organic papers
o2510
Yinet al. C24H28Cl4N2 doi:10.1107/S1600536806019167 Acta Cryst.(2006). E62, o2510–o2511
Acta Crystallographica Section E Structure Reports Online
ISSN 1600-5368
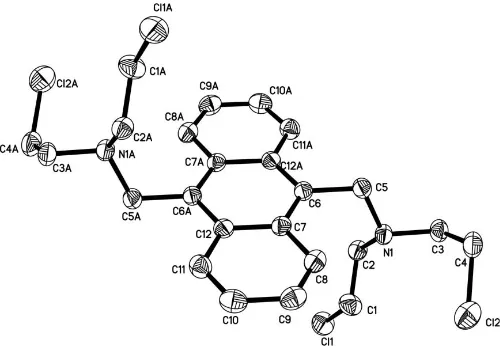
9,10-Bis(dichloroethylaminomethyl)anthracene
Li-Na Yin, Qing-Xiang Liu,* Xiu-Mei Wu, Feng-Jin Cheng and Jian-Hua Guo
College of Chemistry and Life Science, Tianjin Normal University, Tianjin 300074, People’s Republic of China
Correspondence e-mail: qxliu@eyou.com
Key indicators
Single-crystal X-ray study T= 293 K
Mean(C–C) = 0.003 A˚ Rfactor = 0.032 wRfactor = 0.089
Data-to-parameter ratio = 15.3
For details of how these key indicators were automatically derived from the article, see http://journals.iucr.org/e.
Received 26 April 2006 Accepted 23 May 2006
#2006 International Union of Crystallography All rights reserved
The title compound, C24H28Cl4N2, was obtained by the
reaction of 9,10-dichloromethylanthracene and dichloroethyl-amine. The molecule is centrosymmetric, with the substituents above and below the anthracene plane.
Comment
Fluorescent chemosensors constitute an important aspect of supramolecular chemistry (de Silver et al., 1997). Many of these fluorescent sensors with an anthracene ring system have been designed and investigated (Gunnlaugsson et al., 2002). Amine compounds with anthracene chromophores have attracted considerable attention in the past decade owing to their capability to monitor acidic changes of water in rivers (Luigi & Antonio, 1995). We report here the synthesis and crystal structure of the title compound, (2).
[image:1.610.211.509.368.484.2] [image:1.610.207.458.539.712.2]The molecule of (2) is centrosymmetric, with the substit-uents above and below the anthracene ring plane (Fig. 1). The
Figure 1
N—C bond lengths [1.459 (2), 1.452 (2) and 1.466 (2) A˚ ] are similar to those [1.509 (6) and 1.494 (6) A˚ ] of 9,10-dibenzyl-aminemethylanthracene (Changet al., 2000).
Experimental
To a toluene (100 ml) solution of 9,10-dichloromethyl-anthracene (5.00 g, 0.018 mol) were added dichloroethylamine (15.49 g, 0.109 mol) and K2CO3 (24.80 g, 0.180 mol) and the
solution was stirred for 6 h at 353 K. The toluene solution was washed with water (150 ml) and dried with anhydrous MgSO4,
then concentrated to 40 ml; hexane (5 ml) was added to give a yellow solid (yield: 7.20 g, 81.4%; m.p. 535–537 K). Crystals of (2) suitable for X-ray diffraction were obtained by slow evaporation of a methanol solution at room temperature. Analysis calculated for C24H28Cl4N2: C 59.28, H 5.80, Cl 29.16,
N 5.76%; found C 59.11, H 5.34, Cl 28.96, N 5.42%.
Crystal data
C24H28Cl4N2
Mr= 486.28
Monoclinic,P21=c
a= 13.3505 (12) A˚
b= 10.6953 (10) A˚
c= 8.3412 (8) A˚
= 92.879 (1)
V= 1189.52 (19) A˚3
Z= 2
Dx= 1.358 Mg m 3
MoKradiation
= 0.51 mm 1
T= 293 (2) K Block, yellow 0.220.200.16 mm
Data collection
Bruker SMART CCD area-detector diffractometer
’and!scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin= 0.891,Tmax= 0.921
6278 measured reflections 2093 independent reflections 1669 reflections withI> 2(I)
Rint= 0.017
max= 25.0
Refinement
Refinement onF2 R[F2> 2(F2)] = 0.032
wR(F2) = 0.089
S= 1.07 2093 reflections 137 parameters
H-atom parameters constrained
w= 1/[2
(Fo2) + (0.0356P)2
+ 0.408P]
whereP= (Fo2+ 2Fc2)/3
(/)max< 0.001
max= 0.25 e A˚ 3
min= 0.25 e A˚ 3
Extinction correction:SHELXL97
Extinction coefficient: 0.0134 (15)
All H atoms were initially located in a difference Fourier map, but were then constrained to an ideal geometry, with C—H = 0.93–0.97 A˚ andUiso(H) = 1.2Ueq(C), and refined as riding on their parent atoms. Data collection:SMART(Bruker, 2000); cell refinement:SAINT (Bruker, 2000); data reduction:SAINT; program(s) used to solve structure:SHELXS97(Sheldrick, 1990); program(s) used to refine structure: SHELXL97 (Sheldrick, 1997a); molecular graphics: SHELXTL(Sheldrick, 1997b); software used to prepare material for publication:SHELXTL.
This project was supported by Tianjin Normal University Personnel Division (No. 5rl036).
References
Bruker (2000).SMARTandSAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
Chang, T., Heiss, A. M., Cantrill, S. J., Fyfe, M. C. T., Pease, A. R., Rowan, S. J., Stoddart, J. F. & Williams, D. J. (2000).Org. Lett.2, 2943–2946.
Gunnlaugsson, T., Bichell, B. & Nolan, C. (2002).Tetrahedron Lett.43, 4989– 4992.
Luigi, F. & Antonio, P. (1995).Chem. Soc. Rev.24, 197–202. Sheldrick, G. M. (1990).Acta Cryst.A46, 467–473.
Sheldrick, G. M. (1996).SADABS. University of Go¨ttingen, Germany. Sheldrick, G. M. (1997a).SHELXL97. University of Go¨ttingen, Germany. Sheldrick, G. M. (1997b).SHELXTL. Version 5.10 for Windows NT. Bruker
AXS Inc., Madison, Wisconsin, USA.
supporting information
sup-1 Acta Cryst. (2006). E62, o2510–o2511
supporting information
Acta Cryst. (2006). E62, o2510–o2511 [https://doi.org/10.1107/S1600536806019167]
9,10-Bis(dichloroethylaminomethyl)anthracene
Li-Na Yin, Qing-Xiang Liu, Xiu-Mei Wu, Feng-Jin Cheng and Jian-Hua Guo
9,10-Bis(dichloroethylaminomethyl)anthracene
Crystal data
C24H28Cl4N2 Mr = 486.28
Monoclinic, P21/c
a = 13.3505 (12) Å
b = 10.6953 (10) Å
c = 8.3412 (8) Å
β = 92.879 (1)°
V = 1189.52 (19) Å3
Z = 2
F(000) = 508
Dx = 1.358 Mg m−3
Melting point: 162 K
Mo Kα radiation, λ = 0.71073 Å
Cell parameters from 2134 reflections
θ = 3.1–25.1°
µ = 0.51 mm−1
T = 293 K
Block, yellow
0.22 × 0.20 × 0.16 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Radiation source: fine-focus sealed tube Graphite monochromator
φ and ω scans
Absorption correction: multi-scan (SADABS; Sheldrick, 1996)
Tmin = 0.891, Tmax = 0.921
6278 measured reflections 2093 independent reflections 1669 reflections with I > 2σ(I)
Rint = 0.017
θmax = 25.0°, θmin = 3.1°
h = −15→15
k = −12→12
l = −7→9
Refinement
Refinement on F2
Least-squares matrix: full
R[F2 > 2σ(F2)] = 0.032 wR(F2) = 0.089
S = 1.07
2093 reflections 137 parameters 0 restraints
Primary atom site location: structure-invariant direct methods
Secondary atom site location: difference Fourier map
Hydrogen site location: inferred from neighbouring sites
H-atom parameters constrained
w = 1/[σ2(F
o2) + (0.0356P)2 + 0.408P]
where P = (Fo2 + 2Fc2)/3
(Δ/σ)max < 0.001
Δρmax = 0.25 e Å−3
Δρmin = −0.25 e Å−3
Extinction correction: SHELXL97, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4
Special details
Experimental. 1H NMR (300 MHZ, CDCl
3, δ, p.p.m.): 3.483 (t, 8H, CH2), 3.992 (t, 8H, CH2), 4.70 (s, 4H, CH2), 7.50–
7.55 (m, 4H, AnH), 8.34–8.39 (m, 4H, AnH).
Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes.
Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2,
conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used
only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2
are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger.
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
x y z Uiso*/Ueq
Cl1 0.31311 (4) 0.58536 (6) 0.11368 (6) 0.0739 (2)
Cl2 0.43369 (5) 0.28429 (6) 0.63139 (8) 0.0828 (2)
N1 0.28120 (10) 0.51499 (14) 0.58006 (17) 0.0439 (4)
C1 0.30892 (18) 0.5031 (2) 0.2998 (2) 0.0631 (6)
H1A 0.2608 0.4354 0.2882 0.076*
H1B 0.3742 0.4670 0.3268 0.076*
C2 0.28016 (14) 0.58725 (18) 0.4317 (2) 0.0480 (4)
H2A 0.3271 0.6562 0.4430 0.058*
H2B 0.2137 0.6212 0.4078 0.058*
C3 0.37449 (14) 0.5251 (2) 0.6764 (2) 0.0539 (5)
H3A 0.3685 0.5925 0.7531 0.065*
H3B 0.4280 0.5467 0.6069 0.065*
C4 0.40266 (15) 0.4070 (2) 0.7658 (3) 0.0649 (6)
H4A 0.3471 0.3806 0.8283 0.078*
H4B 0.4596 0.4234 0.8395 0.078*
C5 0.19394 (12) 0.53235 (19) 0.6777 (2) 0.0470 (4)
H5A 0.1969 0.6153 0.7247 0.056*
H5B 0.1974 0.4722 0.7648 0.056*
C6 0.09445 (12) 0.51724 (16) 0.5842 (2) 0.0413 (4)
C7 0.06525 (12) 0.39764 (16) 0.5287 (2) 0.0417 (4)
C8 0.12792 (14) 0.29072 (18) 0.5557 (2) 0.0513 (5)
H8 0.1903 0.3007 0.6089 0.062*
C9 0.09895 (16) 0.17556 (19) 0.5063 (2) 0.0587 (5)
H9 0.1411 0.1075 0.5266 0.070*
C10 0.00580 (17) 0.15792 (19) 0.4244 (3) 0.0593 (5)
H10 −0.0134 0.0783 0.3903 0.071*
C11 −0.05615 (15) 0.25555 (18) 0.3946 (2) 0.0527 (5)
H11 −0.1177 0.2416 0.3403 0.063*
C12 −0.03016 (12) 0.38025 (16) 0.4440 (2) 0.0415 (4)
Atomic displacement parameters (Å2)
U11 U22 U33 U12 U13 U23
supporting information
sup-3 Acta Cryst. (2006). E62, o2510–o2511
Cl2 0.0764 (4) 0.0783 (4) 0.0935 (5) 0.0169 (3) 0.0014 (3) 0.0056 (3)
N1 0.0356 (8) 0.0567 (9) 0.0393 (8) −0.0015 (7) 0.0006 (6) 0.0055 (7)
C1 0.0807 (15) 0.0655 (13) 0.0428 (11) 0.0061 (11) 0.0008 (10) 0.0045 (9)
C2 0.0444 (10) 0.0538 (11) 0.0459 (10) 0.0017 (8) 0.0045 (8) 0.0063 (9)
C3 0.0380 (10) 0.0707 (13) 0.0526 (11) −0.0048 (9) −0.0024 (8) −0.0009 (10)
C4 0.0466 (11) 0.0925 (17) 0.0547 (12) 0.0057 (11) −0.0056 (9) 0.0074 (11)
C5 0.0412 (10) 0.0599 (11) 0.0399 (10) −0.0022 (8) 0.0012 (7) −0.0007 (9)
C6 0.0366 (9) 0.0512 (10) 0.0362 (9) −0.0028 (8) 0.0039 (7) 0.0007 (8)
C7 0.0382 (9) 0.0486 (10) 0.0387 (9) 0.0001 (8) 0.0065 (7) 0.0024 (8)
C8 0.0462 (10) 0.0543 (11) 0.0535 (11) 0.0043 (9) 0.0034 (8) 0.0044 (9)
C9 0.0625 (13) 0.0511 (12) 0.0631 (13) 0.0113 (10) 0.0090 (10) 0.0051 (10)
C10 0.0723 (14) 0.0438 (11) 0.0623 (13) −0.0035 (10) 0.0084 (11) −0.0035 (10)
C11 0.0512 (11) 0.0528 (11) 0.0541 (12) −0.0083 (9) 0.0026 (9) −0.0029 (9)
C12 0.0396 (9) 0.0469 (10) 0.0385 (9) −0.0036 (8) 0.0074 (7) −0.0006 (8)
Geometric parameters (Å, º)
Cl1—C1 1.787 (2) C5—H5A 0.970
Cl2—C4 1.788 (2) C5—H5B 0.970
N1—C3 1.452 (2) C6—C12i 1.405 (2)
N1—C2 1.459 (2) C6—C7 1.409 (2)
N1—C5 1.466 (2) C7—C8 1.428 (3)
C1—C2 1.487 (3) C7—C12 1.437 (2)
C1—H1A 0.970 C8—C9 1.349 (3)
C1—H1B 0.970 C8—H8 0.930
C2—H2A 0.970 C9—C10 1.401 (3)
C2—H2B 0.970 C9—H9 0.930
C3—C4 1.505 (3) C10—C11 1.347 (3)
C3—H3A 0.970 C10—H10 0.930
C3—H3B 0.970 C11—C12 1.433 (3)
C4—H4A 0.970 C11—H11 0.930
C4—H4B 0.970 C12—C6i 1.405 (2)
C5—C6 1.515 (2)
C3—N1—C2 113.68 (14) N1—C5—C6 113.66 (14)
C3—N1—C5 111.51 (14) N1—C5—H5A 108.8
C2—N1—C5 115.48 (14) C6—C5—H5A 108.8
C2—C1—Cl1 111.44 (15) N1—C5—H5B 108.8
C2—C1—H1A 109.3 C6—C5—H5B 108.8
Cl1—C1—H1A 109.3 H5A—C5—H5B 107.7
C2—C1—H1B 109.3 C12i—C6—C7 119.79 (15)
Cl1—C1—H1B 109.3 C12i—C6—C5 121.04 (16)
H1A—C1—H1B 108.0 C7—C6—C5 119.15 (15)
N1—C2—C1 108.39 (16) C6—C7—C8 121.65 (16)
N1—C2—H2A 110.0 C6—C7—C12 120.20 (15)
C1—C2—H2A 110.0 C8—C7—C12 118.15 (16)
N1—C2—H2B 110.0 C9—C8—C7 121.73 (18)
H2A—C2—H2B 108.4 C7—C8—H8 119.1
N1—C3—C4 113.46 (16) C8—C9—C10 120.38 (19)
N1—C3—H3A 108.9 C8—C9—H9 119.8
C4—C3—H3A 108.9 C10—C9—H9 119.8
N1—C3—H3B 108.9 C11—C10—C9 120.46 (19)
C4—C3—H3B 108.9 C11—C10—H10 119.8
H3A—C3—H3B 107.7 C9—C10—H10 119.8
C3—C4—Cl2 111.43 (15) C10—C11—C12 122.05 (18)
C3—C4—H4A 109.3 C10—C11—H11 119.0
Cl2—C4—H4A 109.3 C12—C11—H11 119.0
C3—C4—H4B 109.3 C6i—C12—C11 122.74 (16)
Cl2—C4—H4B 109.3 C6i—C12—C7 120.02 (15)
H4A—C4—H4B 108.0 C11—C12—C7 117.23 (16)
C3—N1—C2—C1 95.39 (19) C5—C6—C7—C12 178.09 (15)
C5—N1—C2—C1 −133.89 (17) C6—C7—C8—C9 178.25 (18)
Cl1—C1—C2—N1 −178.33 (13) C12—C7—C8—C9 −1.0 (3)
C2—N1—C3—C4 −146.04 (17) C7—C8—C9—C10 0.7 (3)
C5—N1—C3—C4 81.3 (2) C8—C9—C10—C11 −0.3 (3)
N1—C3—C4—Cl2 67.44 (19) C9—C10—C11—C12 0.2 (3)
C3—N1—C5—C6 −177.53 (15) C10—C11—C12—C6i −179.14 (18)
C2—N1—C5—C6 50.7 (2) C10—C11—C12—C7 −0.5 (3)
N1—C5—C6—C12i −112.04 (18) C6—C7—C12—C6i 0.3 (3)
N1—C5—C6—C7 69.6 (2) C8—C7—C12—C6i 179.56 (15)
C12i—C6—C7—C8 −179.54 (16) C6—C7—C12—C11 −178.41 (16)
C5—C6—C7—C8 −1.2 (2) C8—C7—C12—C11 0.9 (2)
C12i—C6—C7—C12 −0.3 (3)