0095-1137/07/$08.00⫹0 doi:10.1128/JCM.01831-06
Copyright © 2007, American Society for Microbiology. All Rights Reserved.
Catabacter hongkongensis
gen. nov., sp. nov., Isolated from Blood
Cultures of Patients from Hong Kong and Canada
䌤
Susanna K. P. Lau,
1,2,3Alan McNabb,
4Gibson K. S. Woo,
1Linda Hoang,
4,5Ami M. Y. Fung,
1Liliane M. W. Chung,
1Patrick C. Y. Woo,
1,2,3* and Kwok-Yung Yuen
1,2,3*
Department of Microbiology1and Research Center of Infection and Immunology,2The University of Hong Kong, and State Key
Laboratory of Emerging Infectious Diseases, The University of Hong Kong,3Hong Kong, Hong Kong, and Laboratory Services,
British Columbia Centre for Disease Control,4and Department of Pathology and Laboratory Medicine,
University of British Columbia,5Vancouver, British Columbia, Canada
Received 4 September 2006/Returned for modification 13 October 2006/Accepted 7 November 2006
Four bacterial isolates were recovered from the blood cultures of four patients, two of whom were from Hong Kong and two of whom were from Canada. The two Hong Kong strains were isolated from a 48-year-old man with intestinal obstruction and secondary sepsis (strain HKU16T
) and from a 39-year-old man with acute appendicitis (strain HKU17), while the two Canadian strains were isolated from a 74-year-old man with biliary sepsis (strain CA1) and from a 66-year-old woman with metastatic carcinoma and sepsis (strain CA2). While the first three patients survived, the last patient died 2 weeks after the episode of bacteremia. All four isolates are strictly anaerobic, nonsporulating, gram-positive coccobacilli that were unidentified by conventional phenotypic tests and commercial identification systems. They grow on sheep blood agar as nonhemolytic pinpoint colonies after 48 h of incubation at 37°C in an anaerobic environment. All are catalase positive and motile, with flagella. They produce acid from arabinose, glucose, mannose, and xylose. They do not produce indole or reduce nitrate. They are sensitive to penicillin, vancomycin, and metronidazole but resistant to cefotaxime. 16S rRNA gene sequence analysis showed 16.0%, 16.8%, and 21.0% base differences from
Clostrid-ium propionicum,Clostridium neopropionicum, andAtopobium minutum, respectively. The GⴙC content of strain
HKU16T
is 40.2% ⴞ 2.2%. Based on their phylogenetic affiliation, unique GⴙC content, and phenotypic characteristics, we propose a new genus and species,Catabacter hongkongensisgen. nov., sp. nov., to describe the bacterium, for which HKU16 is the type strain, and suggest that it be assigned to a new family,
Catabac-teriaceae. The gastrointestinal tract was probably the source of the bacterium for at least three of the four
patients. The isolation of a catalase-positive, motile, nonsporulating, anaerobic gram-positive bacillus in clinical laboratories should raise the possibility ofC. hongkongensis.Further studies should be performed to ascertain the epidemiology and other disease associations of this bacterium.
Medically important anaerobic gram-positive bacilli are a heterogeneous group of bacteria that include members of the
generaClostridium,Actinomyces, Bifidobacterium,Eggerthella,
Eubacterium,Lactobacillus, andPropionibacterium. The genus
classification of these bacteria is traditionally based on pheno-typic characteristics, such as spore formation, biochemical pro-files, and analyses of cell wall fatty acids and metabolic end products. However, analyses of cell wall fatty acids and meta-bolic end products by gas-liquid chromatography are often limited by the lack of special equipment and expertise in most clinical microbiology laboratories and the limited database. Based on 16S rRNA gene analysis as a new standard for clas-sification of bacteria (17, 18), many of these bacteria have undergone taxonomic revisions, with new genera and species
being introduced (8, 9, 12, 15, 23–26). For example,
Eubacte-rium lentumhas been reclassified under a new genus as
Egg-erthella lentain the familyCoriobacteriaceaeby its phylogenetic
position and high G⫹C content (9). Two novel species under
the new genus, i.e.,Eggerthella hongkongensisandEggerthella
sinensis, were also subsequently identified using 16S rRNA
gene analysis (12).
In this study, we report the isolation and characterization of four bacterial isolates from the blood cultures of four patients.
Two of the isolates (strains HKU16T and HKU17) were
re-covered from two patients in Hong Kong, one with intestinal obstruction and secondary sepsis and the other with acute appendicitis. The other two isolates (strains CA1 and CA2) were recovered from two patients in Canada, one of whom developed biliary sepsis after stent removal and the other of whom had metastatic carcinoma of the lung and sepsis syn-drome. The four isolates exhibited similar phenotypic charac-teristics that do not fit into the pattern for any known genus and species. Based on 16S rRNA gene analysis and the unique phenotypic characteristics, we propose a novel genus and
spe-cies,Catabacter hongkongensis gen. nov., sp. nov., to describe
this bacterium.
MATERIALS AND METHODS
Patients and microbiological methods.The BACTEC 9240 blood culture system (Becton Dickinson, MD) was used. Bacterial strains were identified by standard conventional methods (7, 14). Flagellum staining was performed by the method described by Kodaka et al. (10). Testing for spores was performed by malachite green staining and phase-contrast microscopy. In addition, a Vitek system (ANI; bioMerieux Vitek), an ATB expression system (ID32A;
* Corresponding author. Mailing address: Department of Microbiol-ogy, The University of Hong Kong, University Pathology Building, Queen Mary Hospital, Hong Kong, Hong Kong. Phone: (852) 28554892. Fax: (852) 28551241. E-mail for Patrick C. Y. Woo: pcywoo@hkucc.hku.hk. E-mail for Kwok-Yung Yuen: hkumicro@hkucc.hku.hk.
䌤Published ahead of print on 22 November 2006.
395
on May 16, 2020 by guest
http://jcm.asm.org/
bioMerieux Vitek), and an API system (20A; bioMerieux Vitek) were used for the identification of the bacterial isolates in this study. Antibiotic susceptibility tests were performed by Etest, and results were interpreted according to the CLSI (formerly NCCLS) criteria for anaerobic bacteria (16). All tests were performed in triplicate with freshly prepared media on separate occasions.
Scanning electron microscopy.Scanning electron microscopy was performed as described previously (27, 28). Polycarbonate membranes (Nuclepore) with a pore size of 5m were used.
Bacterial DNA extraction, PCR, and sequencing of 16S rRNA genes.Bacterial DNA extraction, PCR amplification, and DNA sequencing of the 16S rRNA genes were performed according to previous published protocols (11, 25, 26). LPW57 (5⬘-AGTTTGATCCTGGCTCAG-3⬘) and LPW205 (5⬘-CTTGTTACG ACTTCACCC-3⬘) (Gibco BRL, Rockville, MD) were used as the PCR primers. Both strands of the PCR products were sequenced twice, using the PCR primers (LPW57 and LPW205) and additional sequencing primers, LPW284 (5⬘-GTTT ACAACCCGAAGGCC-3⬘) and LPW306 (5⬘ -TGAGATGTTGGGTTAAGT-3⬘), designed from the first round of sequencing results.
Phylogenetic characterization.The sequences of the PCR products were com-pared with known 16S rRNA gene sequences in GenBank by multiple sequence alignment using the CLUSTAL W program (22). Their phylogenetic relation-ships to other closely related gram-positive rods were determined using Clustal X, version 1.81 (6), and the neighbor-joining method in GrowTree (Genetics Computer Group, Inc., San Diego, CA). A total of 1,311 nucleotide positions were included in the analysis.
GⴙC content determination. The G⫹C content of the genomic DNA of HKU16T
was determined by thermal denaturation, as described previously (27, 28). The melting temperature (Tm) of the DNA was defined as the temperature at 50% hyperchronicity. The G⫹C content of the genomic DNA was calculated by the following formula: (G⫹C)%⫽2.44Tm⫺169.
Nucleotide sequence accession number.The 16S rRNA gene sequence of HKU16Thas been lodged within the GenBank sequence database under acces-sion no. AY574991.
RESULTS
Patients. (i) Case 1.A 48-year-old Chinese man was admit-ted to the hospital in Hong Kong in 1999 because of fever, repeated vomiting, abdominal distension, and constipation for 2 days. He had end-stage renal disease of unknown etiology. He had been on hemodialysis for 11 years after discontinuation of continuous ambulatory peritoneal dialysis as a result of tuberculous peritonitis. Since the episode of tuberculous peri-tonitis, he had had recurrent episodes of intestinal obstruction, which were treated conservatively. On admission, he was de-hydrated and had generalized abdominal distension and hy-peractive bowel sounds. An abdominal radiograph revealed a dilated small bowel with multiple air-fluid levels in the central abdomen. The large bowel was air filled with fecal matter. A diagnosis of incomplete small-bowel obstruction and second-ary sepsis was made. Blood culture was performed, and intra-venous cefuroxime and metronidazole treatment was com-menced. On day 3 postincubation, the anaerobic blood culture bottle turned positive for a gram-positive bacillus (strain
HKU16T). His fever responded to intravenous antibiotics, and
the intestinal obstruction resolved with conservative treatment. He was discharged after 19 days of hospitalization.
(ii) Case 2.A 39-year-old Chinese man was admitted to the hospital in Hong Kong in 2003 because of central colicky abdominal pain radiating to the right lower quadrant for 1 day. He also complained of vomiting, fever, chills, and rigor. His past medical history was unremarkable. Examination of his abdomen revealed tenderness, guarding, and rebound tender-ness over the right lower quadrant. Blood cultures were per-formed. A clinical diagnosis of acute appendicitis was made, and empirical intravenous cefuroxime and metronidazole treatment was commenced. Emergency laparoscopic
appen-dectomy was performed. At operation, an acutely inflamed appendix with perforation near the base was found, with a small amount of surrounding purulent fluid. Histology of the appendix showed transmural inflammation with marked neu-trophil infiltration and local peritonitis. On day 3 postincuba-tion, the anaerobic blood culture bottle turned positive for a gram-positive bacillus (strain HKU17). He recovered unevent-fully after the operation and was discharged with oral antibi-otics 2 days after admission.
(iii) Case 3.A 74-year-old man was admitted to the hospital in British Columbia, Canada, for removal of a biliary stent in 2004. He had a 4-year history of plasmacytoma complicated by biliary obstruction, which was managed by a combination of radiation therapy and multiple biliary stenting, with the last replacement done in 2002. He was afebrile on admission but developed fever in the evening after complete removal of the biliary stent. Blood cultures were performed, which recovered an anaerobic, gram-positive bacillus (strain CA1). Empirical oral ciprofloxacin treatment was commenced. His fever sub-sided, and he was discharged home with no subsequent com-plications.
(iv) Case 4.A 66-year-old female was admitted to the hos-pital in British Columbia, Canada, because of fever and sepsis syndrome. She was under palliative care for metastatic carci-noma of the lung. Blood cultures were obtained, and the pa-tient was started empirically on cefuroxime and ciprofloxacin. On day 5 postincubation, the anaerobic blood culture bottle turned positive for a gram-positive bacillus with variable stain-ing (strain CA2). The patient died 2 weeks after admission.
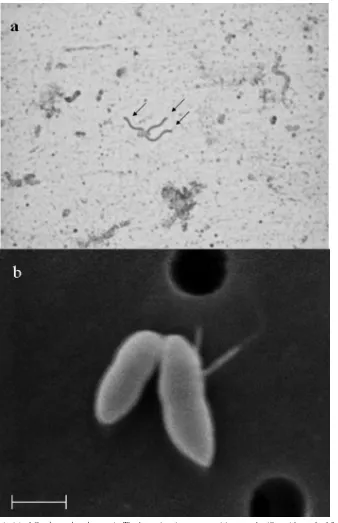
Phenotypic characteristics. HKU16T, HKU17, CA1, and CA2 exhibit similar phenotypic characteristics. They are all nonsporulating, gram-positive coccobacilli. They are motile, with flagella (Fig. 1a). They grow on sheep blood agar as nonhemolytic pinpoint colonies after 48 h of incubation at 37°C in an anaerobic environment. They do not grow in aero-bic or microaerophilic environments. They produce catalase
when tested with 15% H2O2 but do not produce indole or
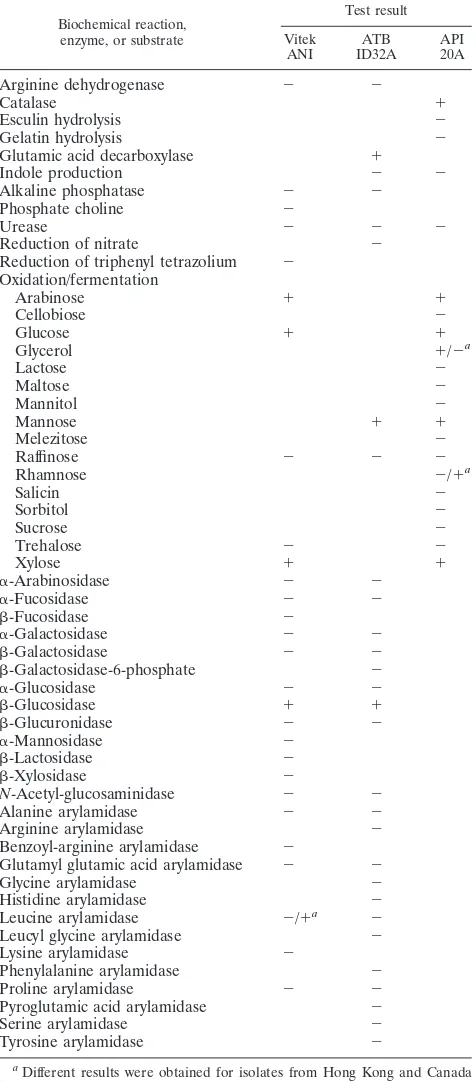
reduce nitrate. Their biochemical profiles are shown in Table 1. They all produce acid from arabinose, glucose, mannose, and xylose. The two isolates from Canada differ from the two isolates from Hong Kong by being negative for glycerol fer-mentation and positive for rhamnose ferfer-mentation and leucine arylamidase. All four isolates were “unidentified” by all three commercially available identification systems used. The code profile for the four isolates by the ATB expression system
(ID32A) was 0012000010. The code profile for HKU16Tand
HKU17 by the API system (20A) was 40415042, and that for CA1 and CA2 was 40414052. All four isolates were susceptible to bile and kanamycin but resistant to colistin. The MICs of cefotaxime, vancomycin, and metronidazole for all four strains
were⬎32g/ml, 2g/ml, and⬍0.016g/ml, respectively. The
MICs of penicillin were 0.75 g/ml for strain HKU16T, 0.5
g/ml for strain HKU17, and 4g/ml for strains CA1 and CA2.
Scanning electron microscopy. A scanning electron
micro-graph of strain HKU16Tis shown in Fig. 1b. The bacterial cells
are short straight rods tapered at both ends, with a tuft of flagella inserted on one side.
Molecular characterization by 16S rRNA gene sequencing and phylogenetic characterization.PCRs to amplify the 16S rRNA genes of all four isolates showed bands of about 1,400
396 LAU ET AL. J. CLIN. MICROBIOL.
on May 16, 2020 by guest
http://jcm.asm.org/
bp. The 16S rRNA gene sequences were identical. There was a 16.0% difference in the 16S rRNA gene sequences of the four
isolates from that ofClostridium propionicum (GenBank
ac-cession no. X77841), a 16.8% difference from that of
Clostrid-ium neopropionicum(GenBank accession no. 76746), a 21.0%
difference from that ofAtopobium minutum(GenBank
acces-sion no. X67148), a 21.9% difference from that ofEggerthella
lenta(GenBank accession no. AF292375), a 22.2% difference
from that ofBifidobacterium dentium(GenBank accession no.
D86183), a 21.7% difference from that ofPropionibacterium
acnes(GenBank accession no. AB097215), and a 21.8%
dif-ference from that ofActinomyces odontolyticus(GenBank
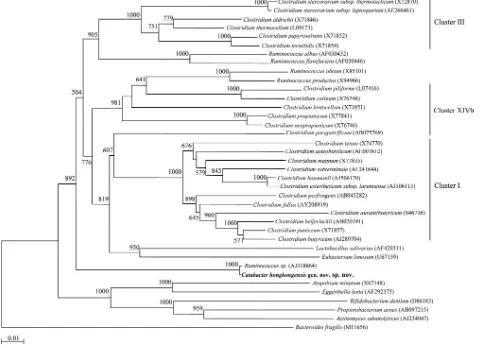
ac-cession no. AJ234047) (Fig. 2). Although the four sequences
[image:3.585.123.461.67.590.2]had⬎99% identity to “Ruminococcussp.” strain CCUG 37327
FIG. 1. Flagellum stain (a) ofCatabacter hongkongensis. The bacterium is a gram-positive coccobacillus with a tuft of flagella inserted on one side (arrows). (b) Scanning electron micrograph ofCatabacter hongkongensis. The bacterium is short and straight and tapered at both ends. Cells vary in length from 0.71 to 1.12m and in diameter from 0.36 to 0.48m (mean, 0.96m by 0.42m;n⫽20). Bar, 0.5m.
on May 16, 2020 by guest
http://jcm.asm.org/
(GenBank accession no. AJ318864), a ruminococcus-like or-ganism from a human clinical source in the United Kingdom,
the true identity of this “Ruminococcus sp.” has not been
validated or published. Moreover, this “Ruminococcus sp.”
possessed a phylogenetic position distant from the known
Ruminococcusspecies, suggesting that it is unlikely to belong
to the genusRuminococcusand may have been misidentified,
since our present four isolates formed small, short bacilli which may be mistaken as cocci (Fig. 2). Based on phylogenetic affiliation, the four isolates form a distinct lineage among the anaerobic gram-positive bacilli and are only peripherally asso-ciated with clusters I, III, and XIVb of the clostridia (2). We propose that they be assigned a novel genus and species,
Catabacter hongkongensis, under a new family,
Catabacteri-aceae. The name “Catabacter,” which did not exist previously,
was constructed as an arbitrary name as an abbreviation for “catalase-positive bacterium” in order to avoid an unusually long genus name.
GⴙC content determination. The G⫹C content of strain
HKU16T(mean⫾standard deviation) was 40.2%⫾2.2%.
DISCUSSION
In this study, we report the isolation of a novel anaerobic gram-positive bacillus from the blood cultures of four patients, two from Hong Kong and two from Canada. The bacterium was likely to be clinically significant in all four patients, as evidenced by its isolation from blood in pure culture and the patients’ systemic responses to the bacteremia. The isolation of the bacterium from patients in both Asia and North America suggests that it is widely distributed. The historical failure to recognize this new genus and species is likely a reflection of the difficulties in accurately identifying anaerobic gram-positive bacilli. While spore formation may not be obvious in isolates primarily recovered from clinical specimens, analysis of cell wall fatty acids and metabolic end products by gas-liquid chro-matography requires special equipment and expertise, which are generally not available in clinical microbiology laborato-ries. Therefore, many of the anaerobic gram-positive bacilli in clinical laboratories are not identified even to the genus level. Application of 16S rRNA gene analysis of suspected isolates is
likely to uncover more strains ofC. hongkongensisand to help
to better define its epidemiology, clinical disease associations,
and pathogenicity. The presence ofC. hongkongensisin Asia,
America, and probably Europe implies that it is a bacterium of
global importance. The possibility ofC. hongkongensisshould
be considered when a catalase-positive, motile,
nonsporulat-ing, anaerobic gram-positive bacillus is encountered. SinceC.
hongkongensisdemonstrates variable susceptibility to penicillin
(MICs range from 0.5 to 4 g/ml), metronidazole should be
considered the drug of choice forC. hongkongensisinfections.
While there were no localizing symptoms or signs in the last patient, we speculate that the source of the bacteremia in the first three patients was the gastrointestinal tract. It has been documented for both animals and humans that intestinal ob-struction (present in case 1) promotes gastrointestinal tract translocation of bacteria (5, 13, 20). Acute appendicitis (present in case 2) is also recognized as being associated with anaerobic bacteremia as a result of bacterial translocation through inflamed intestinal mucosa (1). Moreover, it is well known that biliary sepsis (present in case 3) is usually due to ascending infection by bacteria from the gut through the am-pulla of Vater. In fact, many other nonsporulating anaerobic
[image:4.585.43.281.99.640.2]gram-positive bacilli, including Bifidobacterium, Eggerthella,
TABLE 1. Biochemical profiles of the four blood culture isolates by the Vitek system (ANI), ATB expression system (ID32A),
and API system (20A)
Biochemical reaction, enzyme, or substrate
Test result
Vitek ANI
ATB ID32A
API 20A
Arginine dehydrogenase ⫺ ⫺
Catalase ⫹
Esculin hydrolysis ⫺
Gelatin hydrolysis ⫺
Glutamic acid decarboxylase ⫹
Indole production ⫺ ⫺
Alkaline phosphatase ⫺ ⫺
Phosphate choline ⫺
Urease ⫺ ⫺ ⫺
Reduction of nitrate ⫺
Reduction of triphenyl tetrazolium ⫺ Oxidation/fermentation
Arabinose ⫹ ⫹
Cellobiose ⫺
Glucose ⫹ ⫹
Glycerol ⫹/⫺a
Lactose ⫺
Maltose ⫺
Mannitol ⫺
Mannose ⫹ ⫹
Melezitose ⫺
Raffinose ⫺ ⫺ ⫺
Rhamnose ⫺/⫹a
Salicin ⫺
Sorbitol ⫺
Sucrose ⫺
Trehalose ⫺ ⫺
Xylose ⫹ ⫹
␣-Arabinosidase ⫺ ⫺
␣-Fucosidase ⫺ ⫺
-Fucosidase ⫺
␣-Galactosidase ⫺ ⫺
-Galactosidase ⫺ ⫺
-Galactosidase-6-phosphate ⫺
␣-Glucosidase ⫺ ⫺
-Glucosidase ⫹ ⫹
-Glucuronidase ⫺ ⫺
␣-Mannosidase ⫺
-Lactosidase ⫺
-Xylosidase ⫺
N-Acetyl-glucosaminidase ⫺ ⫺
Alanine arylamidase ⫺ ⫺
Arginine arylamidase ⫺
Benzoyl-arginine arylamidase ⫺
Glutamyl glutamic acid arylamidase ⫺ ⫺
Glycine arylamidase ⫺
Histidine arylamidase ⫺
Leucine arylamidase ⫺/⫹a ⫺
Leucyl glycine arylamidase ⫺
Lysine arylamidase ⫺
Phenylalanine arylamidase ⫺
Proline arylamidase ⫺ ⫺
Pyroglutamic acid arylamidase ⫺
Serine arylamidase ⫺
Tyrosine arylamidase ⫺
aDifferent results were obtained for isolates from Hong Kong and Canada
(result for Hong Kong isolates/result for Canadian isolates).
398 LAU ET AL. J. CLIN. MICROBIOL.
on May 16, 2020 by guest
http://jcm.asm.org/
Eubacterium, andLactobacillus, are common flora of the hu-man gastrointestinal tract. Further studies should be carried
out to determine ifCatabacter hongkongensisis also one of our
normal gut commensals.
C. hongkongensisexhibited phenotypic and genotypic
char-acteristics that are very different from those of other closely related medically important bacterial genera (Table 2).
Mem-bers of the genus Clostridium produce spores. However, C.
hongkongensis, like members of theEubacterium, Eggerthella,
Bifidobacterium,Propionibacterium, andActinomycesgenera, is
nonsporulating. MostClostridiumspecies andC. hongkongensis
are motile, whereas members of Eubacterium have variable
motility and those ofEggerthella,Bifidobacterium,
Propionibac-terium, and Actinomyces are nonmotile. C. hongkongensis,
Eubacterium, Eggerthella, and Bifidobacterium are obligately
anaerobic, whereas some members of theClostridium,
Propi-onibacterium, and Actinomyces genera are aerotolerant. C.
hongkongensis, some members of the Eggerthella and
Propi-onibacteriumgenera, andActinomyces viscosus produce
cata-lase, but members of the Clostridium, Eubacterium, and
Bifidobacteriumgenera do not.C. hongkongensisand members
ofEubacterium,Eggerthella,Bifidobacterium, andActinomyces
do not produce indole, but some members ofClostridiumand
Propionibacterium produce indole. C. hongkongensis and
Bifidobacteriumdo not reduce nitrate, but some members of
the Clostridium, Eubacterium, Eggerthella, Propionibacterium,
andActinomyces genera reduce nitrate. Genotypically,
mem-bers of the genusEubacteriumhave low G⫹C contents of 30 to
40 mol%, HKU16Thas a G⫹C content of about 40%,
mem-bers of theEggerthella,Bifidobacterium,Propionibacterium, and
Actinomycesgenera have high G⫹C contents of over 55%, and
those of the genusClostridiumhave highly variable G⫹C
con-tents of 26 to 56%. Furthermore, the 16S rRNA genes ofC.
hongkongensisexhibited⬎16% nucleotide differences from the
16S rRNA genes of all previously described bacteria.
Phyloge-netic analysis showed that C. hongkongensis isolates form a
distinct lineage among the anaerobic gram-positive rods and are only peripherally associated with clusters I, III, and XIVb
of the clostridia (2) (Fig. 2). Although it is closest toC.
propi-onicum, a species that has never been reported to be associated
with human infection, it is well discerned from the clade.
LactobacillusandEubacterium, which are also associated
phy-logenetically with clusters I and XIVb of the clostridia, are
[image:5.585.50.533.72.420.2]classified under different families, i.e., Lactobacillaceae and
FIG. 2. Phylogenetic tree showing the relationships ofCatabacter hongkongensisgen. nov., sp. nov., to related anaerobic gram-positive bacteria. The tree was constructed by using the neighbor-joining method, withBacteroides fragilisas the root. Bootstrap values were calculated from 1,000 trees. Bar, estimated number of substitutions per 100 bases, using the Jukes-Cantor correction. Names and accession numbers are given as cited in the GenBank database.
on May 16, 2020 by guest
http://jcm.asm.org/
Eubacteriaceae, respectively, because of their distinct pheno-typic and genopheno-typic characteristics. Therefore, the deep branch
ofC. hongkongensisin phylogenetic analysis, together with its
unique phenotypic characteristics, is representative of a novel genus, and we propose that it be assigned to a new family,
Catabacteriaceae.
The production of catalase inC. hongkongensismay explain
the relative aerotolerance of the bacterium. In aerobic and facultative bacteria, DNA damage by oxygen radicals gener-ated under oxygen exposure is prevented by the dismutation of
O2⫺to H2O2by superoxide dismutase and the elimination of
H2O2by catalase and peroxidase (3, 4). On the other hand, the
sensitivity to oxygen exposure of anaerobes has been attributed to the lack of these protective mechanisms against oxidative stress. Nevertheless, some anaerobes are more aerotolerant
than others. ForBacteroides fragilis, one of the most
aerotol-erant species, it has been shown that the production of catalase plays a role in the protection against oxidative stress (19).
Moreover, it was found that clinical isolates ofB. fragiliswere
more aerotolerant than fecal isolates, suggesting that
aerotol-erance may be an important virulence factor (21).C.
hongkon-gensisisolates were able to survive in the presence of oxygen
for 72 to 96 h on chocolate agar incubated in 5% CO2, in contrast to only 12 h for a strain of the strict anaerobe
Prevotella melaninogenica (data not shown). Further studies
will be needed to clarify the role of catalase production in the
oxidative stress response and virulence ofC. hongkongensis.
Description of Catabacter hongkongensisgen. nov., sp. nov.
Catabacter(Ca.ta.bac⬘ter. arbitrary name; N.L.cata-
[abbrevi-ation for catalase positive], derived from Gr.kata, down; N.L.
masc. n.bacter, rod; N.L. masc. n.Catabacter, catalase-positive
rod);hongkongensis(hong.kong.en⬘sis. N.L. fem. adj. in honor
of Hong Kong, the place where the type strain was isolated). Cells are obligately anaerobic, gram-positive coccobacilli or short, straight rods. The bacterium grows on sheep blood agar as nonhemolytic pinpoint colonies after 48 h of incubation at 37°C in an anaerobic environment. It does not grow in aerobic or microaerophilic environments. The organism does not pro-duce spores but is motile, with flagella. It propro-duces catalase but does not produce indole or reduce nitrate. It produces acid from arabinose, glucose, mannose, and xylose (Table 1). The
moles percent G⫹C content of the DNA of the type strain,
HKU16, is 40.2%⫾2.2%. The organism was isolated from the
blood cultures of three patients with acute gastrointestinal
compromises and one with acute sepsis. The type strain ofC.
hongkongensisis strain HKU16.
ACKNOWLEDGMENTS
This work was partly supported by the University Development Fund, a University Research Grant Council grant (HKU 7236/02 M), a Committee for Research and Conference grant, The University of Hong Kong, and the William Benter Infectious Disease Fund.
We thank Hans G. Tru¨per (Institut fu¨r Mikrobiologie & Biotech-nologie, Rheinische Friedrich-Wilhelms-Universita¨t Bonn, Germany) for advice on the nomenclature of the novel bacterial genus and spe-cies, Pam Kibsey (Fraser Health, Royal Columbian Hospital, New Westminster, British Columbia, Canada) for providing the case infor-mation on the Canadian isolates, and Kathy Adie and Nancy Kopp (Laboratory Services, British Columbia Center for Disease Control, Vancouver, British Columbia, Canada) for their expert technical as-sistance. TABLE 2. Comparison of characteristics of Catabacter hongkongensis and those of medically important, closely related anaerobic gram-positive rods Characteristic Description Catabacter hongkongensis (HKU16 T) Clostridium Eubacterium Eggerthella Bifidobacterium Propionibacterium Actinomyces Gram smear appearance Coccobacilli or short, straight rods Straight or curved rods that are rounded, tapered, or blunt ended Short rods Short rods or coccobacilli Rods with or without one bifurcated end, ends may appear club-like Variable, may be diphtheroidal or club shaped, with one end round and the other end tapered, or coccoid, bifid, or branched Variable, straight or slightly curved rods with true branching, with or without clubs; may be diphtheroidal or pleomorphic Spore formation No Yes No No No No No Motility Motile All but a few species are motile Variable Nonmotile Nonmotile Nonmotile Nonmotile Oxygen requirement Obligately anaerobic Obligately anaerobic or aerotolerant Obligately anaerobic Obligately anaerobic Obligately anaerobic Variable Variable Catalase ⫹⫺ (rarely weakly positive) ⫺ Variable ⫺ Variable ⫺ (except for A. viscosus ) Indole production ⫺ Variable ⫺⫺⫺ Variable ⫺ Nitrate reduction ⫺ Variable Variable Variable ⫺ Variable Variable G ⫹ C content (mol %) 40.2 ⫾ 2.2 26–56 30–40 62 57–64 59–66 55–68
400 LAU ET AL. J. CLIN. MICROBIOL.
on May 16, 2020 by guest
http://jcm.asm.org/
REFERENCES
1.Brook, I.2002. Clinical review: bacteremia caused by anaerobic bacteria in children. Crit. Care6:205–211.
2.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genusClostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol.44:812–826.
3.Fridovich, I.1978. The biology of oxygen radicals. Science201:875–880. 4.Hassan, H. M., and I. Fridovich.1978. Regulation of the synthesis of catalase
and peroxidase inEscherichia coli. J. Biol. Chem.253:6445–6450. 5.Horgan, A. F., R. C. Stuart, E. M. O’Shaughnessy, B. Cryan, and W. O.
Kirwan.1994. Bacterial translocation during peroperative colonic lavage of the obstructed rat colon. Br. J. Surg.81:1796–1798.
6.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson.
1998. Multiple sequence alignment with ClustalX. Trends Biochem. Sci.
10:403–405.
7.Jousimies-Somer, H. R., P. Summanen, D. M. Citron, E. J. Baron, H. M. Wexler, and S. M. Finegold. 2002. Wadsworth anaerobic bacteriology manual, 6th ed. Star Publishing, Belmont, CA.
8.Kageyama, A., and Y. Benno.2000.Coprobacillus catenaformisgen. nov., sp. nov., a new genus and species isolated from human feces. Microbiol. Immu-nol.44:23–28.
9.Kageyama, A., Y. Benno, and T. Nakase.1999. Phylogenetic evidence for the transfer ofEubacterium lentumto the genusEggerthellaasEggerthella lenta gen. nov., comb. nov. Int. J. Syst. Bacteriol.49:1725–1732.
10.Kodaka, H., A. Y. Armfield, G. L. Lombard, and V. R. Dowell, Jr.1982. Practical procedure for demonstrating bacterial flagella. J. Clin. Microbiol.
16:948–952.
11.Lau, S. K. P., P. C. Y. Woo, H. Tse, K. W. Leung, S. S. Y. Wong, and K. Y. Yuen.2003. InvasiveStreptococcus iniaeinfections outside North America. J. Clin. Microbiol.41:1004–1009.
12.Lau, S. K. P., P. C. Y. Woo, G. K. S. Woo, A. M. Y. Fung, K. M. Wong, K. M. Chan, S. F. Tang, and K. Y. Yuen.2004.Eggerthella hongkongensissp. nov.
andEggerthella sinensissp. nov., two novelEggerthellaspecies, account for
half of the cases ofEggerthellabacteremia. Diagn. Microbiol. Infect. Dis.
49:255–263.
13.Merrett, N. D., J. Jorgenson, P. Schwartz, and D. R. Hunt.1994. Bacteremia associated with operative decompression of a small bowel obstruction. J. Am. Coll. Surg.179:33–37.
14.Murray, P. R., E. J. Baro, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.).2003. Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, DC.
15.Nakazawa, F., S. E. Poco, T. Ikeda, M. Sato, S. Kalfas, G. Sundqvist, and E. Hoshino.1999.Cryptobacterium curtumgen. nov., sp. nov., a new genus of gram-positive anaerobic rod isolated from human oral cavities. Int. J. Syst. Bacteriol.49:1193–1200.
16.National Committee for Clinical Laboratory Standards.2003. Methods for
antimicrobial susceptibility testing of anaerobic bacteria. Approved standard M11-A5, 5th ed. National Committee for Clinical Laboratory Standards, Wayne, PA.
17.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins.
1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med.323:1573–1580.
18.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow.1992. Identification of the uncultured bacillus of Whipple’s disease. N. Engl. J. Med.327:293–301.
19.Rocha, E. R., T. Selby, J. P. Coleman, and C. J. Smith.1996. Oxidative stress response in an anaerobe,Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J. Bacteriol.178:6895–6903.
20.Sagar, P. M., J. MacFie, P. Sedman, J. May, B. Mancey-Jones, and D. Johnstone.1995. Intestinal obstruction promotes gut translocation of bacte-ria. Dis. Colon Rectum38:640–644.
21.Tally, F. P., P. R. Stewart, V. L. Sutter, and J. E. Rosenblatt.1975. Oxygen tolerance of fresh clinical anaerobic bacteria. J. Clin. Microbiol.1:161–164. 22.Thompson, J. D., D. G. Higgins, and T. J. Gibson.1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res.22:4673–4680.
23.Wade, W. G., J. Downes, D. Dymock, S. J. Hiom, A. J. Weightman, F. E. Dewhirst, B. J. Paster, N. Tzellas, and B. Coleman. 1999. The family
Coriobacteriaceae: reclassification ofEubacterium exiguum(Poco et al. 1996)
andPeptostreptococcus heliotrinreducens(Lanigan 1976) asSlackia exigua
gen. nov., comb. nov., andSlackia heliotrinireducensgen. nov., comb. nov.,
andEubacterium lentum(Prevot 1938) asEggerthella lentagen. nov., comb.
nov. Int. J. Syst. Bacteriol.49:595–600.
24.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, J. L. L. Teng, B. H. L. Wong, M. K. M. Wong, E. Hon, G. W. K. Tang, and K. Y. Yuen.2003.Actinomyces
hongkongensissp. nov. A novelActinomycesspecies isolated from a patient
with pelvic actinomycosis. Syst. Appl. Microbiol.26:518–522.
25.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, G. Y. Wong, and K. Y. Yuen.2002. Diagnosis of pelvic actinomycosis by 16S ribosomal RNA gene sequencing and its clinical significance. Diagn. Microbiol. Infect. Dis.43:113–118. 26.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, and K. Y. Yuen.2002.
Identifi-cation by 16S ribosomal RNA gene sequencing ofLactobacillus salivarius bacteremic cholecystitis. J. Clin. Microbiol.40:265–267.
27.Woo, P. C. Y., D. M. W. Tam, K. W. Leung, S. K. P. Lau, J. L. L. Teng, M. K. M. Wong, and K. Y. Yuen.2002.Streptococcus sinensissp. nov., a novel
Streptococcus species isolated from a patient with infective endocarditis.
J. Clin. Microbiol.40:805–810.
28.Yuen, K. Y., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, M. K. M. Wong, and S. K. P. Lau.2001.Laribacter hongkongensisgen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol.39:4227–4232.