The
groEL
Gene Is a Promising Target for
Species-Level Identification of
Tsukamurella
Jade L. L. Teng,aYing Tang,aTsz Ho Chiu,aCandy L. W. Cheung,a Antonio H. Y. Ngan,aCheung Ngai,aSamson S. Y. Wong,aTak-Lun Que,b Susanna K. P. Lau,aPatrick C. Y. Wooa
Department of Microbiology, The University of Hong Kong, Hong Kong, Chinaa; Department of Pathology, Tuen Mun Hospital, Hong Kong, Chinab
KEYWORDS groEL, gene, species, identification,Tsukamurella
D
ifferentiation of Tsukamurella from the other related genera, such as Nocardia,Rhodococcus, andGordonia, and species-level identification within these genera
have been difficult in most clinical microbiology laboratories, as they share similar phenotypic properties. Among various molecular methods, 16S rRNA gene sequenc-ing has been the most widely used for bacterial identification, especially in cases where bacterial isolates are difficult to identify by phenotypic tests. However, previous studies showed that most Tsukamurella species share highly similar 16S rRNA gene sequences (⬎99% nucleotide identities), and thus this gene target cannot be confidently used for the identification of these species (1). Examples of alternative gene targets, such asssrA(stable small RNA),secA(the secretion ATPase),
rpoB (beta-subunit of RNA polymerase), and groEL (heat shock protein 60), have been used successfully for species-level identification in other bacterial genera (2–5). However, the usefulness of these gene targets for species-level identification
ofTsukamurellais not known.
In this study, we sequenced five gene targets (16S rRNA,ssrA,secA,rpoB, and
groEL) from 16 type and reference strains of all currently recognized Tsukamurella
species and evaluated their usefulness for species-level identification (Table 1). The complete set of primer sequences and the PCR conditions are shown in Table S1 in the supplemental material. Among the five gene targets,ssrA,secA, andrpoBfailed to differentiate between strains of different species or to show the same clustering for strains of the same species (results were shown in Table S1 and Fig. S1 in the supplemental material). Since only 16S rRNA andgroELgene sequences were able to show correct species assignments, the usefulness of these two gene targets was further evaluated by determining their sequences in 34 additional clinical (n⫽18) and veterinary (n⫽ 16) isolates, and their species identities (15 were T.
tyrosino-solvensand 19 wereT. pulmonis) were confirmed by DNA-DNA hybridization (DDH)
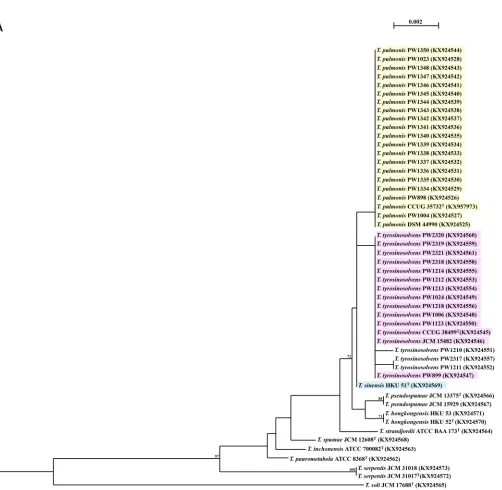
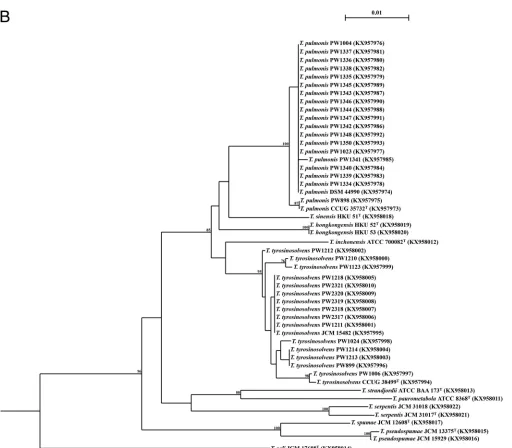
(6, 7). The interspecies similarities of 16S rRNA sequences of all 50 Tsukamurella
isolates (16 type and reference strains and 34 clinical isolates) ranged from 96.7 to 99.9%, whereas those ofgroELgene sequences ranged from 91.1% to 98.2% (Fig. 1). The intraspecies identities of 16S rRNA ranged from 99.9% to 100%, whereas those of the groELgene ranged from 98.7% to 100.0% (Fig. 1). Although both the 16S rRNA and groELgene sequences were able to correctly identify the 16 type and reference strains, when the 34 additional clinical isolates were included to evaluate their usefulness, the 16S rRNA gene failed to differentiate T. sinensis from some strains of T. pulmonis and T. tyrosinosolvens, showing up to 99.9% sequence similarities between two differentTsukamurellaspecies. On the other hand, using a
Accepted manuscript posted online14 December 2016
CitationTeng JLL, Tang Y, Chiu TH, Cheung CLW, Ngan AHY, Ngai C, Wong SSY, Que T-L, Lau SKP, Woo PCY. 2017. ThegroELgene is a promising target for species-level identification ofTsukamurella. J Clin Microbiol 55:649 – 653.
https://doi.org/10.1128/JCM.02260-16.
EditorPaul Bourbeau
Copyright© 2017 American Society for Microbiology.All Rights Reserved. Address correspondence to Susanna K. P. Lau, skplau@hku.hk, or Patrick C. Y. Woo, pcywoo@hku.hk.
J.L.L.T. and Y.T. contributed equally to this article.
crossm
on May 16, 2020 by guest
http://jcm.asm.org/
threshold value of 98.2% based on thegroELgene sequence, species identities of all 50Tsukamurella strains were completely concordant with those determined by the gold standard, DDH, suggesting that this threshold value may be useful for species-level identification inTsukamurella.
TABLE 1Bacterial strains and accession numbers
Species and strain
GenBank accession no.
16S rRNA ssrA secA rpoB groEL
T. pulmonis
CCUG 35732T KX924524 KX931996 KX931980 KX932012 KX957973
DSM 44990 KX924525 KX931997 KX931981 KX932013 KX957974
PW898 KX924526 KX957975
PW1004 KX924527 KX957976
PW1023 KX924528 KX957977
PW1334 KX924529 KX957978
PW1335 KX924530 KX957979
PW1336 KX924531 KX957980
PW1337 KX924532 KX957981
PW1338 KX924533 KX957982
PW1339 KX924534 KX957983
PW1340 KX924535 KX957984
PW1341 KX924536 KX957985
PW1342 KX924537 KX957986
PW1343 KX924538 KX957987
PW1344 KX924539 KX957988
PW1345 KX924540 KX957989
PW1346 KX924541 KX957990
PW1347 KX924542 KX957991
PW1348 KX924543 KX957992
PW1350 KX924544 KX957993
T. tyrosinosolvens
CCUG 38499T KX924545 KX931998 KX931982 KX932014 KX957994
JCM 15482 KX924546 KX931999 KX931983 KX932015 KX957995
PW899 KX924547 KX957996
PW1006 KX924548 KX957997
PW1024 KX924549 KX957998
PW1123 KX924550 KX957999
PW1210 KX924551 KX958000
PW1211 KX924552 KX958001
PW1212 KX924553 KX958002
PW1213 KX924554 KX958003
PW1214 KX924555 KX958004
PW1218 KX924556 KX958005
PW2317 KX924557 KX958006
PW2318 KX924558 KX958007
PW2319 KX924559 KX958008
PW2320 KX924560 KX958009
PW2321 KX924561 KX958010
T. paurometabolaATCC 8368T KX924562 KX932000 KX931984 KX932016 KX958011
T. inchonensisATCC 700082T KX924563 KX932001 KX931985 KX932017 KX958012
T. strandjordaeATCC BAA-173T KX924564 KX932002 KX931986 KX932018 KX958013
T. soliJCM 17688T KX924565 KX932003 KX931987 KX932019 KX958014
T. pseudospumae
JCM 13375T KX924566 KX932004 KX931988 KX932020 KX958015
JCM 15929 KX924567 KX932005 KX931989 KX932021 KX958016 T. spumaeJCM 12608T KX924568 KX932006 KX931990 KX932022 KX958017
T. sinensisHKU 51T KX924569 KX932007 KX931991 KX932023 KX958018
T. hongkongensis
HKU 52T KX924570 KX932008 KX931992 KX932024 KX958019
HKU 53 KX924571 KX932009 KX931993 KX932025 KX958020 T. serpentis
JCM 31017T KX924572 KX932010 KX931994 KX932026 KX958021
JCM 31018 KX924573 KX932011 KX931995 KX932027 KX958022
on May 16, 2020 by guest
http://jcm.asm.org/
In conclusion, we showed that the performance of groEL gene sequencing for species-level identification of Tsukamurella was better than that of 16S rRNA gene sequencing. Further study using additional clinical isolates belonging to species other
thanT. pulmonis andT. tyrosinosolvens will be necessary to thoroughly evaluate the
usefulness of thegroELgene and its threshold value (98.2%) for species-level identifi-cation ofTsukamurella.
Accession number(s).The sequence data of the 16S rRNA (1,221 nucleotide posi-tions) and groEL(677 nucleotide positions) genes obtained in this study have been submitted to GenBank with accession no. KX924524to KX924573andKX957973to KX958022, respectively. The sequence data ofssrA(366 nucleotide positions),secA(607
FIG 1Phylogenetic trees showing the relationship of the 50Tsukamurellaisolates (16 type and reference strains and 34 clinical isolates) inferred from partial 16S rRNA (1,221 nucleotide positions) (A) andgroEL(677 nucleotide positions) (B) sequence data by the maximum likelihood method using the model GTR⫹I⫹G andMycobacterium smegmatisMC2155 (CP001663.1) as the outgroup. The scale bar indicates the estimated number of substitutions
per base. Numbers at nodes indicate levels of bootstrap support calculated from 1,000 trees and expressed as percentages. Species shaded in the same color represent high sequence similarities (ⱖ99.9%) between two differentTsukamurellaspecies. All sequences obtained from this study and accession numbers are given as cited in the GenBank database.
on May 16, 2020 by guest
http://jcm.asm.org/
[image:3.585.49.551.54.544.2]nucleotide positions), andrpoB (284 nucleotide positions) obtained in this study (see Fig. S1A to C in the supplemental material) have been submitted to GenBank with accession no. KX931996 to KX932011, KX931980 to KX931995, and KX932012 to KX932027, respectively.
SUPPLEMENTAL MATERIAL
Supplemental material for this article may be found at https://doi.org/10.1128/ JCM.02260-16.
TEXT S1,PDF file, 0.2 MB.
ACKNOWLEDGMENTS
This work was partly supported by the Strategic Research Theme Fund, the Small Project Fund, The University of Hong Kong, and by a Croucher Senior Medical Research Fellowship, Croucher Foundation, Hong Kong.
FIG 1(Continued)
on May 16, 2020 by guest
http://jcm.asm.org/
[image:4.585.41.553.63.511.2]REFERENCES
1. Woo PC, Ng KH, Lau SK, Yip KT, Fung AM, Leung KW, Tam DM, Que TL, Yuen KY. 2003. Usefulness of the MicroSeq 500 16S ribosomal DNA-based bacterial identification system for identification of clinically signif-icant bacterial isolates with ambiguous biochemical profiles. J Clin Microbiol 41:1996 –2001. https://doi.org/10.1128/JCM.41.5.1996-2001 .2003.
2. Conville PS, Zelazny AM, Witebsky FG. 2006. Analysis of secA1 gene sequences for identification of Nocardia species. J Clin Microbiol 44: 2760 –2766.https://doi.org/10.1128/JCM.00155-06.
3. Osawa K, Shigemura K, Shirai H, Kato A, Okuya Y, Jikimoto T, Arakawa S, Fujisawa M, Shirakawa T. 2015. Bacterial identification usingssrA encod-ing transfer-messenger RNA. Southeast Asian J Trop Med Public Health 46:720 –727.
4. Adékambi T, Shinnick TM, Raoult D, Drancourt M. 2008. CompleterpoB
gene sequencing as a suitable supplement to DNA-DNA hybridization for bacterial species and genus delineation. Int J Syst Evol Microbiol 58: 1807–1814.https://doi.org/10.1099/ijs.0.65440-0.
5. Hossain MT, Kim EY, Kim YR, Kim DG, Kong IS. 2012. Application ofgroEL
gene for the species-specific detection ofVibrio parahaemolyticusby PCR. Lett Appl Microbiol 54:67–72.
6. Teng JL, Tang Y, Wong SS, Ngan AH, Huang Y, Tsang CC, Choi GK, Lau SK, Woo PC. 2016.Tsukamurella hongkongensissp. nov. andTsukamurella sinensissp. nov., isolated from patients with keratitis, catheter-related bacteraemia and conjunctivitis. Int J Syst Evol Microbiol 66:391–397.
https://doi.org/10.1099/ijsem.0.000733.
7. Seong CN, Kim YS, Baik KS, Choi SK, Kim MB, Kim SB, Goodfellow M. 2003.
Tsukamurella sunchonensissp. nov., a bacterium associated with foam inactivated sludge. J Microbiol 41:83– 88.