Copyright © 2002, American Society for Microbiology. All Rights Reserved.
Enterococcus gilvus
sp. nov. and
Enterococcus pallens
sp. nov.
Isolated from Human Clinical Specimens
Gregory J. Tyrrell,
1,2* LeeAnn Turnbull,
1Lúcia M. Teixeira,
3Johanne Lefebvre,
4Maria da Glória S. Carvalho,
3Richard R. Facklam,
5and Marguerite Lovgren
1The National Centre for Streptococcus—Canada, The Provincial Laboratory for Public Health, Northern Alberta,1and
Department of Laboratory Medicine and Pathology, University of Alberta,2Edmonton, Alberta, and Institut Laboratorie
de Sante Publique du Quebec, National de Sante Publique du Quebec, Ste. Anne de Bellevue, Quebec,4Canada;
Instituto de Microbiologia, Universidade Federal do Rio de Janeiro, Rio de Janerio, RJ, 21941, Brazil3; and
Respiratory Diseases Branch, Centers for Disease Control and Prevention, Atlanta, Georgia 303335
Received 3 October 2001/Returned for modification 10 December 2001/Accepted 14 December 2001
Light yellow-pigmented (strain PQ1) and yellow-pigmented (strain PQ2), gram-positive, non-spore-forming, nonmotile bacteria consisting of pairs or chains of cocci were isolated from the bile of a patient with cholecystitis (PQ1) and the peritoneal dialysate of another patient with peritonitis (PQ2). Morphologically and biochemically, the organisms phenotypically belonged to the genusEnterococcus. Whole-cell protein (WCP) analysis and sequence analysis of a segment of the 16S rRNA gene suggested that they are new species within the genusEnterococcus. PQ1 and PQ2 displayed less than 70% identities to other enterococcal species by WCP analysis. Sequence analysis showed that PQ1 shared the highest level of sequence similarity withEnterococcus raffinosusandE. malodoratus(sequence similarities of 99.8% to these two species). Sequence analysis of PQ2 showed that it had the highest degrees of sequence identity with the group I enterococciE. malodoratus(98.7%), E. raffinosus(98.6%),E. avium(98.6%), andE. pseudoavium(98.6%). PQ1 and PQ2 can be differentiated from the otherEnterococcusspp. in groups II, III, IV, and V by their phenotypic characteristics: PQ1 and PQ2 produce acid from mannitol and sorbose and do not hydrolyze arginine, placing them in group I. The yellow pigmentation differentiates these strains from the other group I enterococci. PQ1 and PQ2 can be differentiated from each other since PQ1 does not produce acid from arabinose, whereas PQ2 does. Also, PQ1 isEnterococcus Accuprobe assay positive and pyrrolidonyl--naphthylamide hydrolysis positive, whereas PQ2 is negative by these assays. The nameEnterococcus gilvussp. nov. is proposed for strain PQ1, and the nameEnterococcus pallenssp. nov. is proposed for strain PQ2. Type strains have been deposited in culture collections asE. gilvus ATCC BAA-350 (CCUG 45553) andE. pallensATCC BAA-351 (CCUG 45554).
Species identification of enterococci in the clinical labora-tory has gained importance in the last decade due primarily to this organism’s ability to acquire new antibiotic resistance de-terminants, including resistance to vancomycin (12). There are five recognized groups of enterococci (groups I to V), which include a total of 21 species (4, 7, 21, 23, 24, 26). The majority of these can be identified to the species level by conventional identification techniques. Classic species identification of en-terococci involves assays for a combination of biochemical and morphological characteristics of the unknown organism. Bio-chemical characteristics refer to the ability of enterococci to utilize approximately 10 or more different substrates, resulting in a characteristic pattern for a particular species (6, 7). This identification scheme also involves assays for two morpholog-ical characteristics, motility and yellow pigmentation. There
are only two motile enterococci,Enterococcus gallinarumand
E. casseliflavus. They can be differentiated from each other on
the basis of their pigmentation;E. casseliflavusis yellow andE.
gallinarumis not. There are only two yellow-pigmented
entero-cocci of clinical significance to humans, E. mundtii and E.
casseliflavus, and they can be distinguished from each other by
their motilities.E. flavescenswas proposed as a new species of
motile yellow-pigmented enterococci; however, further work
has suggested that this species is a variant ofE. casseliflavus
and does not warrant designation as a separate species (17, 19,
23). A third yellow-pigmented enterococcal species isE.
sulfu-reus(14). The environmental niche for this organism appears
to be plants, whereasE. mundtiiandE. casseliflavusare animal
derived (14, 22). Besides E. casseliflavus (E. flavescens), E.
mundtii, andE. sulfureus, there have been no reports of other species of yellow-pigmented enterococci.
We report on the isolation and identification of two new
yellow-pigmented Enterococcus species that we have
desig-natedEnterococcus gilvussp. nov. andEnterococcus pallenssp.
nov.
MATERIALS AND METHODS
Bacterial strains.The unknown enterococci (strains PQ1 and PQ2) were referred to the National Centre for Streptococcus (Edmonton, Alberta, Canada) for species identification. PQ1 was cultured from the bile of a patient with cholecystitis, and PQ2 was cultured from a peritoneal dialysate of another pa-tient who developed peritonitis from a perforated intestine. PQ1 and PQ2 have been deposited with the American Type Culture Collection (ATCC) and the Culture Collection of the University of Göteborg (CCUG). Type strains (ob-tained from ATCC) representing the enterococcal species that were physiolog-ically most related to the two isolates were also included in the study. These were
E. aviumATCC 14025T,E. casseliflavusATCC 25788T,E. faecalisATCC 19433T,
E. malodoratusATCC 43197T,E. mundtiiATCC 43186T,E. raffinosusATCC
49427T,E. pseudoaviumATCC 49372T,E. saccharolyticusATCC 43076T, andE.
sulfureusATCC 49903T.
* Corresponding author. Mailing address: 2B3.13 Walter Mackenzie Centre, 8440-112 St., Edmonton, Alberta, Canada T6G 2J2. Phone: (780) 407-8949. Fax: (780) 407-3864. E-mail: g.tyrrell@provlab.ab.ca.
1140
on May 15, 2020 by guest
http://jcm.asm.org/
Morphological and biochemical analysis.Strains PQ1 and PQ2 were grown on Trypticase soy sheep blood agar (TSA-SB) incubated at 35°C without CO2.
Cellular morphology was observed after Gram staining of a smear prepared from a culture grown in thioglycolate broth, air dried, and fixed with methanol. Group-ing was done by the Lancefield hot acid extraction method (9). Lancefield acid extracts were examined for lines of identity with group-specific antisera. Pheno-typic characterization was carried out by conventional tests (6, 7). These included catalase production; susceptibility to vancomycin as determined with a 30-g vancomycin disk; pyrrolidonyl--naphthylamide (PYR) and leucine--naphthyl-amide (LAP) hydrolysis; hydrolysis of esculin in the presence of 40% bile; arginine hydrolysis; esculin hydrolysis; growth at 10 and 45°C; growth in 2.0, 4.0, and 6.5% NaCl; motility; pigment production; pyruvate utilization; hippurate hydrolysis; reduction of tetrazolium; growth on potassium tellurite agar; and acid production from the carbohydrates listed in Table 1. Pigment production was visually assayed by growing the bacteria on Luria-Bertani (LB) agar or TSA-SB for 24 h and scraping off the growth with a white cotton swab (20). Biochemical reactions were read after 7 days of incubation; however, bile esculin hydrolysis and hippurate hydrolysis tests were read at 48 h, and the test for the production of acid from methyl-␣-D-glucopyranoside was read at 72 h. Motility was assayed
with 0.03% semisolid agar with tetrazolium chloride at 30°C for 7 days. Tests for growth in 2.0, 4.0, and 6.5% NaCl and at 10 and 45°C were performed in duplicate in heart infusion broth with 0.1% bromocresol purple indicator. The results were recorded after 24 h, 48 h, and 7 days. TheEnterococcusAccuprobe assay (Gen-Probe, San Diego, Calif.) was carried out according to the instruc-tions of the manufacturer. TheEnterococcusAccuprobe assay is a nonsubjective method for the identification ofEnterococcusspecies. The assay uses a single-stranded DNA probe with a chemiluminescent label that is complementary to specific rRNA sequences unique toEnterococcus.
Long-chain fatty acid analysis.Isolates were grown on brain heart infusion agar at 35°C for 24 h. The organisms were harvested into 1 ml of NaOH in aqueous methanol and were incubated at 100°C for 0.5 h. The tubes were then cooled, and 2 ml of HCl in aqueous methanol was added. This mixture was then incubated at 80°C for 10 min and cooled rapidly. The fatty acids were extracted into hexane and methyl-tert-butyl ether and washed with dilute NaOH. The extracts were run on a Hewlett-Packard model HP5890 Series II gas chromato-graph (MIDI, Inc., Newark, Del.). The MIDI Microbial Identification System consists of the Hewlett-Packard gas chromatograph linked to a computer system with Sherlock and ChemStation software (MIDI, Inc.).
Whole-cell protein preparation and analysis.Preparation of whole-cell ex-tracts and analysis of profiles by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were performed as described previously (11, 15), with a few modifications. Briefly, the strains were grown on plates containing TSA-SB. Bacterial cells were removed from the surface of the plate with an inoculating loop and suspended in 5 ml of sterile saline solution in order to obtain a turbidity equal to that of a no. 8 McFarland density standard, centri-fuged, and resuspended in 0.25 ml of an aqueous lysozyme solution (10 mg/ml). The protein profiles of the type strains of the different species were compared according to their percentages of similarity, estimated by use of the Dice coef-ficient and clustered by the unweighted pair group method with averages by using the Molecular Analyst Fingerprinting Plus software package, version 1.12, of the Image Analysis System (Bio-Rad Laboratories, Hercules, Calif.).
16S rRNA gene sequencing.The methodologies for extraction of chromosomal DNA and performance of the PCR are described in detail elsewhere (18, 25). Oligonucleotides were purchased from Gibco BRL (Burlington, Ontario, Can-ada). PCR amplifications were performed with a model 9600 thermocycler (Per-kin-Elmer, Norwalk, Conn.). The oligonucleotides used as primers for PCR and for sequencing were as described previously (9). The DNA sequences of the 16S rRNA gene (rDNA) amplicons were determined directly by use of the oligonu-cleotide primers as sequencing primers, the DYEnamic terminator cycle se-quencing premix kit (Amersham Pharmacia Biotech Inc., Cleveland, Ohio), and an ABI 373 sequencer. The amplicons were sequenced in both directions.
Sequence and phylogenetic analyses.The sequences obtained were compared with the sequences of strains belonging to other related enterococcal species retrieved from the GenBank database by the CLUSTAL method with the Expert Sequences Analysis software of the DNASTAR program (DNASTAR Inc., Madison, Wis.). Consensus sequences were determined and then grouped into clusters according to the sequence distances between all pairs. Clusters were aligned as pairs and then collectively as sequence groups to produce the overall alignment. After the multiple-sequence alignment was completed, the neighbor-joining method was used to construct a dendrogram showing the phylogenetic relationships (8).
Nucleotide sequence accession numbers.The partial 16S rDNA sequences of strains PQ1 and PQ2 have been submitted to GenBank and can be found under accession numbers AY033814 and AY033815, respectively.
RESULTS AND DISCUSSION
Initial isolation and cultural characteristics.Strain PQ1 was
isolated from the bile of a patient with cholecystitis. E. faecium
andE. casseliflavuswere also isolated from the same specimen. Strain PQ2 was isolated from peritoneal dialysate fluid of an-other patient. Initial characterization placed PQ1 and PQ2 in
the genusEnterococcus. When grown on TSA-SB or LB agar
or in broth, the cells of PQ1 and PQ2 were gram positive and spherical, occurred in chains, and were nonmotile. PQ1 pro-duced a light yellow pigment, and PQ2 propro-duced a bright yellow pigment. Both strains grew in 6.5% NaCl and at 10, 35, and 45°C. PQ1 and PQ2 were catalase negative, reacted with group D antiserum, and were positive for hydrolysis of esculin and esculin with 40% bile. PQ1 and PQ2 were LAP hydrolysis
positive and utilized pyruvate. PQ1 was Enterococcus
Accu-probe assay positive and PYR hydrolysis positive, whereas PQ2 was Accuprobe assay negative and PYR negative. Currently, only three species of enterococci are Accuprobe assay negative
and PYR hydrolysis negative:E. cecorum,E. columbae, andE.
saccharolyticus(7). Both PQ1 and PQ2 were sensitive to
van-comycin by the 30-g vancomycin disk assay, and both strains
were tellurite negative. Both strains produced acid from man-nitol and sorbose and were arginine negative, suggesting that they are members of the group I enterococci (Table 1) (7). PQ1 and PQ2 also produced acid from glycerol, lactose, meli-bose, raffinose, rimeli-bose, salicin, sorbitol, sucrose, and trehalose.
They did not produce acid from inulin, methyl-␣-D
-glucopyr-anoside, or xylose. Also, PQ1 did not produce acid from arab-inose or hydrolyze hippurate, whereas PQ2 did. PQ1 reduced tetrazolium, whereas PQ2 did not. Biochemically, PQ1 most
closely resemblesE. malodoratus.However, unlikeE.
malodo-ratus, PQ1 was pigmented yellow, although the color was duller
than the bright yellow pigment typical ofE. casseliflavusorE.
mundtii. PQ2 most closely resemblesE. raffinosus; however,
PQ2 is pigmented a bright yellow, whereasE. raffinosusis not.
Also, PQ2 does not hydrolyze PYR, unlikeE. raffinosus.
Pig-ment production was easily visualized when the organisms were grown on LB agar or a blood agar plate and then scraped off with a cotton swab. The pigmentation phenotype was pre-served upon subculture.
Three enterococci are currently classified as having yellow
pigmentation. These areE. casseliflavus(group II),E. mundtii
(group II), andE. sulfureus(Group IV).
Long-chain fatty acid analysis. The following long-chain fatty acids were detected in strain PQ1: 12:0, 14:0, 15:0, 16:0,
17:0, 18:0, 18:19c, 18:17c, and cyclo-C19 (Table 2). Other
major fatty acids which were present in PQ1 (but which could not be quantitated owing to the poor resolution of the
chro-matogram) were 16:17c and i15:0 2-OH, 18:26,9c and 18:0,
and 19:0 cyclo10c and 196. The following long-chain fatty
acids were detected in strain PQ2: 14:0, 15:0, 16:0, 17:0, 18:0,
18:19c, 18:17c, and cyclo-C19(Table 2). Other major fatty
acids which were present in PQ2 (but which could not be quantitated owing to the poor resolution of the
chromato-gram) were 16:17c/il15:0 2-OH, 18:26, 9c/18:0, and 19:0
on May 15, 2020 by guest
http://jcm.asm.org/
cyclo10c/196. The long-chain fatty acid profiles of PQ1 and PQ2 do not match those of any of the group I enterococci. However, the long-chain fatty acid profile of PQ1 does most
closely resemble that ofE. raffinosus, suggesting that it is most
related to this species, and the long-chain fatty acid profile of
PQ2 most closely resembles that ofE. avium.
[image:3.587.45.539.93.470.2]Whole-cell protein analysis. SDS-PAGE analysis of whole-cell proteins showed that neither PQ1 nor PQ2 had an identity
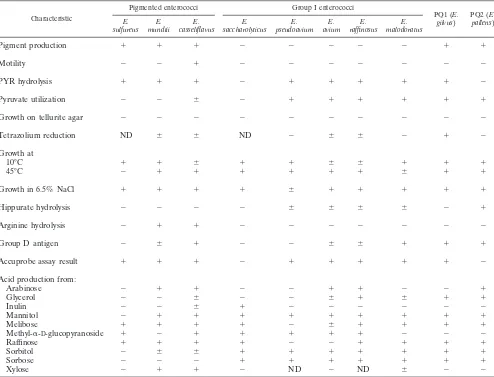
TABLE 1. Differential characteristics of PQ1 and PQ2 and comparison with those of other pigmented enterococci and related group I enterococcal speciesa
Characteristic
Pigmented enterococci Group I enterococci
PQ1 (E.
gilvus) PQ2 (pallensE.)
E.
sulfureus mundtiiE. casseliflavusE. saccharolyticusE. pseudoaviumE. aviumE. raffinosusE. malodoratusE.
Pigment production ⫹ ⫹ ⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫹ ⫹
Motility ⫺ ⫺ ⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺
PYR hydrolysis ⫹ ⫹ ⫹ ⫺ ⫹ ⫹ ⫹ ⫹ ⫹ ⫺
Pyruvate utilization ⫺ ⫺ ⫾ ⫺ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹
Growth on tellurite agar ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺
Tetrazolium reduction ND ⫾ ⫾ ND ⫺ ⫾ ⫾ ⫺ ⫹ ⫺
Growth at
10°C ⫹ ⫹ ⫾ ⫹ ⫹ ⫾ ⫾ ⫹ ⫹ ⫹
45°C ⫺ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹ ⫾ ⫹ ⫹
Growth in 6.5% NaCl ⫹ ⫹ ⫹ ⫹ ⫾ ⫹ ⫹ ⫹ ⫹ ⫹
Hippurate hydrolysis ⫺ ⫺ ⫺ ⫺ ⫾ ⫾ ⫾ ⫾ ⫺ ⫹
Arginine hydrolysis ⫺ ⫹ ⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺
Group D antigen ⫺ ⫾ ⫹ ⫺ ⫺ ⫾ ⫾ ⫹ ⫹ ⫹
Accuprobe assay result ⫹ ⫹ ⫹ ⫺ ⫹ ⫹ ⫹ ⫹ ⫹ ⫺
Acid production from:
Arabinose ⫺ ⫹ ⫹ ⫺ ⫺ ⫹ ⫹ ⫺ ⫺ ⫹
Glycerol ⫺ ⫺ ⫾ ⫺ ⫺ ⫾ ⫹ ⫾ ⫹ ⫹
Inulin ⫺ ⫺ ⫾ ⫹ ⫺ ⫺ ⫺ ⫺ ⫺ ⫺
Mannitol ⫺ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹
Melibose ⫹ ⫹ ⫹ ⫹ ⫺ ⫾ ⫹ ⫹ ⫹ ⫹
Methyl-␣-D-glucopyranoside ⫹ ⫺ ⫹ ⫹ ⫹ ⫹ ⫹ ⫺ ⫺ ⫺
Raffinose ⫹ ⫹ ⫹ ⫹ ⫺ ⫺ ⫹ ⫹ ⫹ ⫹
Sorbitol ⫺ ⫾ ⫾ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹
Sorbose ⫺ ⫺ ⫺ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹ ⫹
Xylose ⫺ ⫹ ⫹ ⫺ ND ⫺ ND ⫾ ⫺ ⫺
aCharacteristics are based on those obtained in the present study and/or reported elsewhere (2, 6, 7, 10, 14, 19, 25).⫹, strains are positive with rare exceptions;⫺,
strains are negative with rare exceptions;⫾, strains may be either positive or negative; ND, not determined.
TABLE 2. Long-chain fatty acid compositions ofE. gilvus, andE. pallens, and related group I enterococcal species
Species
Composition (%)
C12:0 C14:0 C15:0 C16:0 C17:0 C18:0 C18:19c C18:17c Cyclo-C19
Summed featuresa
3 5 7
E. saccharolyticus 0.5 9.2 0.3 17.1 0 2.4 3.1 41.8 0 23.0 2.3 0 E. pseudoavium 2.4 40.7 0.5 15.0 0.5 10.2 7.8 11.8 0 7.9 3.3 0 E. avium 1.0 34.6 0.6 14.2 0.5 8.8 4.0 13.5 7.0 9.0 3.1 2.7 E. raffinosus 0.5 29.4 0.4 13.8 0.4 8.2 2.7 11.8 12.8 11.9 2.7 3.1 E. malodoratus 1.5 24.3 0.7 14.6 0.4 5.9 1.8 12.5 17.5 16.5 2.4 0
E. gilvus 0.3 27.9 0.9 17.7 1.2 8.9 3.0 13.9 11.6 8.3 2.9 3.0 E. pallens 0 26.0 0.6 17.0 0.5 9.4 5.2 13.6 5.3 15.5 3.6 2.3
aSummed features represent groups of two or three fatty acids that could not be separated by gas-liquid chromatography with the MIDI system. Summed feature
3 was for fatty acids 16:17c and 15:0 iso 2OH. Summed feature 5 was for fatty acids 18:26, 9c, and 18:0 ANTE. Summed feature 7 was for fatty acids 19:0cyclo10c and 196.
on May 15, 2020 by guest
http://jcm.asm.org/
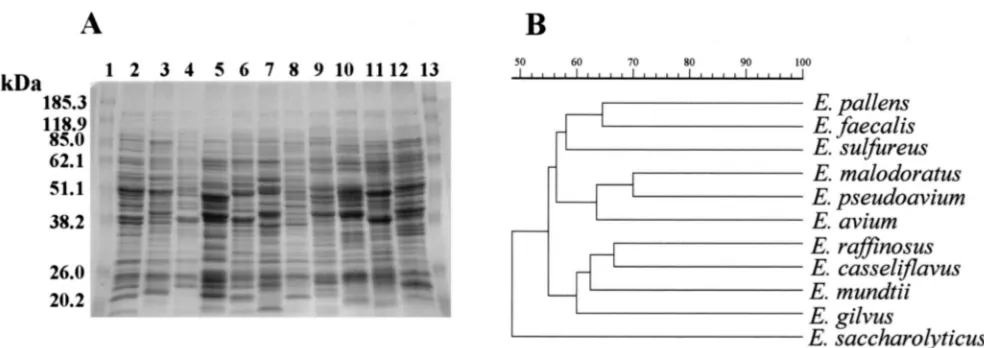
[image:3.587.41.543.580.702.2]of 70% of greater to any of the enterococcal species examined, including those to which they were most physiologically related (Fig. 1), suggesting that PQ1 and PQ2 are distinct species of enterococci. The whole-cell protein profiles of PQ1 and PQ2 were easily distinguishable from each other. PQ1 most closely
resemblesE. raffinosus,E. casseliflavus, andE. mundtii(60%
identity); and PQ2 most closely resembles E. faecalis (64%
identity).
16S rDNA sequence analysis. To assess the genealogical affinities between PQ1 and PQ2 and their relationship with other enterococci, comparative 16S rRNA gene sequencing analysis was performed. Amplification of the 16S rRNA gene sequences yielded the expected PCR products. PCR of the 16S rDNAs of isolates PQ1 and PQ2 generated sequences of 1,295 and 1,294 bp, respectively. The first base corresponds to posi-tion 7 and the last base corresponds to posiposi-tion 1342 of the
Escherichia coli16S rRNA gene (5). The sequenced DNA was compared to those of the other enterococcal species obtained from GenBank or published previously (17). The accession numbers of these sequences are shown in Fig. 2.
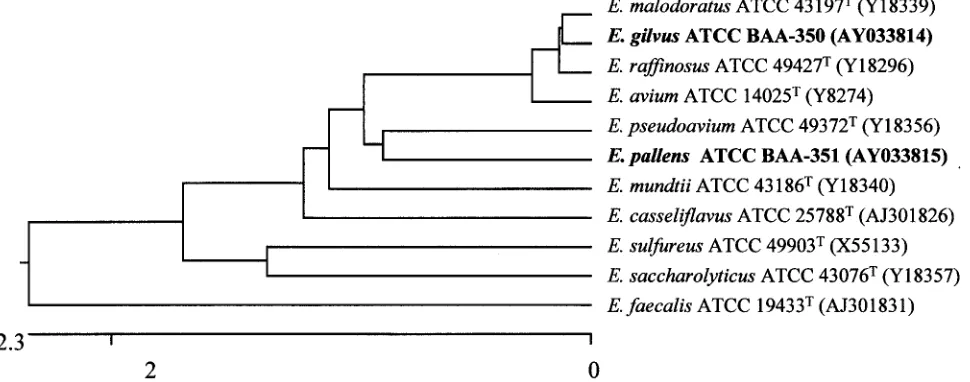
Sequence searches of the GenBank and Ribosomal Data-base Project data libraries revealed that PQ1 and PQ2 are phylogenetically most closely associated with species of the
genusEnterococcus. A tree depicting the phylogenetic affinity
of PQ1 and PQ2 with other enterococci is shown in Fig. 2. The PQ1 sequence analyzed most closely matched those of
E. malodoratusandE. raffinosus. PQ1 is 99.8% similar toE. raffinosus and 99.8% similar to E. malodoratus. There are 3
nucleotide differences betweenE. raffinosusand PQ1 and
be-tween E. malodoratus and PQ1 in the 1,295 bp of the 16S
rDNA sequenced. This suggests that these three species are very closely related from an evolutionary perspective. The next
most closely related species areE. pseudoavium(99.5%
iden-tity) andE. avium(99.5% identity).
Analysis of the sequenced segment of the 16S rDNA of PQ2
showed that PQ2 is 98.7% similar toE. malodoratusand 98.6%
similar toE. raffinosus,E. avium, andE. pseudoavium. In the
1,294 bp examined, there were 20 nucleotide differences
be-tween PQ2 andE. malodoratus, 21 nucleotide differences
be-tween PQ2 andE. raffinosus, 22 nucleotide differences between
PQ2 andE. avium, and 25 nucleotide differences between PQ2
andE. pseudoavium.
The 16S rDNA sequence differences together with the pro-duction of yellow pigment, the biochemical utilization patterns, and the results of whole-cell protein and long-chain fatty acid analyses strongly suggest that PQ1 and PQ2 are new species of
the genusEnterococcusfor which the namesEnterococcus
gil-vusandEnterococcus pallens, respectively, are proposed. Strains PQ1 and PQ2 are the only yellow-pigmented entero-cocci in group I. This is the first report of pigmented group I enterococci. It is possible that these bacteria have mistakenly been identified in the past as one of the other yellow-pig-mented enterococci and have therefore gone unrecognized as new species. Studies in the early 1970s indicated that the yellow pigment in enterococci was the result of carotenoid production by the organism (13, 22). Carotenoids are naturally occurring pigments found in a wide variety of bacteria, cyanobacteria, algae, fungi, higher plants, crustaceans, insects, fish, and birds (1). In bacteria, carotenoids act as quenchers of potentially toxic oxygen radicals and as light-gathering pigments in pho-tosynthesis (1, 3, 13, 16).
Also, while strain PQ1 (E. gilvus) was isolated from the bile
of a patient suffering from cholecystitis and PQ2 (E. pallens)
was isolated from the peritoneal dialysate of another patient
with peritonitis, it is unclear what pathogenic role E. gilvus
and/orE. pallenshas in causing infection in humans.
Description ofE. gilvussp. nov.Enterococcus gilvus(gil.vus⬘. L. adj., pale yellow, referring to the pale yellow pigmenta-tion of the bacterium). Cells are gram-positive cocci and spherical and mostly occur in short chains. The cells are pigmented a light yellow color in comparison to the other
pigmented enterococci,E. casseliflavus,E. mundtii, andE.
sul-FIG. 1. (A) SDS-PAGE profiles of whole-cell protein extracts ofE. gilvus,E. pallens, and strains belonging to related enterococcal species. Lanes 1 and 13, molecular mass markers; lane 2,E. aviumATCC 14025T; lane 3,E. malodoratusATCC 43197T; lane 4,E. pseudoaviumATCC
49372T; lane 5,E. saccharolyticusATCC 43076T; lane 6,E. raffinosusATCC 49427T; lane 7,E. pallens(PQ2); lane 8,E. gilvus(PQ1); lane 9,E.
casseliflavusATCC 25788T; lane 10,E. mundtiiATCC 43186T; lane 11,E. sulfureusATCC 49903T; lane 12,E. faecalisATCC 19433T. (B)
Den-drogram resulting from computer-assisted analysis of the protein profiles in panel A. The scale represents average percent similarities. A 0.6% tolerance value was used to construct the dendrogram.
on May 15, 2020 by guest
http://jcm.asm.org/
[image:4.587.45.537.71.245.2]fureus. The organism is nonmotile and catalase negative. It is Lancefield group D antigen positive. It grows at 10 and 35°C and has delayed growth at 45°C. It grows in 2.0, 4.0, and 6.5% NaCl. It produces acid from glycerol, lactose, mannitol, meli-bose, raffinose, rimeli-bose, salicin, sorbitol, sormeli-bose, sucrose, and trehalose. It does not produce acid from arabinose, inulin,
methyl-␣-D-glucopyranoside, or xylose. It is positive for esculin
hydrolysis in the presence of 40% bile. Black colonies were not produced when grown on tellurite-containing media. It is pos-itive for pyruvate utilization and reduction of tetrazolium. The strain is positive for LAP hydrolysis and PYR hydrolysis and negative for arginine dihydrolase hydrolysis and hippurate
hy-drolysis. It is Enterococcus Accuprobe assay positive. The
strain can also be differentiated from other enterococcal spe-cies by its unique whole-cell protein profile and the sequence of its 16S rRNA gene. The type strain is ATCC BAA-350 (CCUG 45553; PQ1).
Description ofE. pallenssp. nov.Enterococcus pallens(pall.
ens⬘. L. adj., yellowish, referring to the yellow pigmentation of
the bacterium). Cells are gram-positive cocci and spherical and mostly occur in short chains. The bacterium is pigmented a bright yellow color. The organism is nonmotile and catalase negative. It is Lancefield group D antigen positive. It grows at 10, 35, and 45°C. It grows in 2.0, 4.0, and 6.5% NaCl. It produces acid from arabinose, glycerol, lactose, mannitol, melibose, raffinose, ribose, salicin, sorbitol, sorbose, sucrose,
and trehalose. It does not produce acid from inulin, methyl-␣
-D-glucopyranoside, or xylose. It is positive for esculin
hydroly-sis in the presence of 40% bile and for hippurate hydrolyhydroly-sis. It is also positive for pyruvate utilization and negative for reduc-tion of tetrazolium. The strain is positive for LAP hydrolysis and negative for both PYR hydrolysis and arginine hydrolysis.
Testing by theEnterococcusAccuprobe assay is negative. The
strain can also be differentiated from the other species of
Enterococcusby its unique whole-cell protein profile and the sequence of its 16S rRNA gene. The type strain is ATCC BAA-351 (CCUG 45554; PQ2).
REFERENCES
1.Armstrong, G., and J. Hearst.1996. Genetics and molecular biology of carotenoid pigment biosynthesis. FASEB J.10:228–237.
2.Carvalho, M. D. G. S., L. M. Teixeria, and R. R. Facklam.1998. Use of tests for acidification of methyl-␣-D-glucopyranoside and susceptibility to
efromy-cin for differentiation of strains ofEnterococcusand some related genera. J. Clin. Microbiol.36:1584–1587.
3.Chamovitz, D., N. Misawa, G. Sandman, and J. Hirschberg.1992. Molecular cloning and expression inEscherichia coliof a cyanobacterial gene coding for phytoene synthase, a carotenoid biosynthesis enzyme. FEBS Lett.296:305– 310.
4.deVaux, A., G. Laguerre, C. Divies, and H. Prevost.1998.Enterococcus asini
sp. nov. isolated from the caecum of donkeys (Equus asinus). Int. J. Syst. Bacteriol.48:383–387.
5.Ehresmann, C., P. Stiegler, P. Fellner, and J. P. Ebel.1972. The determi-nation of the primary structure of the 16S ribosomal RNA ofEscherichia coli. 2. Nucleotide sequences of products from partial enzymatic hydrolysis. Bio-chimie54:901–967.
6.Facklam, R. R., and M. D. Collins.1989. Identification of Enterococcus
species isolated from human infections by a conventional test scheme. J. Clin. Microbiol.27:731–734.
7.Facklam, R. R., D. F. Sahm, and L. M. Teixeira.1999. Enterococcus, p. 297–305.InP. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
8.Higgins, D. G., and P. M. Sharp.1988. CLUSTAL: a package for performing multiple alignment on a microcomputer. Gene73:237–244.
9.Johnson, J. L.1994. Similarity analysis of rRNAs, p. 683–700.InP. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. ASM Press, Washington, D.C.
10.Koneman, E., S. Allen, W. Janda, P. Schreckenbreger, and W. Winn, Jr. 1997. The gram-positive cocci: part II: streptococci, enterococci and the
Streptococcus-like bacteria, p. 431–466.InE. Koneman, S. Allen, W. Janda, P. Schreckenbreger, and W. Winn, Jr. (ed.), Color atlas and textbook of diagnostic microbiology, 5th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
11.Laemmli, U. K.1970. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature227:680–685.
[image:5.587.54.539.80.271.2]12.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin.1988. Plasmid-mediated resistance to vancomycin and teicoplanin inEnterococcus faecium. N. Engl. J. Med.319:157–161.
FIG. 2. Phylogenetic relationships ofE. gilvus,E. pallens, and related enterococcal species based on 16S rRNA gene sequencing analysis. The dendrogram is based on the sequence identities of 1,294 nucleotides of the 16S rRNA gene. The dendrogram was constructed by the CLUSTAL method with the Expert Sequences Analysis software of the DNASTAR program. The neighbor-joining method was used to construct a dendrogram showing the phylogenetic relationships. The scale units indicate the distance between sequence pairs. The ATCC strain numbers and the GenBank accession numbers of the 16S rRNA gene sequences (in parentheses) are indicated for each species used to construct the dendrogram.
on May 15, 2020 by guest
http://jcm.asm.org/
13.Liaaen-Jensen, S.1965. Biosynthesis and function of carotenoid pigments in microorganisms. Annu. Rev. Microbiol.19:163–182.
14.Martinez-Murica, A. J., and M. D. Collins.1991.Enterococcus sulfureus, a new yellow-pigmentedEnterococcusspecies. FEMS Microbiol. Lett.80:69– 74.
15.Merquior, V. L. C., J. M. Peralta, R. R. Facklam, and L. M. Teixeira.1994. Analysis of electrophoretic whole-cell protein profiles as a tool for charac-terization ofEnterococcusspecies. Curr. Microbiol.28:149–153.
16.Misawa, N., M. Nakagawa, K. Kobayashi, S. Yamano, Y. Izawa, K. Naka-mura, and K. Harashima.1990. Elucidation of theErwinia uredovora carot-enoid biosynthetic pathway by functional analysis of gene products expressed inEscherichia coli. J. Bacteriol.172:6704–6712.
17.Patel, R., K. E. Piper, M. S. Rouse, J. M. Steckelberg, J. R. Uhl, P. Kohner, M. K. Hopkins, F. R. Cockerill III, and B. C. Kline.1998. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotileEnterococcus gallinarumisolates. J. Clin. Microbiol.36:3399– 3407.
18.Pitcher, D., N. Saunders, and R. Owens.1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol.8:151– 156.
19.Pompei, R., F. Berlutti, M. Thaller, A. Ingianni, G. Cortis, and B. Dainelli. 1992.Enterococcus flavescenssp. nov., a new species of enterococci of clinical origin. Int. J. Syst. Bacteriol.42:365–369.
20.Sambrook, J., E. F. Fritsch, and T. Maniatas.1989. Molecular cloning: a
laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
21.Svec, P., L. A. Devriese, I. Sedlácek, M. Baele, M. Vancanneyt, F. Haese-brouck, J. Swings, and J. Doskar.2000.Enterococcus haemoperoxidussp. nov. andEnterococcus moraviensissp. nov., isolated from water. Int. J. Syst. Evol. Microbiol.51:1567–1574.
22.Taylor, R., M. Ikawa, and W. Chesbro.1971. Carotenoids in yellow-pig-mented enterococci. J. Bacteriol.105:676–678.
23.Teixeira, L. M., M. D. G. S. Carvalho, V. C. C. Merquior, A. G. Steigerwalt, D. G. M. Teixeira, D. J. Brenner, and R. R. Facklam.1997. Recent ap-proaches on the taxonomy of the enterococci and some related microorgan-isms. Adv. Exp. Med. Biol.418:397–400.
24.Teixeira, L. M., M. D. G. S. Carvalho, M. M. B. Espinola, A. G. Steigerwalt, M. P. Douglas, D. J. Brenner, and R. R. Facklam.2001.Enterococcus por-cinussp. nov. andEnterococcus rattisp. nov.: novel species associated with enteric disorders in animals. Int. J. Syst. Evol. Microbiol.51:1737–1743. 25.Tyrrell, G. J., R. N. Bethune, B. Willey, and D. E. Low.1997. Species
identification of enterococci via intergenic ribosomal PCR. J. Clin. Micro-biol.35:1054–1060.
26.Vancanneyt, M., C. Snauwaert, I. Cleenwerck, M. Baele, P. Descheemaeker, H. Goosens, B. Pot, P. Vandamme, J. Swings, F. Haesebrouck, and L. A. Devriese.2001.Enterococcus villorumsp. nov., an enteroadherent bacterium associated with diarrhoea in piglets. Int. J. Syst. Evol. Microbiol.51:393–400.