-Gerdes S.2002. Testing of susceptibility of Mycobacterium tuberculosisto pyrazinamide with the nonradiometric BACTEC MGIT 960 system. J. Clin. Microbiol.40:1670 –1674. 17. Cui Z, Wang J, Lu J, Huang X, Hu Z.2011. Association of mutation
patterns ingyrA/Bgenes and ofloxacin resistance levels inMycobacterium tuberculosisisolates from East China in 2009. BMC Infect. Dis.11:78. doi: 10.1186/1471-2334-11-78.
18. Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, III, Wang H, Zhang W, Zhang Y.2011. Pyrazinamide inhibits trans-translation in Mycobac-terium tuberculosis. Science333:1630 –1632.
on May 16, 2020 by guest
http://jcm.asm.org/
19. Wayne LG.1974. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am. Rev. Respir. Dis.109:147–151. 20. Morlock GP, Crawford JT, Butler WR, Brim SE, Sikes D, Mazurek GH,
Woodley CL, Cooksey RC.2000. Phenotypic characterization ofpncA mutants ofMycobacterium tuberculosis. Antimicrob. Agents Chemother.
44:2291–2295.
21. Rivoire N, Ravololonandriana P, Rasolonavalona T, Martin A, Portaels F, Ramarokoto H, Rasolofo Razanamparany V.2007. Evaluation of the resazurin assay for the detection of multidrug-resistantMycobacterium tuberculosisin Madagascar. Int. J. Tuberc. Lung Dis.11:683– 688. 22. CLSI.2011. Susceptibility testing of mycobacteria, Nocardiae, and other
aerobic actinonycetes; approved standard, 2nd ed. CLSI M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA.
23. Mshana RN, Tadesse G, Abate G, Miörner H.1998. Use of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide for rapid detec-tion of rifampin-resistantMycobacterium tuberculosis. J. Clin. Microbiol.
36:1214 –1219.
24. Iseman MD. 1993. Treatment of multidrug-resistant tuberculosis. N. Engl. J. Med.329:784 –791.
25. Chaulet P, Boulahbal F, Grosset J.1995. Surveillance of drug resistance for tuberculosis control: why and how? Tuber. Lung Dis.76:487– 492. 26. World Health Organization.1994. World Health Organization
Tuber-culosis Programme and International Union Against TuberTuber-culosis and Lung Disease guidelines for surveillance of drug resistance in tuberculosis. WHO/TB/94.178. World Health Organization, Geneva, Switzerland. 27. Chedore P, Bertucci L, Wolfe J, Sharma M, Jamieson F.2010. Potential
for erroneous results indicating resistance when using the Bactec MGIT 960 system for testing susceptibility ofMycobacterium tuberculosisto pyr-azinamide. J. Clin. Microbiol.48:300 –301.
28. Piersimoni C, Mustazzolu A, Giannoni F, Bornigia S, Gherardi G, Fattorini L.2013. Prevention of false resistance results obtained testing susceptibility ofMycobacterium tuberculosisto pyrazinamide with the
Bac-tec MGIT 960 system using a reduced inoculum. J. Clin. Microbiol.51: 291–294.
29. Werngren J, Sturegård E, Juréen P, Angeby K, Hoffner S, Schön T.
2012. Reevaluation of the critical concentration for drug susceptibility testing ofMycobacterium tuberculosisagainst pyrazinamide using wild-type MIC distributions andpncAgene sequencing. Antimicrob. Agents Chemother.56:1253–1257.
30. Heifets LB, Flory MA, Lindholm-Levy PJ.1989. Does pyrazinoic acid as an active moiety of pyrazinamide have specific activity against Mycobac-terium tuberculosis? Antimicrob. Agents Chemother.33:1252–1254. 31. Sun Z, Scorpio A, Zhang Y.1997. ThepncAgene from naturally
pyrazi-namide-resistantMycobacterium aviumencodes pyrazinamidase and con-fers pyrazinamide susceptibility to resistantM.tuberculosiscomplex or-ganisms. Microbiology143:3367–3373.
32. Sharma B, Pal N, Malhotra B, Vyas L, Rishi S.2010. Comparison of MGIT 960 & pyrazinamidase activity assay for pyrazinamide susceptibility testing ofMycobacterium tuberculosis. Indian J. Med. Res.132:72–76. 33. Butler WR, Kilburn JO.1983. Susceptibility ofMycobacterium
tubercu-losisto pyrazinamide and its relationship to pyrazinamidase activity. An-timicrob. Agents Chemother.24:600 – 601.
34. Zhou M, Geng X, Chen J, Wang X, Wang D, Deng J, Zhang Z, Wang W, Zhang XE, Wei H.2011. Rapid colorimetric testing for pyrazinamide susceptibility ofM.tuberculosisby a PCR-basedin-vitrosynthesized pyr-azinamidase method. PLoS One 6:e27654. doi:10.1371/journal.pone .0027654.
35. Quiliano M, Gutierrez AH, Gilman RH, López C, Evangelista W, Sotelo J, Sheen P, Zimic M.2011. Structure-activity relationship in mutated pyrazinamidases fromMycobacterium tuberculosis. Bioinformation
6:335–339.
36. Petrella S, Gelus-Ziental N, Maudry A, Laurans C, Boudjelloul R, Sougakoff W.2011. Crystal structure of the pyrazinamidase of Mycobac-terium tuberculosis: insights into natural and acquired resistance to pyrazi-namide. PLoS One6:e15785. doi:10.1371/journal.pone.0015785.